ABSTRACT
Extensive studies showed the vital function of long noncoding RNAs (lncRNAs) in the pathological and physiological progression of tumors. Previous evidence has indicated that lncRNA MYLK Antisense RNA 1 (MYLK-AS1) acts as an oncogene to facilitate the progression of several tumors. Nevertheless, little is known about its biological role in gastric cancer (GC). Our report intended to probe the underlying mechanism and function of MYLK-AS1 in GC. Results revealed that MYLK-AS1 showed an upregulated level in GC. It was worth mentioning that upregulated MYLK-AS1 caused the unfavorable clinical outcome in GC patients. Functional assays indicated that MYLK-AS1 silencing retarded the proliferation, cell cycle, migration, and invasion in GC. Besides, in vivo assay validated that MYLK-AS1 deficiency also restrained tumor growth. Through in-depth mechanism exploration, MYLK-AS1 was uncovered to bind with wnhancer of zeste homolog 2 (EZH2), an epigenetic inhibitor, to inhibit the level of Large Tumor Suppressor 2 (LATS2), thereby exerting carcinogenicity. Conclusively, our research highlighted the importance of MYLK-AS1 in GC, indicating that MYLK-AS1 might be an effective biomarker for GC.
KEYWORDS:
Introduction
As the third frequent reason for cancer-relevant death, gastric cancer (GC) seriously threatens public health [Citation1]. Global cancer statistics manifests that the occurrence rate and deaths of GC cases are increasing each year [Citation1]. Due to the difficulty in advanced-stage diagnosis and the shortage of effective treatments, the clinical outcome of GC patients is quite unsatisfactory [Citation2]. Thus, it is urgent to enrich the knowledge related to the biology of GC and to identify effective tumor biomarkers in early detection and therapy.
Based on human genome study, just a little fraction of human genes can encode proteins, whereas most of the rest (~97%) is tightly transcribed into noncoding RNA [Citation3,Citation4]. Long noncoding RNAs (lncRNAs) are a new cluster of noncoding RNAs (ncRNAs), and its aberrant expression in tumor tissues attracted extensive researchers’ attention [Citation5,Citation6]. Existing research has indicated that lncRNA is capable of functioning as a tumor suppressor gene [Citation7] or carcinogene [Citation8] in the initiation and progression of tumors. In addition, lncRNAs have increasingly been biomarkers of diagnosis, therapy, or prognosis in tumors, including GC [Citation9,Citation10]. Therefore, to investigate novel therapeutic methods of GC, it is quite necessary to search more novel tumor-associated lncRNAs and to probe their biological role and mechanisms.
As a 814-bp lncRNA, MYLK Antisense RNA 1 (MYLK-AS1) locates at chromosome 3 and is originally identified as possible alternative promoters of human genes by the report from the National Institutes of Health Mammalian Gene Collection (MGC) Program [Citation11,Citation12]. MYLK-AS1 has been reported as an oncogene in various human cancers. For example, MYLK-AS1 facilitated nephroblastoma progression via upregulation of CCNE1 [Citation13]. MYLK-AS1 promoted the tumorigenesis of hepatocellular carcinoma via the EGFR/HER2-ERK1/2 signaling pathway [Citation14]. However, the biological role of MYLK-AS1 in GC is still unclear.
Large Tumor Suppressor 2 (LATS2), a member of LATS family [Citation15,Citation16], can inhibit tumor occurrence and development by regulating cell-cycle progression [Citation17]. For instance, miR-135b promoted cell proliferation, migration and invasion of breast cancer via downregulation of LATS2 expression [Citation18]. LncRNA CCAT2 contributed to GC progression by regulating the expression of E-cadherin and LATS2 [Citation19]. In addition, LATS2 is also a part of Hippo signaling pathway, which can be destroyed by the loss of LATS2 [Citation20]. A number of literatures have reported that lncRNA can interact with Enhancer of zeste homolog 2 (EZH2) to modulate LATS2 expression [Citation21,Citation22]. Therefore, our emphasis was to inspect the function of MYLK-AS1 and its relationship with EZH2 and LATS2.
The present study aimed to investigate the biological role and mechanism of MYLK-AS1 in GC. We hypothesized that MYLK-AS1 might act as an oncogene in GC progression by epigenetically silencing LATS2.
Materials and Methods
Tissue samples
GC tissues (n = 70) and adjacent normal tissues (n = 70) were collected from GC patients at Huaihua First People’s Hospital. Informed consents were acquired from each patient, who has not been received any anticancer therapy before the surgery. After leaving the body, all the tissues were frozen in liquid nitrogen at once and preserved at −80°C for further use. The research was permitted by the Ethics Committee of Huaihua First People’s Hospital and each participant signed the informed consent.
Cell lines and cell culture
GC cell lines (AGS, MKN45, SNU-16 and TMK-1) and human normal gastric mucosal cell line (GES-1) were provided by ATCC (Manassas, VA, USA). The cells were maintained in DMEM (Gibco, MD, USA) at 37°C in a humidified condition containing 5% CO2. The DMEM was supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin.
Cell transfection
Short hairpin (sh) RNA targeting MYLK-AS1 (sh-MYLK-AS1; 5ʹ- GGCAACCAUUACUCACCAATT-3ʹ), EZH2 (sh-EZH2; 5ʹ-GAGGUUCAGACGAGCUGAUUU-3ʹ) or LATS2 (sh-LATS2; 5ʹ- GCCACCCAGAGAUAUUACUTT-3ʹ), and corresponding control shRNA (sh-NC 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ) were commercially obtained from GenePharma (Shanghai, China). Utilizing Lipofectamine 3000 (Invitrogen, Carlsbad, CA), all above plasmids were transfected into the GC cells (AGS and MKN45 cells). Incubated for 48 h, the efficiency of transfection was verified by RT-qPCR, and transfected cells were applied for further assays.
RT-qPCR analysis
The use of TRIzol Reagent (Invitrogen) was for extracting total RNA in transfected cells. The reverse transcription of RNAs into cDNAs was carried out by PrimeScript RT reagent kit (TaKaRa, Japan). Then, based on the direction of SYBR® Green Realtime PCR Master Mix (TaKaRa), RT-qPCR reactions were performed. Finally, normalized to GAPDH, 2−ΔΔCt method [Citation23] was employed for calculating relative RNA levels. The sequences of the primers were as follows: MYLK-AS1: forward, 5ʹ- TCTCCTTGTGCAAACCTCC-3ʹ and reverse, 5ʹ-CCCACATTGAGCGAATGCC-3′; LATS2 forward, 5′-TAGAGCAGAGGGCGCGGAAG-3′ and reverse, 5′- CCAACACTCCACCAGTCACAGA-3′; EZH2 forward, 5′- GTGGAGAGATTATTTCTCAAGATG-3′ and reverse, 5′- CCGACATACTTCAGGGCATCAGCC-3′; GAPDH forward, 5′- GAAGATGGTGATGGGATTTC-3′ and reverse, 5′-GAAGGTGAAGGTCGGAGTC-3′.
CCK-8 assay
After transfection, CCK-8 assay was carried out to evaluate cell viability. Plated in 96-well plates, GC cells were incubated for 0, 24, 48, or 72 h. Then, CCK-8 solution (10 µl/well, Dojindo) was added at each time point to incubate for further 2 h at 37°C. Later, the measurement of optical density wavelength was completed by a microplate reader at 450 nm [Citation24].
Flow cytometry analysis
Cells were harvested after being transfected for 48 h. Through CycleTESTTM PLUS DNA Kit (BD Bioscience, NJ, USA), the transfected cells were colored with propidium iodide (PI), and then analyzed by flow cytometry (FACScan; BD Biosciences) [Citation25]. The cell ratio in each phase was compared for cell-cycle analysis.
Transwell assay
To test cell migratory and invasive abilities, transwell assay was performed. After transfection, cells (2 × 104 cells/well) in 1% FBS medium were added into the top chamber (8 µm pore size; Millipore) with Matrigel-coated membrane (for invasion analysis) or without Matrigel-coated membrane (for migration analysis). The medium supplementing with 10% FBS was placed to the bottom chamber. 24 h later, cells remaining in top chamber were removed, and cells migrating into lower chamber were immobilized by using methanol and stained by using crystal violet. Subsequently, by a microscope (Olympus, Tokyo, Japan), migrated and invaded cells were counted.
Tumor xenografts
Four-week-old BALB/c nude mice (male) were provided by Beijing Vital River Laboratory Animal Technology Company, and fed under standard conditions. GC cells with stable transfection of sh-MYLK-AS1 and sh-NC were inoculated into each nude mouse with subcutaneous injection. Every per week, tumor volume was monitored. 28 days later, the mice were euthanized, and tumors were collected and weighted for subsequent analysis. The Committee on the Ethics of Huaihua First People’s Hospital approved this study.
Immunohistochemistry (IHC)
For tumor sections, tissues from the excised tumors were in fixation by 10% formaldehyde, embedment by paraffin, and cut into sections (4-μm thick). After above procedures, the sections were carried out at an incubation with Ki67 antibody (Abcam, USA) at 4°C overnight. After incubating with secondary antibody, they were stained with diaminobenzidine, and a light microscope was used to take photographs [Citation26].
Western blot analysis
The isolation of total protein was performed via using RIPA buffer (Beyotime, Shanghai, China). The SDS-PAGE was applied to resolve the protein samples, which were later transferred onto PVDF membranes. Then, blocked with 5% fat-free milk for 2 h, the membranes were incubated with primary antibodies against LATS2 (ab110780, Abcam), EZH2 (ab191080, Abcam), E-cadherin (ab1416, Abcam), N-cadherin (ab18203, Abcam), or GAPDH (ab8245, Abcam), and then incubated with secondary antibody (Abcam) after washing. Finally, ECL detection (ThermoScientific) was used to visualize the protein signals.
Luciferase reporter assay
The wild-type or mutant-type LATS2 promoter region was inserted into the pGL3 luciferase reporters (Promega). The pGL3-LATS2 promoter-Wt or pGL3-LATS2 promoter-Mut reporters were co-transfected with sh-MYLK-AS1 or sh-NC into GC cells through using Lipofectamine 2000 (Invitrogen). After 48 h, dual-luciferase reporter assay system was employed to analyze the luciferase activity.
Subcellular fractionation assay
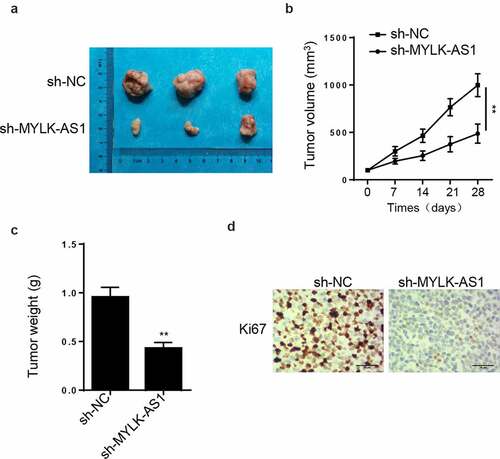
Firstly, AGS and MKN45 cells (1 × 106 cells) were suspended in precooled PBS. Next, the cells were centrifuged in cell fractionation buffer, followed by the collection of cell cytoplasm. Later, cell nuclei were acquired after cells lysed in cell disruption buffer. MYLK-AS1 expression in cytoplasmic and nuclear fractions was assayed by RT-qPCR. GAPDH or U6 were separately used as cytoplasmic or nuclear reference.
RNA immunoprecipitation (RIP) assay
RIP assay was used to validate the interaction between MYLK-AS1 and EZH2 according to the manufacturer’s protocol [Citation27]. GC cells were lysed in RNA lysis buffer, and later incubated with anti-EZH2-conjugated magnetic beads with anti-IgG as the negative control. Later, RIP buffer and PBS were applied to wash material that unbound on the magnetic beads. To purify the bound RNAs, the magnetic beads were resuspended by using TRIzol Reagents (1 ml). Finally, RT-qPCR was utilized for the analysis of the extracted RNAs levels.
ChIP assay
The ChIP Assay Kit (Beyotime) was employed for ChIP assay [Citation28]. In brief, cells were cross-linked with 1% formaldehyde, and later 0.125 mol/L glycine (final concentration) was added to terminate the reaction. After being processed by sonication, DNA fragments were generated and then immunoprecipitated using anti-EZH2 antibody (ab191250, Abcam), and IgG (Abcam) served as the internal control. At last, RT-qPCR was employed for determining the enrichment of precipitated DNAs.
Statistical analysis
Statistical data analysis was conducted by SPSS 20.0 (SPSS, Chicago, IL, USA), and results were shown as the mean ± SD. Kaplan–Meier method was used to calculate the overall survival curve. Student’s t-test or one-way ANOVA was employed to compare the differences between two or more groups. Each assay was repeated at least thrice, and P < 0.05 was considered as statistical significance.
Results
MYLK-AS1 was upregulated and predicted poor prognosis in GC
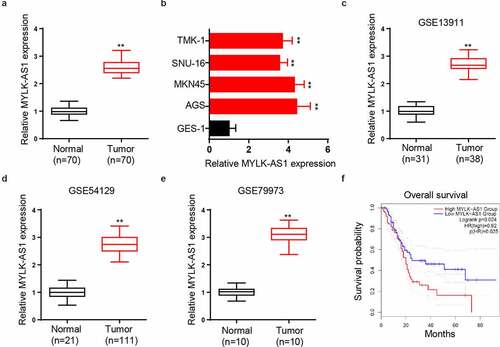
To explore MYLK-AS1 functional role in GC, we first utilized RT-qPCR to analyze MYLK-AS1 level in GC patients. The results indicated that MYLK-AS1 level in GC tissues was considerably upregulated compared to that in adjacent normal tissues (). Besides, MYLK-AS1 expression in GC cells (AGS, MKN45, SNU-16, and TMK-1) was detected by RT-qPCR, and GES-1 cell line was taken as the negative control. Data manifested that MYLK-AS1 expression was higher in GC cells (). In addition, by analyzing data (GSE13911, GSE54129, and GSE79973) from Gene Expression Omnibus (GEO) database, we found that MYLK-AS1 was highly expressed in GC tissues compared with that in normal gastric tissues (). Moreover, Kaplan–Meier survival analysis in another GEO dataset (GSE62254) indicated overall survival time of GC patients with high MYLK-AS1 expression was shorter than those with low MYLK-AS1 expression (). Collectively, MYLK-AS1 presented a high level in GC and predicted an unsatisfactory prognosis.
Figure 1. Relative MYLK-AS1 expression in GC tissues and cells, and its clinical significance. (A) RT-qPCR analysis for MYLK-AS1 expression in GC and adjacent normal tissues. (B) Relative MYLK-AS1 expression in GC cell lines and human normal gastric mucosal cell line by RT-qPCR analysis. (C-E) The expression of MYLK-AS1 was analyzed in GEO datasets (GSE13911, GSE54129 and GSE79973). (F) Overall survival of GC patients with high or low MYLK-AS1 expression by Kaplan-Meier method. **p< 0.01
MYLK-AS1 deficiency inhibited cell proliferation, cell cycle, migration, and invasion
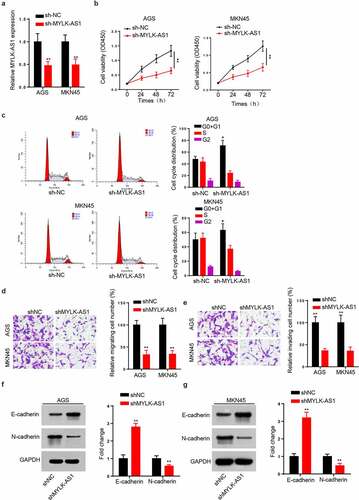
According to the above results, we carried out some in vitro assays to probe MYLK-AS1 function in GC. AGS and MKN45 cell lines, presenting higher MYLK-AS1 expression than SNU-16 and TMK-1 cell lines, were chosen to silence the expression of MYLK-AS1. The results showed that MYLK-AS1 expression was significantly decreased by transfecting sh-MYLK-AS1 (). CCK-8 assay indicated viability of sh-MYLK-AS1-transfected AGS and MKN45 cell lines was reduced with the comparison of the control group (). Moreover, flow cytometry analysis showed that MYLK-AS1 knockdown caused a cell-cycle arrest at G0/G1 phase (). Additionally, transwell assay showed that the migratory and invasive capacities were inhibited by the deficiency of MYLK-AS1 (). Next, the EMT process markers (E-cadherin and N-cadherin) were assessed through western blot. The E-cadherin level was increased, and N-cadherin level was decreased after MYLK-AS1 knockdown ( and G). Based on these results, we concluded that MYLK-AS1 knockdown inhibited GC cell proliferation and invasion in vitro.
Figure 2. Function of MYLK-AS1 on GC cell growth and invasion in vitro. (A) Relative MYLK-AS1 expression in AGS and MKN45 cells transfected with sh-MYLK-AS1 and sh-NC. (B) The viability of AGS and MKN45 cells transfected with sh-MYLK-AS1 and sh-NC was determined by CCK-8 assay. (C) The cell cycle of transfected AGS and MKN45 cells was assessed by flow cytometry analysis. (D and E) Transwell assay was carried out to evaluate the migratory and invasive abilities after AGS and MKN45 cells were transfected with sh-MYLK-AS1 and sh-NC. (F and G) The protein levels of E-cadherin and N-cadherin were measured by western blot. *p < 0.05, **p< 0.01
MYLK-AS1 silencing suppressed tumor growth in vivo
Then, AGS cells transfected with sh-MYLK-AS1 and sh-NC were injected into nude mice to evaluate the function of MYLK-AS1 in vivo in GC tumorigenesis. The result manifested that tumors harvested from MYLK-AS1-silencing group had a smaller size (). Moreover, tumor volume in sh-MYLK-AS1 group was also smaller (). In addition, compared with control group, tumor weight in sh-MYLK-AS1 group was lowered (). Importantly, IHC staining manifested that MYLK-AS1 knockdown decreased Ki67 expression with comparison of control group (). Taken together, MYLK-AS1 played oncogenic role in vivo.
Figure 3. Effect of MYLK-AS1 on GC tumor growth in vivo. (A) The tumors after being removed from the mice. (B) Tumor volume was calculated. (C) Tumor weight was determined after tumors were harvested. (D) IHC staining was conducted to determine Ki67 expression of tumors from sh-MYLK-AS1 and sh-NC groups. **p< 0.01
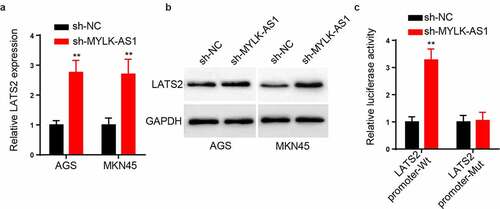
MYLK-AS1 inhibited LATS2 transcription in GC cells
Subsequently, the relationship between MYLK-AS1 and LATS2 was explored. As demonstrated in , compared to control group, the expression of LATS2 mRNA was increased by silencing MYLK-AS1. Moreover, MYLK-AS1 knockdown elevated the level of LATS2 protein by western blot analysis (). For further elucidating the specific mechanism of MYLK-AS1-mediating LATS2 expression, the luciferase reporter assay was carried out. As a result, MYLK-AS1 knockdown augmented the luciferase activity of pGL3-LATS2 promoter-Wt reporters, indicating that MYLK-AS1 retarded the transcription of LATS2 (). The above results suggested that MYLK-AS1 suppressed LATS2 transcription in GC cells.
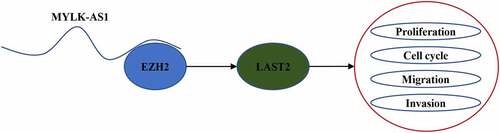
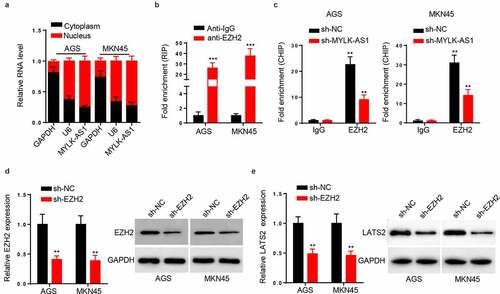
MYLK-AS1 recruited EZH2 to inhibit LATS2 transcription
To further explore the mechanism that MYLK-AS1 regulated LATS2 transcription, we conducted subcellular fractionation assay to assess the localization of MYLK-AS1 in GC cells. As a result, MYLK-AS1 was chiefly distributed in the nuclei of GC cells (). Subsequently, RIP assay demonstrated the considerable enrichment of MYLK-AS1 in the beads conjugated with anti-EZH2 (), suggesting that EZH2 interacted with MYLK-AS1. The next ChIP assay results indicated that MYLK-AS1 deficiency weakened the binding ability of EZH2 to LATS2 promoter (). Extensive reports have confirmed that EZH2 can be the main modulator in GC [Citation29,Citation30]; therefore, we utilized EZH2 shRNA to knockdown EZH2. Results manifested that EZH2 mRNA and protein expressions were both significantly reduced with the transfection of sh-EZH2 (), suggesting the successful transfection efficiency of sh-EZH2 in GC cells. In addition, we observed that EZH2 silencing downregulated the mRNA and protein expressions of LATS2 (). To be concluded, MYLK-AS1 inhibited LATS2 transcriptional expression by recruiting EZH2 in GC.
Figure 5. MYLK-AS1 decreased LATS2 transcriptional expression through recruiting EZH2. (A) MYLK-AS1 distribution in AGS and MKN45 cells was confirmed by subcellular fractionation assay. (B) RIP assay was performed in AGS and MKN45 cells, and the relative enrichment of MYLK-AS1 was detected by RT-qPCR. (C) The sh-MYLK-AS1 plasmid was transfected into AGS and MKN45 cells, and ChIP assay were conducted by the use of specific anti-EZH2 antibodies. (D) EZH2 mRNA and protein levels were tested by RT-qPCR and western blot in AGS and MKN45 cells transfected with sh-EZH2 or sh-NC. (E) RT-qPCR and western blot analyses for LATS2 mRNA and protein levels in transfected AGS and MKN45 cells. **p < 0.01, ***p< 0.001
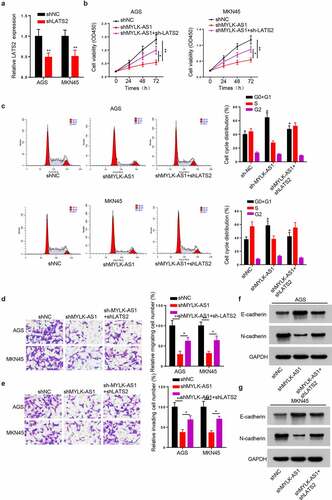
LATS2 silencing reversed the effect of MYLK-AS1 deficiency on GC cell growth
Finally, to verify whether MYLK-AS1 regulated GC cell growth via silencing LATS2 expression, we performed a series of restoration assays. RT-qPCR was employed to validate the transfection efficiency of sh-LATS2. The results manifested that LATS2 knockdown effectively decreased LATS2 expression in GC cells (). Then, CCK-8 assay results indicated that decreased LATS2 expression rescued sh-MYLK-AS1-damaged cell viability (). In addition, flow cytometry analysis revealed that LATS2 silencing countervailed the G0/G1 arrest caused by MYLK-AS1 deficiency (). Moreover, transwell assay indicated that MYLK-AS1 silencing-mediated inhibition in cell migration and invasion was reversed with sh-LATS2 transfection (). In addition, the increased E-cadherin expression and the decreased N-cadherin expression induced by MYLK-AS1 knockdown were attenuated by LATS2 silencing ( and G). Taken together, LATS2 knockdown involved the oncogenic effect of MYLK-AS1 in GC.
Figure 6. The effect of MYLK-AS1 knockdown on GC cell growth and invasion was rescued by silencing LATS2. (A) The transfection efficiency of sh-LATS2 in AGS and MKN45 cells. (B) Cell viability was evaluated by CCK-8 assay in AGS and MKN45 cells transfected with sh-NC, sh-MYLK-AS1 or sh-MYLK-AS1+sh-LATS2. (C) The cell cycle of transfected AGS and MKN45 cells. (D and E) Transwell assay was performed to assess the migration and invasion of transfected AGS and MKN45 cells. (F and G) The protein levels of E-cadherin and N-cadherin were measured by western blot. *p < 0.05, **p< 0.01
Discussion
In recent years, increasing lncRNAs were found by RNA-sequencing technology with the continuous improvement in experimental technology, and most of them were validated to play a necessary role in tumor occurrence and development [Citation31–33]. Although studies have reported numerous differently-expressed lncRNAs, which can regulate the progression of GC [Citation34–36], more lncRNAs linked to GC progression are still needed to be explored. MYLK-AS1, as a 814-bp lncRNA, has been found to facilitate the invasion and angiogenesis in hepatocellular carcinoma by acting as miR-424-5p sponge to activate VEGFR-2 signaling pathway [Citation37]. Nevertheless, function and mechanism of MYLK-AS1 is little known in GC pathogenesis and progression. Herein, we discovered upregulation of MYLK-AS1 in GC tissues and cells. The silencing of MYLK-AS1 in GC cells restrained cell viability, cell cycle, migration, and invasion. It was also validated that MYLK-AS1 knockdown inhibited tumor growth through in vivo experiments.
LATS2 has been confirmed as a tumor suppressor and exerts anti-tumor effect by multiple signaling pathways [Citation38,Citation39]. As the core kinase of Hippo pathway, LATS2 was also reported to act as inhibitor gene in diverse cancers, including prostate cancer [Citation40], lung cancer [Citation41], leukemia [Citation42], and breast cancer [Citation43] via regulating phosphorylation of YAP and TAZ (transcriptional co-activator with PDZ domain) [Citation44,Citation45]. Additionally, the shortage of LATS2 was confirmed to associate with elevated resistance of chemotherapeutic drug in leukemia [Citation46] and prostate cancer [Citation47]. Our luciferase reporter assay results showed that MYLK-AS1 knockdown increased LATS2 promoter activity.
EZH2, a subunit of PRC2, is identified as highly conserved histone methyltransferase that exerts regulatory function via triggering H3K27me3 trimethylation [Citation48]. It is also reported to restrain translation of numerous downstream genes and modulate cell cycle, proliferation, differentiation, apoptosis, aging, and tumorigenesis [Citation49,Citation50]. Existing reports indicated that lncRNAs can bind with EZH2 to play its biological role via modulating downstream gene expression [Citation51,Citation52]. Our study revealed that MYLK-AS1 directly bound with EZH2 and promoted cell proliferation and invasion by decreasing LATS2 expression.
Conclusions
Our research was the first to explore biological role and mechanism of MYLK-AS1 in GC, and results showed the upregulated level of MYLK-AS1 in GC cells. Further, it was discovered that MYLK-AS1 silenced LATS2 expression to exert oncogenic function by binding with EZH2. These findings might offer a novel insight into the exploration of GC diagnosis and treatment. In the future study, we will enlarge the sample size of animal experiments to investigate the biological role of MYLK-AS1 in GC.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the author(s).
Acknowledgements
Not applicable.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. .
- Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641.
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874.
- Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652).
- Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268–277.
- Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74(23):6890–6902.
- Huang J, Zhou N, Watabe K, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5(1):e1008.
- Yin D, Lu X. Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered. 2021;12(1):225–239.
- Wang H, Di X, Bi Y, et al. Long non-coding RNA LINC00649 regulates YES-associated protein 1 (YAP1)/Hippo pathway to accelerate gastric cancer (GC) progression via sequestering miR-16-5p. Bioengineered. 2021;12(1):1791–1802. .
- Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–16903.
- Kimura K, Wakamatsu A, Suzuki Y, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16(1):55–65.
- Zhu S, Zhang J, Gao X, et al. Silencing of long noncoding RNA MYLK-AS1 suppresses nephroblastoma via down-regulation of CCNE1 through transcription factor TCF7L2. J Cell Physiol. 2021;236(8):5757–5770.
- Liu J, Zhao SY, Jiang Q, et al. Long noncoding RNA MYLK-AS1 promotes growth and invasion of hepatocellular carcinoma through the EGFR/HER2-ERK1/2 signaling pathway. Int J Biol Sci. 2020;16(11):1989–2000.
- Abe Y, Ohsugi M, Haraguchi K, et al. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580(3):782–788.
- McPherson JP, Tamblyn L, Elia A, et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. Embo J. 2004;23(18):3677–3688.
- Yabuta N, Okada N, Ito A, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 2007;282(26):19259–19271.
- Hua K, Jin J, Zhao J, et al. miR-135b, upregulated in breast cancer, promotes cell growth and disrupts the cell cycle by regulating LATS2. Int J Oncol. 2016;48(5):1997–2006.
- Wang YJ, Liu JZ, Lv P, et al. Long non-coding RNA CCAT2 promotes gastric cancer proliferation and invasion by regulating the E-cadherin and LATS2. Am J Cancer Res. 2016;6(11):2651–2660.
- Tschöp K, Conery AR, Litovchick L, et al. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011;25(8):814–830.
- Li Q, Pan X, Zhu D, et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316.
- Jin L, Cai Q, Wang S, et al. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018;9(10):1017.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Hong Y, Liang H, Uzair Ur R, et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421.
- Zhang L, Kang W, Lu X, et al. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17(15):1886–1900.
- Chen L, Hu N, Wang C, et al. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17(3):319–329.
- Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782.
- Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202.
- Lu Y, Hou K, Li M, et al. Exosome-Delivered LncHEIH promotes gastric cancer progression by upregulating EZH2 and stimulating methylation of the GSDME Promoter. Front Cell Dev Biol. 2020;8:571297.
- Han H, Wang S, Meng J, et al. Long noncoding RNA PART1 restrains aggressive gastric cancer through the epigenetic silencing of PDGFB via the PLZF-mediated recruitment of EZH2.Oncogene. 2020;39(42):6513–6528.
- Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746–2755.
- Barnhill LM, Williams RT, Cohen O, et al. High expression of CAI2, a 9p21-embedded long noncoding RNA, contributes to advanced-stage neuroblastoma. Cancer Res. 2014;74(14):3753–3763.
- Prensner JR, Chen W, Iyer MK, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74(6):1651–1660.
- Cao Y, Xiong JB, Zhang GY, et al. Long Noncoding RNA UCA1 Regulates PRL-3 expression by sponging microRNA-495 to promote the progression of gastric cancer. Mol Ther Nucleic Acids. 2020;19:853–864.
- Li ZT, Zhang X, Wang DW, et al. Overexpressed lncRNA GATA6-AS1 Inhibits LNM and EMT via FZD4 through the Wnt/β-catenin signaling pathway in GC. Mol Ther Nucleic Acids. 2020;19:827–840.
- Dong Y, Li X, Lin Z, et al. HOXC-AS1-MYC regulatory loop contributes to the growth and metastasis in gastric cancer. J Exp Clin Cancer Res. 2019;38(1):502.
- Teng F, Zhang JX, Chang QM, et al. Correction to: lncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):277.
- Gao Y, Yi J, Zhang K, et al. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36(1):161.
- Wu T, Hu H, Zhang T, et al. miR-25 promotes cell proliferation, migration, and invasion of non-small-cell lung cancer by targeting the LATS2/YAP signaling pathway. Oxid Med Cell Longev. 2019;2019:9719723.
- Powzaniuk M, McElwee-Witmer S, Vogel RL, et al. The LATS2/KPM tumor suppressor is a negative regulator of the androgen receptor. Mol Endocrinol. 2004;18(8):2011–2023.
- Strazisar M, Mlakar V, Glavac D. LATS2 tumour specific mutations and down-regulation of the gene in non-small cell carcinoma. Lung Cancer. 2009;64(3):257–262.
- Jiménez-Velasco A, Román-Gómez J, Agirre X, et al. Downregulation of the large tumor suppressor 2 (LATS2/KPM) gene is associated with poor prognosis in acute lymphoblastic leukemia. Leukemia. 2005;19(12):2347–2350.
- Takahashi Y, Miyoshi Y, Takahata C, et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11(4):1380–1385.
- Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J Biol Chem. 2008;283(41):27534–27546.
- Lei QY, Zhang H, Zhao B, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28(7):2426–2436.
- Kawahara M, Hori T, Chonabayashi K, et al. Kpm/Lats2 is linked to chemosensitivity of leukemic cells through the stabilization of p73. Blood. 2008;112(9):3856–3866.
- Guo Y, Cui J, Ji Z. miR-302/367/LATS2/YAP pathway is essential for prostate tumor-propagating cells and promotes the development of castration resistance.Oncogene. 2017;36(45):6336–6347.
- Fillmore CM, Xu C, Desai PT, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to topoII inhibitors. Nature. 2015;520(7546):239–242.
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313.
- Joshi P, Carrington EA, Wang L, et al. Dominant alleles identify SET domain residues required for histone methyltransferase of Polycomb repressive complex 2. J Biol Chem. 2008;283(41):27757–27766.
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914.
- Chen L, Hu N, Wang C, et al. Long Noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-catenin pathway by scaffolding EZH2. Clin Cancer Res. 2018;24(3):684–695.