ABSTRACT
In this study, the expression of PTAR in cervical cancer tissues and cells was quantified by real-time PCR. Then, the roles of PTAR in HeLa cell proliferation and cell cycle were analyzed by a CCK-8 assay and flow cytometry, respectively.The effects of PTAR on cell migration and invasion were checked by Transwell and wound healing assays.The effect of PTAR on HeLa cell apoptosis was analyzed using annexin V/FITC staining. Finally, the interaction between PTAR and miR-101 in uterine cancer was verified through a dual-luciferase reporter assay and correlation analysis. The results showed that PTAR expression was aberrantly ascended in cervical cancer tissues and cell lines (Caski, SW756, SiHa, C33A and HeLa cells). Overexpressed PTAR could promote cell proliferation, migration and invasion in HeLa cells, which were suppressed by PTAR knockdown. Moreover, cell cycle progression stalled at the G1-G0 phase could be released with PTAR overexpression. The transfection of a PTAR vector inhibited apoptosis, while si-PTAR transfection increased apoptosis. Furthermore, PTAR could act as an endogenous sponge by directly binding to miR-101 and downregulating miR-101 expression. In conclusion, lncRNAPTAR plays a vital role and may be an effective target for the diagnosis and therapy of cervical cancer.
Introduction
Uterine cancer is the most common and highest fatality rates malignant tumor of the female reproductive system in developing countries. During primary surgical treatment, common sites of uterine cancer metastasis, such as adnexa, peritoneal surfaces, and pelvic and para-aortic lymph nodes, are identified [Citation1]. It is estimated that there are approximately 500,000 new cases occur and 275,000 cases died each year worldwide [Citation2]. Thus, a clear molecular mechanism that induce cell proliferation and migration would benefit the treatment of uterine cancer.
In recent decades, noncoding RNAs (ncRNAs) have been increasingly investigated in the field of molecular oncology because they involve in various pathway activation in the pathological process of tumors [Citation3]. The association between ncRNAs and human disease has been widely investigated in the context of microRNA (miRNA) [Citation4].However, long noncoding RNAs (lncRNAs) regulate gene expression in many manners, including transcription, chromosome remodeling,and post-transcriptional processing, which are nonprotein-coding RNAs that are longer than 200 nucleotides, resulting in human diseases and cancers [Citation5,Citation6]. Moreover, it has been confirmed that lncRNAs act as competitive RNA (ceRNA) for microRNAs (miRNAs) to regulate tumor occurrence, cell invasion and migration [Citation7,Citation8]. PTAR is a newly identified lncRNA that regulate ETM transition in ovarian cancer by regulating ZEB1 through binding miR-101-3p [Citation7]. However, it is still not clear about the exact role of PTAR in uterine cancer. In addition, in uterine cancer, miR-101 expression is obviously inhibited [Citation9]. Therefore, it is reasonable to speculate that PTAR may promote uterine cancer by binding miR-101.
In the present study, we hypothesized that lncRNA PTAR would be a potential biomarker for cervical cancer diagnosis and as a sponge to bind with downstream miRNA. We first analyzed PTAR expression in cervical cancer tissues, para-carcinoma tissues and various uterine cancer cell lines. Then, the effects of PTAR on cellbiology about proliferation, migration, invasion and apoptosis were analyzed in uterine cancer. Finally, the interaction between PTAR and miR-101 was clarified.
Materials and methods
Ethics statement. This study was performed according to the guidelines of the Qingdao University Institutional Review Board (IRB).
Tissue collection. Cancer tissues and corresponding para-carcinoma tissues were collected from Songshan Hospital, Affiliated Hospital of Qingdao University (Qingdao, China) from February 2017 to January 2018. Patients aged ≥18 years with histologically confirmed advanced or recurrent uterine cervical cancer, or STS not curable by surgical or radiation therapy were eligible. Other main inclusion criteria were: ≥1 previous chemotherapy regimen for advanced/recurrent uterine cervical, or ≥2 previous chemotherapy regimens for advanced/recurrent STS; ≥1 measurable lesion as defined in the RECIST guidelines, version 1.1; ECOG performance status score of 0 or 1; and adequate hematological, hepatic, and renal function.
Cell culture. The Caski, SW756, SiHa, C33A and HeLa human cervical cancer cell lines were cultured using RMPI-1640 medium (HyClone, LA, USA). 293 T cells were cultured using DMEM medium (HyClone, LA, USA) supplemented with 10% fetal bovine serum (FBS, HyClone, LA, USA) and 1% penicillin and streptomycin (HyClone, LA, USA) and were incubated at 37°C with 5% CO2.
Plasmid construction and transfection. A pcDNA3.1 vector cDNA was used for encoding PTAR subcloning. PTAR siRNA was synthesized by Sigma-Aldrich (MO, USA) for PTAR knockdown (HeLa-si-PTAR cells). Vectors of hsa-miR-101 mimics or inhibitors were to the plates and incubated for 48 h, after 6 h transfection.
Dual-luciferase procedure. PTAR with a wild-type miR-101-3p binding site and PTAR with a mutatedmiR-101-3p binding site were generated and fused to the luciferase reporter vector psi-CHECKTM-2 (Promega, WI, USA). A dual-luciferase reporter assay kit (Promega, WI, USA) was used for the luciferase assays. Briefly, the harvested cells were washed once with cold PBS. Cells were lysed with passive lysis buffer (100 μL), and the supernatant was collected by centrifugation at 12,000 × g for 30s. Promega GloMax (WI, USA) was used to determine the relative luciferase expression values. The results are presented as the relative luciferase activity (firefly luciferase/Renilla luciferase).
Real-time PCR procedure. Total RNA of tissues and cells was extracted using TRIzol reagent (Invitrogen, CA, USA) and reverse transfected using cDNA reverse transcription kits (Promega, WI, USA). The 7500 Fast RT-PCR instrument was used for detection. The PCR amplification process included the following parameters: 95°C for 2 min, 40 cycles at 95°C for 15 s, and 60°C for 32 s. The relative mRNA expression was detected by the 2-ΔΔCt method, using GAPDH orU6 as the reference genes. The PCR primer sequences are shown in .
Table 1. PCR primer sequences used in this study
Cell proliferation assay. The CCK-8 kit was chosen for cell proliferation evaluation according to previous instructions. Briefly, cells were plated and cultured in 96-well plates. Then, 10 μL of CCK-8 reagent was added for viable cell staining. After an additional incubation for 4 h, the spectrophotometric absorbance at 450 nm was recorded. Cell proliferation was calculated using the following equation proliferation (%) = (1-ODassessment time/OD0 h) ×100%.
Cell cycle analysis. Cells were cultured to 75–80% confluence. First, PBS was used to wash the cells three times. Then, the cells were trypsinized, collected and washed with PBS. Next, the cells were fixed in 70% cold ethanol and stored at −20°C. After fixation, the cells were washed with PBS and stained with 50 μg/mL PI in the dark for 30 min at 4°C. The cells were subsequently analyzed using flow cytometry.
Annexin V-FITC flow cytometry. Analysis of apoptosis was performed using an Annexin V-FITC Apoptosis Detection Kit (Keygen, Nanjing, China). Briefly, the cells were trypsinized and collected after they were treated. After washing, the cells twice with PBS, the cells were stained with PI and annexin V-FITC before flow cytometry analysis.
Western blot analysis. Total proteins were extracted and separated by electrophoresis (Bio-Rad) and then electrophoreticallytransferred into the NC membranes. Then, the NC membranes were blocked with 3% milk (Sangon Biotech), and incubated in primary antibody solution overnight at 4°C. After washing with TBST for 3 times, the corresponding secondary antibody was used for membrane incubation for 1 h at RT temperature. Blots were then developed with an ECL detection system as reported methods. Rabbit anti-BCL-W, anti-BCL-2, anti-BCL-XL and anti-BAX antibodies were purchased from Cell Signaling Technology (MA, USA). A rabbit anti-caspase-3 antibody was purchased from Abcam (MA, USA). A rabbit anti-GAPDH antibody was obtained from Akasomics (Shanghai, China). IgG-HRP secondary antibodies were purchased from Southern Biotech (AL, USA).
Cell migration and invasion. For the invasion assay, the upper chamber was coated with a mixture of MEM (HyClone, LA, USA) and Matrigel (BD Biosciences, CA, USA). Non-invading cells on the upper side were removed using a cotton swab. Cells that migrated through the filters were stained with 0.5% crystal violet (Sigma-Aldrich, MO, USA). Images of the cells were acquired at 200× magnification.
Wound healing assay. HeLa cells were seeded in six-well plates (1 × 106 cells/mL) (Corning, NY, USA) overnight to form a monolayer. Confluent monolayers (100%) were scratched with 10 μL micropipette tips to produce a wound are that did not contain cells. Growth medium deprived of FBS was added to the plates. After transfection, wound healing images were obtained by microscopy at 0, 6, 24 and 48 h. Wound size was quantified using Image-Pro Plus 6.0 and calculated using the following equation migration (%) = (1-wound width assessment time/wound width0 h)×100%.
Statistical analysis.All data was shown as mean ± SD. For comparison between 2 groups, Student’s t-test was used for statistical analysis.Statistically significance was set at P < 0.05 using GraphPad Prism 6.0 software.
Results
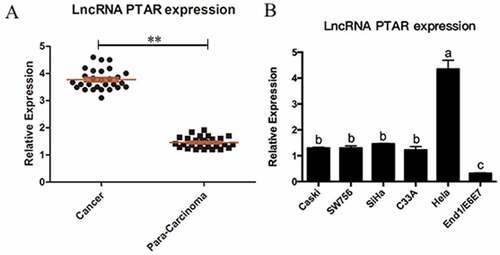
The expression of PTAR lncRNA in cervical cancer. In order to investigate the biological function of PTAR in cervical cancer, we collected 28 pairs of uterine cancer tissues and corresponding para-carcinoma tissues. Then, we determined the expression level of PTAR by real-time PCR. The results indicated that the expression level of PTAR in the uterine cancer tissues was significantly greater than that in the para-carcinoma tissues (P < 0.05) (). Next, PTAR expression was also analyzed in 5 types of cervical cancer cell lines, including Caski, SW756, SiHa, C33A and HeLa cells. The PTAR expression level was significantly increased in all investigated cervical cancer cell lines compared with the End1/E6E7 normal cervical cell line (P < 0.05). Moreover, the PTAR expression level in HeLa cells was significantly greater than that in Caski, SW756, SiHa and C33A cells (P < 0.01) (). Therefore, for the subsequent experiments, we used HeLa cells to further explore the role of PTAR in uterine cancer.
Figure 1. LncRNA PTAR expression was increased in uterine cancer tissues and Hela cell line. (a) Expression of lncRNA PTAR was detected by qRT-PCR and using GAPDH for normalization. The lncRNA PTAR expression in 28 pairs of uterine cancer compared with corresponding para-carcinoma specimens, **P < 0.01 vs Control. (b) PTAR expression was examined by qRT-PCR in uterine cancer cell lines and the normal cervical cell line End1/E6E7. N = 3, Letters indicate P < 0.05 vs Control
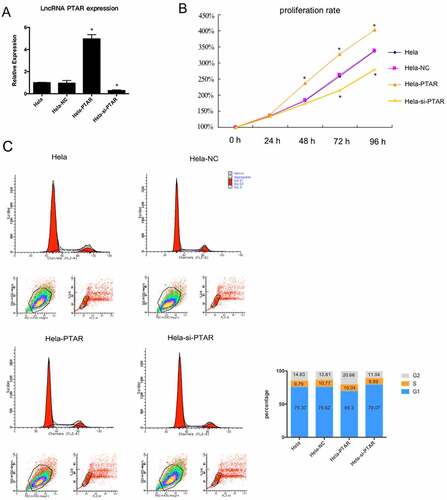
PTAR lncRNA increased uterine carcinoma cell proliferation. To examine the effects of PTAR on uterine carcinoma cell proliferation over 96 h, a PTAR overexpression vector and si-PTAR were transfected into HeLa cells. The PTAR overexpression vector significantly increased the PTAR expression level (P < 0.05), while si-PTAR markedly decreased the PTAR expression level (P < 0.05) (). Then, a CCK-8 assay was performed to evaluate HeLa cell proliferation with PTAR overexpression and knockdown. The results showed that beginning on day 2, the proliferation rate of HeLa cells overexpressing PTAR was significantly increased (P < 0.05); in contrast, PTAR knockdown significantly contributed to a decrease in cell proliferation beginning on day 3 (P < 0.05) (). Then, flow cytometry was used to further investigate the cell cycle. displayed that cell cycle progression stalled at the G1-G0 phase could be released with PTAR overexpression.
Figure 2. LncRNA PTAR promoted Hela cell proliferation. (a) PTAR expression in Hela cell line was examined by qRT-PCR after transfection with si-PTAR or PTAR vector. (b) Proliferation rate of the Hela cells was determined by CCK-8 assay after transfection with si-PTAR or PTAR for 0 h, 24 h, 48 h, 72 h and 96 h. (c) Cell cycle of Hela cells was analyzed by flow cytometry using PI staining. n = 3, *P < 0.05 vs control
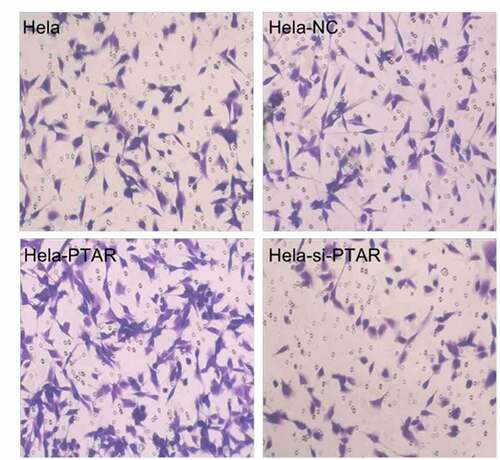
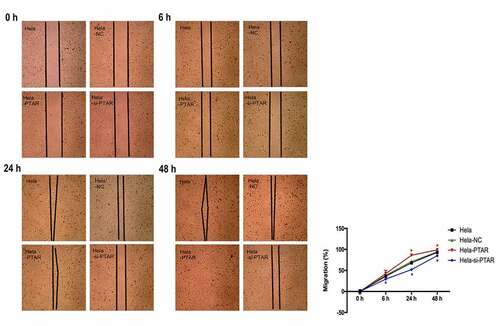
PTAR lncRNA promoted uterine carcinoma cell migration and invasion. Transwell assays were performed to evaluate the migration and invasion abilities of HeLa cells with PTAR overexpression and knockdown. shows that PTAR overexpression significantly enhanced the migration and invasion abilities of HeLa cells (P < 0.01); however, si-PTAR led to a decrease in cell migration and invasion abilities (P < 0.01). Then, a scratch wound healing assay was carried out to further confirm the role of PTAR on cell migration. As shown in , at 24 and 48 h after the wound was established, compared with HeLa-NC cells, HeLa-PTAR cells were observed to migrate toward the open wound to close the scratched wound and significantly accelerated the wound healing process (P < 0.05). However, HeLa-si-PTAR cells showed a negative trend toward wound healing at 6, 24 and 48 h (P < 0.05).
Figure 3. LncRNA PTAR promoted Hela cell migration and invasion. The cell (a) migration and (b) invasion ability was determined by transwell assay after transfection with si-PTAR or PTAR vector. n = 4, **P < 0.01 vs control. Images of the cells were photographed at 200 x magnification
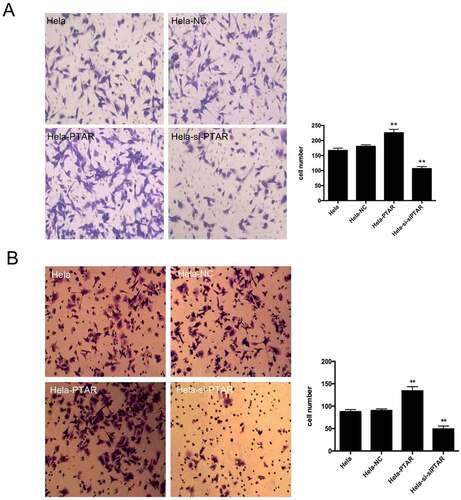
Figure 4. LncRNA PTAR promoted Hela cell wound healing. The cell migration ability after transfection with si-PTAR or PTAR vector for 0 h, 6 h, 24 h and 48 h were further determined by wound healing assay. n = 4, *P < 0.05 vs control. The black thick lines indicate the wound edges.Images of the cells were photographed at 200 x magnification
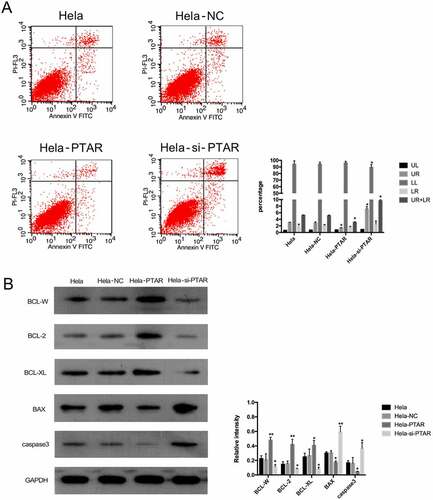
PTAR lncRNA inhibited the apoptosis of uterine carcinoma cells. To further investigate the role of PTAR in cell apoptosis, we detected the apoptotic cells by flow cytometry. The percentage of apoptotic HeLa cells is shown in by flow cytometry under the same culture conditions. PTAR overexpression contributed to decreases in early and late apoptotic cells (P < 0.05). In contrast, for PTAR knockdown, the ratio of viable cells to total cells was decreased, while the percentages of late apoptotic cells were increased (P < 0.05). To further confirm the apoptosis of HeLa cells, Western blotting was performed to analyze the expression of antiapoptotic and proapoptotic proteins. As shown in , the expression levels of the antiapoptotic BCL-2 family members BCL-W, BCL-2 and BCL-XL were significantly upregulated by PTAR overexpression (P < 0.05) but significantly downregulated by PTAR knockdown (P < 0.05). In contrast, the expression levels of BAX and caspase-3, which are two proapoptotic proteins, were significantly decreased in HeLa-PTAR cells (P < 0.05) and increased in HeLa-si-PTAR cells (P < 0.05).
Figure 5. LncRNA PTAR inhibited Hela cell apoptosis. (a)The cell apoptosis after transfection with si-PTAR or PTAR vector were determined by Annexin V/FITC using flow cytometry. (b) Protein levels of BCL-W, BCL-2, BCL-XL, BAX, caspase3 and GAPDH in Hela cells were determined using Abs recognizing total protein. The expression of the apoptosis-related proteins was normalized to GAPDH expression.N = 3, Letters indicate P < 0.05, **P < 0.01 vs control
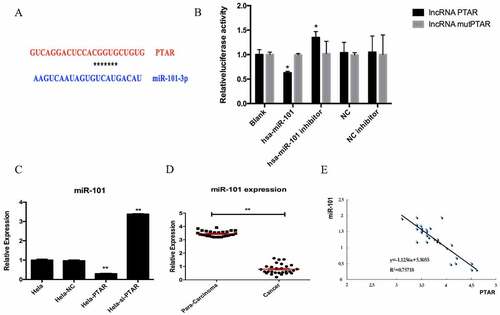
PTAR lncRNA negatively regulated miR-101 expression. In order to check the molecular mechanism of PTAR, we analyzed the downstream miRNA of PTAR. According to the prediction results, there is a binding site for miR-101 in PTAR (). It is observed that hsa-miR-101 reduced the luciferase activities of the wild-type (WT) PTAR vector. However, the luciferase activity in 293 T cells transfected with the miR-101 inhibitor was increased, indicating the direct binding between PTAR and miR-101 (). Then, the miR-101 expression level in HeLa cells was analyzed. The results showed that miR-101 expression was significantly downregulated by PTAR overexpression (P < 0.01) but was significantly upregulated by PTAR knockdown (p < 0.01) (). Moreover, the para-carcinoma tissues had a higher miR-101 expression level than the cancer tissues (P < 0.01) (), and the correlation analysis suggested that the expression of miR-101 was negatively correlated with the expression of PTAR in cancer tissues (R2 = 0.75718) ().
Figure 6. PTAR reduced miR-101 expression in uterine cancer. (a) Putative miR-101 binding sequence of PTAR. (b) The normalized luciferase activity detection results in this study. (c) miR-101 expression was detected after transfection with si-PTAR or PTAR vector. (d) Expression of miR-101 was analyzed and normalized to GAPDH expression, in 28 pairs of uterine cancer compared with corresponding para-carcinoma specimens. (e) Correlation analysis between PTAR expression and miR-101 expression.n = 3, *P < 0.05,**P < 0.01 vs control
Discussion
Uterine cancer is the main cause of death for female tumor. In recent decades, augmenting evidence has shown that lncRNAs provide a cellular growth advantage, leading to progressive and uncontrolled tumor growth [Citation10,Citation11]. Therefore, efforts should be made to clarify the biological and molecular mechanisms of lncRNAs in cervical cancer.
In the present study, we demonstrated that the expression of PTAR, which is a novel lncRNA, was significantly upregulated in uterine cancer tissues and uterine cancer cell lines (). Specifically, an increased PTAR expression level was shown to promote HeLa cell proliferation, migration and invasion and decrease cell apoptosis (). However, the knockdown of PTAR expression led to the significant inhibition of cell proliferation, migration and invasion and led to a significant increase in cell apoptosis (). These findings suggest that PTAR plays a direct role in the modulation of cell proliferation and uterine cancer progression and could be a useful novel prognostic and progression marker for uterine cancer.
Although most lncRNAs have been demonstrated to play crucial biological roles in human cancers, accurate molecular alterations modulated by lncRNAs remain largely unknown. Commonly, ceRNAs refer to all transcripts that may become the targets of miRNA such as lncRNA, pseudogene RNA, and circular RNA [Citation12]. Increasing reports have suggested that interactions between noncoding RNAsplay key roles in cancer progression regulation. For example, UICLM lncRNA acts as a ceRNA for miR-215 to promote colorectal cancer metastasis [Citation13]. In gallbladder cancer, PAGBC lncRNA promotes tumor growth by competitively binding to miR-133b and miR-511 [Citation14]. In a recent study, PTAR lncRNA was found to promote epithelial-to-mesenchymal transition and tumor cell invasion and metastasis in malignant ovarian cancer by binding miR-101-3p [Citation7]. In the present study, we also identified miR-101-3p as a target of PTAR in uterine cancer by bioinformatics prediction based on sequence complementarity and dual-luciferase assays. Since PTAR was highly expressed in uterine cancer tissue, we further investigated the effects of PTAR in uterine cancer and its relationship with miR-101.In HeLa cells, overexpression of PTAR decreased miR-101 level, while silencing of PTAR increased miR-101 expression, because PTAR may promote uterine cancer by regulating miR-101 targeted mRNAs. Similar results can be found in the study of Zhao et al., in which the knockdown of lncRNA ANRIL could increase the expression of its target miRNA, let-7a [Citation15]. Moreover, in the present study, we also observed that the miR-101 expression level in para-carcinoma tissues was significantly greater than that in cancer tissues (), suggesting that miR-101-3p is downstream of PTAR. The inverse correlation between the expression levels of PTAR and miR-101-3p in clinical uterine cancer samples further validated the targeted association between the two noncoding RNAs (). The involvement of miR-101 in cancer development has been investigated by many groups. For example, downregulation of miR-101 expression in gastric and colon cancers is correlated with tumor growth [Citation16,Citation17]. Forced expression of miR-101 inhibits prostate cancer cell growth [Citation18]. Moreover, in uterine cancer, miR-101 has been reported to suppress the proliferation and invasion of aggressive cancer cells [Citation19]. Similarly, our study also suggested the tumor-promoting role of miR-101.Overall, these data indicate that PTAR may serve as an endogenous sponge of miR-101 and that miR-101 facilitates uterine cancer.
In summary, our work showed that PTAR functions as an oncogene by promoting the proliferation, invasion, and migration of uterine cancer cells and inhibiting the apoptosis of uterine cancer cells. Notably, mechanistic analysis uncovers that PTAR-miR-101 pathway plays an important role in uterine cancer. The role of PTAR in uterine cancer suggest that PTAR may be an effective target for uterine cancer therapies.However, limitations exist in this study. For example, data were absent on additional cell lines besides HeLa cells, and the evaluation of apoptosis can be more persuasive if detected under certain stress. Moreover, the possible downstream targets of miR-101 were not included. Since PTAR is a newly found lncRNA that can regulate ovarian cancer, it is aimed to figure out the effect of PTAR in uterine cancer and its possible mechanism of action. So here we only analyzed the binding site between PTAR and miR-101. In some literatures, miR-101 was discovered to regulate many downstream targets, such as EZH2, MCL-1, FOS, ZEB1 and TRIM44 [Citation19–21]. Thus, in the future study, the possible downstream targets of PTAR-miR-101 will be investigated in cervical cancer. There are still limitations in this study, we enrolled participants histologically confirmed advanced or recurrent uterine cervical cancer, and we did not divide the participants into different groups according to histological types (squamous cell carcinoma, adenocarcinoma, or endometrial cancer). Therefore, we did not measure the pTAR expression in different groups. We will perform further investigation in this direction.
Conclusion
In this study, we investigated the biological functions of lncRNA PTAR in cervical cancer. Those novel findings suggest that PTAR plays a direct role in the modulation of cell proliferation and uterine cancer progression and could be a useful novel prognostic and progression marker for uterine cancer.
Highlight
(1) LncRNA PTAR was ascended in cervical cancer tissues and cell lines;
(2) LncRNA PTAR as sponge by directly binding to miR-101;
(3) LncRNA PTAR could be used as potential therapy target.
Authors’ contributions
The work was designed by XW. The experiments were performed by HS and SZ. The statistical analyses were performed by XW. The manuscript was prepared by XW.
Ethics statement
The project was approved by theSongshan Hospital Affiliated to Qingdao University (No. QDUEC201601237).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Walker JL, Piedmonte MR, NM S, et al. Pearl ML and Sharma SK: recurrence and survival after random assignment to laparoscopy versus laparotomy for comprehensive surgical staging of uterine cancer: gynecologic Oncology Group LAP2 Study. J Clin Oncol. 2012;30(7):695–700.
- Peng L, Peng W. Hu P and Zhang HF: clinical significance of expression levels of serum ADRA1A in hysterocarcinoma patients. Oncol Lett. 2018;15(6):9162–9166.
- Calle AS, Kawamura Y, Yamamoto Y. Takeshita F and Ochiya T: emerging roles of long non‐codingRNAin cancer. Cancer Sci. 2018;109(7):2093–2100.
- Paul P, Chakraborty A, Sarkar D, et al. Malakar AK and Chakraborty S. Interplay between miRNAs and Human Diseases: a Review. J Cell Physiol. 2007-2018, 2018;233: 2007–2018.
- Shi X, Sun M, Liu H. Yao Y and Song Y: long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166.
- Guo X, Xiao H, Guo S, et al. Chen J and Lou G: long noncoding RNA HOTAIR knockdown inhibits autophagy and epithelial-mesenchymal transition through the Wnt signaling pathway in radioresistant human cervical cancer HeLa cells. J Cell Physiol. 2008;234(4):3478–3489.
- Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 2018;17(1):1–13. .
- Tan X, Banerjee P, Liu X, et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest. 2018;128(4):1267–1282. .
- Ratner ES, Tuck D, Richter C, et al. Putherford TJ and Weidhaas JB: microRNA signatures differentiate uterine cancer tumor subtypes. Gynecol Oncol. 2018;118(3):251–257.
- Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci US A. 2009;106(28):11667–11672. .
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719.
- Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 viaceRNA mediated miRNA evasion. Nat Commun. 2017;8:289: 1–17.
- Chen D, Lu Y, Zhang J, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7(19):4836–4849.
- Wu X, Wang F, Li H, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 1837-1853, 2017;18: 1837–1853.
- Zhao B, Yang Y, Hu LB, et al. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-β1/Smad signaling pathway. Cancer Biomark. 2017;21(3):613–620. .
- He X, Shao Y, Li X, et al. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 2012;279(22):4201–4212. .
- Strillacci A, Griffoni C, Sansone P, et al. downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315(8):1439–1447. MiR-101. .
- Hao Y, Gu X, Zhao Y, et al. Enforced Expression of miR-101 Inhibits Prostate Cancer Cell Growth by Modulating the COX-2 Pathway In Vivo. Cancer Prev Res. 2011;4(7):1073–1083.
- Konno Y, Dong P, Xiong Y, et al. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget. 2015;51:S12–S12.
- Han L, Chen W, Xia Y, et al. inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1. Am J Transl Res. 2018;10(4): 1172–1183. MiR-101.
- Li L, Shao MY, Zou SC, et al. MiR-101-3p inhibits EMT to attenuate proliferation and metastasis in glioblastoma by targeting TRIM44. J Neurooncol 2019;141(1): 19–30