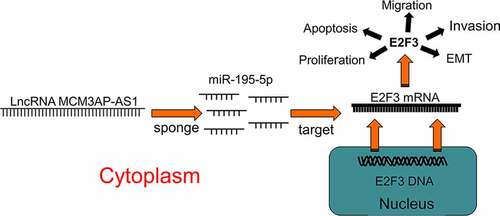
ABSTRACT
Lung cancer (LC) ranks first among all causes of cancer-related death, with non-small cell lung cancer (NSCLC) taking up 85% of lung cancer cases. Although lncRNA MCM3AP antisense RNA 1 (MCM3AP-AS1) has been reported to be an oncogenic factor in NSCLC, its detailed mechanism in NSCLC is unknown. In this study, quantitative real-time polymerase chain reaction (qRT-PCR) was performed to determine MCM3AP-AS1, microRNA (miR)-195-5p and E2F transcription factor 3 (E2F3) mRNA expressions in NSCLC tissues and cells. Western blot was utilized to determine the expression levels of E2F3, BCL2-associated X protein (Bax), B-cell lymphoma-2 (Bcl-2), E-cadherin and N-cadherin. CCK-8 and Transwell assays were conducted to examine cell proliferation, migration and invasion, respectively. Dual-luciferase reporter assay and RNA immunoprecipitation experiments were used to determine the regulatory relationships between MCM3AP-AS1 and miR-195-5p, and miR-195-5p and E2F3. We demonstrated that MCM3AP-AS1 was overexpressed in NSCLC tissues and cells, and MCM3AP-AS1 overexpression accelerated the proliferation, migration and invasion of NSCLC cells. In addition, MCM3AP-AS1 overexpression markedly up-modulated Bcl-2 expression and repressed Bax expression; MCM3AP-AS1 overexpression also significantly up-regulated N-cadherin expression and suppressed E-cadherin expression in NSCLC cells. What is more, in NSCLC cells, miR-195-5p was a target of MCM3AP-AS1, and the latter worked as a molecular sponge for miR-195-5p to regulate E2F3 expression. Collectively, MCM3AP-AS1, serving as a competitive endogenous RNA (ceRNA) to regulate miR-195-5p/E2F3 axis, promotes NSCLC progression, which is a promising therapeutic target for NSCLC.
KEYWORDS:
1. Introduction
Lung cancer (LC) is the commonest malignancy in the world with the highest mortality [Citation1]. Non-small cell lung cancer (NSCLC), as the main pathological subtype of LC, accounts for more than 80% of cases of LC [Citation2]. The treatment strategies of NSCLC mainly include surgery, radiotherapy, chemotherapy, targeted therapy and immunotherapy [Citation3]. Although a lot of efforts have been made to improve the therapy of NSCLC, the five-year survival rate of the patients is only about 18% [Citation4,Citation5]. Therefore, it is necessary to have a better understanding of the molecular mechanisms underlying NSCLC development to further improve the prognosis of the patients.
In recent years, accumulating studies find that long non-coding RNAs (lncRNAs) are aberrantly expressed in diverse tumors, which is involved in tumorigenesis and disease development [Citation6,Citation7]. For instance, lncRNA TPTEP1, SPRY4-IT1 and ZEB2-AS1 are reported to be aberrantly expressed in NSCLC and modulate biological processes such as proliferation and metastasis of cancer cells [Citation8–10]. Previous researches show that lncRNA MCM3AP antisense RNA 1 (MCM3AP-AS1) is associated with the progression of some tumors, such as prostate carcinoma, gastric carcinoma and cervical carcinoma [Citation11–13]. Reportedly, in NSCLC, MCM3AP-AS1 expression is also up-regulated, and the proliferation, migration and angiogenesis of NSCLC cells are impeded by MCM3AP-AS1 knockdown [Citation14]. However, the detailed mechanism of its effects in NSCLC is vague.
Some previous studies report that microRNA-195-5p (miR-195-5p) is a tumor suppressor in multiple cancers including NSCLC [Citation15–17]. Interestingly, our bioinformatics data suggested that MCM3AP-AS1 contained the binding sequence for miR-195-5p. We supposed that MCM3AP-AS1 could probably contribute to NSCLC progression via interacting with miR-195-5p. In the current research, we validated that MCM3AP-AS1 expression was up-regulated in NSCLC tissues and MCM3AP-AS1 enhanced the proliferation and metastasis of NSCLC cells; mechanistically, it was unmasked that MCM3AP-AS1 exerted tumor-promoting effects by modulating the miR-195-5p/E2F transcription factor 3 (E2F3) axis.
2. Materials and methods
2.1. Sample collection
From June 2017 to August 2018, sixty-three pairs of matched NSCLC tissues and tissues of resection margin were taken from the tissue bank of the Cancer Hospital of the University of Chinese Academy of Sciences. All patients were histologically confirmed as NSCLC by pathologists at the Cancer Hospital of the University of Chinese Academy of Sciences and were not suffering from any other malignancies, and no neoadjuvant therapy was given prior to surgery. Among the patients, there are 38 males and 25 females, aged from 41–74 years old (average: 52.4). Patients receiving neoadjuvant therapy, patients with mixed NSCLC/small cell histology or CNS metastases were excluded. The surgically resected NSCLC tissues and paracancerous tissues (at least 5 cm away from the tumor margin) were collected and stored in liquid nitrogen immediately after resection during surgery. All specimens were confirmed as NSCLC or normal lung tissue by postoperative pathological examination. All patients who provided tissues signed informed consent before the surgery, and all of them agreed to the use of their samples in scientific research. This research was endorsed by the Research Ethics Committee of Cancer Hospital of the University of Chinese Academy of Sciences (Approval Number: 2017–05).
2.2. Cell culture
NSCLC cells (A549, H358, H1299, H460 and H226) and human lung epithelial cells BEAS-2B were available from China Center for Type Culture Collection (CCTCC, Wuhan, China). The cells were cultured in RPMI-1640 medium (Biosharp, Shanghai, China) containing 10% fetal bovine serum (FBS, Biosharp, Shanghai, China) at 37°C in 5% CO2.
2.3. Cell transfection
MiR-195-5p mimics (5ʹ-UAGCAGCACAGAAAUAUUGGC-3ʹ) and its control miR NC (5ʹ-UCACAACCUCCUAGAAAGAGUAGA-3ʹ), miR-195-5p inhibitors (5ʹ-GCCAAUAUUUCUGUGCUGCUA-3ʹ) and its control inh NC (5ʹ-UUGUACUACACAAAAGUACUG-3ʹ), two different shRNAs targeting MCM3AP-AS1 (sh-MCM3AP-AS1#1: 5ʹ-GCTCACAATGATGGCACTA-3ʹ and sh-MCM3AP-AS1#2:
5ʹ-GGACAGAGGGAACATGGAT-3ʹ), its control sh-NC (5ʹ-GCTTAACGTAGACGACCTA-3ʹ), MCM3AP-AS1 overexpression plasmid and their negative controls (GenePharma, Shanghai, China) were designed and synthesized. In transfection, the final concentration of oligonucleotides was 50 nM. Transfection was conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) under the guidance of the protocol provided by the manufacturer. After 24 h following transfection, transfection efficiency was determined using quantitative real-time polymerase chain reaction (qRT-PCR) or used for subsequent experiments.
2.4. Bioinformatics analysis
In this study, GEPIA database (http://gepia.cancer-pku.cn/) was searched to analyze MCM3AP-AS1 expression in NSCLC samples [Citation18]; Kaplan Meier-plotter database (https://kmplot.com/) was employed to analyze the prognostic prediction value of MCM3AP-AS1 [Citation19]; StarBase database (http://starbase.sysu.edu.cn/) was used to validate the correlations among MCM3P-AS1, miR-195-5p and E2F3 [Citation20]. LncBase Predicted v.2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/) [Citation21] and TargetScan databases (http://www.targetscan.org/vert_72/) [Citation22] were applied to look for the binding sites between miR-195-5p and MCM3AP-AS1, E2F3 3ʹUTR.
2.5. qRT-PCR
Total RNA from tissues and cells was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was reversely transcribed into cDNA using the reverse transcription kit (Applied Biosystems, Foster City, CA, USA). cDNA was used as a template, and qRT-PCR was conducted using SYBR® Premix-Ex-Taq™ (Takara, Tokyo, Japan) on ABI7300 PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Additionally, the relative expression of miR-195-5p was normalized to U6, and the relative expressions of MCM3AP-AS1 and E2F3 mRNA were normalized to GAPDH, with 2−ΔΔCt method [Citation23]. Primer sequences were shown in .
Table 1. Sequences used for qRT-PCR
2.6. Cell viability
A549 and H226 cells were inoculated into 96-well plates at 1.0 × 103/well before 10 μL of cell counting kit (CCK-8) reagent (MedChem Express, Monmouth Junction, New Jersey, USA) was added into the wells at the 1st, 2nd, and 3rd d, respectively, and OD 450 values were measured on a microplate reader after 2 h of incubation.
2.7. EdU assay
A549 and H226 cells were inoculated into 24-well plates, respectively, and cultured for 24 h. After that, the cells were incubated with 200 μL of 50 μmol/L EdU medium (RiboBio Co., LTD, Guangzhou, China) for 2 h, rinsed with PBS and then fixed with paraformaldehyde for 10 min. Following that, the cells were incubated for 5 min with 200 μL of 2 mg/ml of glycine and rinsed with PBS for 5 min. One hundred microliters of PBS with 0.5% Triton X-100 was added to each well, and the cells were placed on a shaker for 10 min and then washed with PBS for 5 min. Subsequently, the cells were stained with Apollo® solution in the dark for 30 min, and nuclear DNA was counterstained with DAPI. Then, the cells were observed, and the images were collected using a fluorescent microscope in the dark.
2.8. Transwell assay
Migration assays were performed using Transwell chambers (8 µM pore size; Corning, Beijing, China). After dispersing A549 and H226 cells with 0.25% trypsin, the cells were centrifuged and resuspended with serum-free medium. 5 × 104 cells were inoculated into each chamber before the complete medium was added in the 24-well plate. After 24 h incubation at 37°C, the unmigrated cells were removed. The cells on the Transwell membranes were fixed with 4% paraformaldehyde for 10 min and stained with 0.5% crystal violet solution. After being rinsed with tap water, the migrated cells were counted under an inverted microscope. Invasion experiments were performed with Transwell chambers coated with Matrigel, and the remaining procedures were the same as the migration assay.
2.9. Western blot
Cellular protein was extracted using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China), and protein concentration was determined using a BCA kit (Beyotime Biotechnology, Shanghai, China). After SDS-PAGE, the protein samples were transferred to PVDF membranes and blocked with 5% skimmed milk for 1 h at room temperature. The corresponding primary antibodies were then added to interact with the proteins at 4°C overnight, and then the membrane was rinsed with tris buffered saline with Tween 20 (TBST). Next, the secondary antibodies were added to incubate the membrane for 2 h at room temperature before the membrane was rinsed with TBST again, and then ECL chemiluminescent kit (Millipore, Billerica, MA, USA) was added onto the membrane to develop the bands. The antibodies used in this study, including anti-E2F3 (ab50917, 1:500), anti-Bax (ab32503, 1:500), anti-Bcl-2 (ab185002, 1:500), anti-E-cadherin (ab11512, 1:500), anti-N-cadherin (ab18203, 1:500) and anti-β-actin (ab179467, 1:2000), were all bought from Abcam (Shanghai, China).
2.10. Dual-luciferase reporter gene experiment
Wild-type MCM3AP-AS1 and mutant MCM3AP-AS1 sequence, or wild-type E2F3 3′-UTR and mutant E2F3 3′-UTR sequence were inserted into a pGL3 vector (Promega, Madison, WI, USA) to construct MCM3AP-AS1-WT1, MCM3AP-AS1-WT2, MCM3AP-AS1-WT3, MCM3AP-AS1-MUT1, MCM3AP-AS1-MUT2, MCM3AP-AS1-MUT3, E2F3-WT and E2F3-MUT luciferase reporter vectors. The luciferase reporter experiment was conducted using the dual-luciferase reporter assay kit (Promega, Madison, WI, USA). MCM3AP-AS1-WT, MCM3AP-AS1-MUT, E2F3-WT and E2F3-MUT were co-transfected into HEK-293 T cells with miR-195-5p mimic (5ʹ-UAGCAGCACAGAAAUAUUGGC-3ʹ), miR-NC (5ʹ-UCACAACCUCCUAGAAAGAGUAGA-3ʹ), miR-195-5p inhibitors (5ʹ-GCCAAUAUUUCUGUGCUGCUA-3ʹ) and inh-NC (5ʹ-UUGUACUACACAAAAGUACUG-3ʹ) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), respectively, and the luciferase activity of the reporter of each group was measured 24 h later. The relative luciferase activity was normalized with renilla luciferase activity.
2.11. RIP experiment
A549 and H226 cells were transfected with pcDNA3.1-MCM3AP-AS1 vector or pcDNA3.1 vector. Forty-eight hours later, the cells were subjected to RIP assays using anti-Ago2 antibody and the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instruction. qRT-PCR was conducted to detect MCM3AP-AS1 and miR-195-5p expressions in the immunoprecipitate.
2.12. Lung metastasis model in vivo
The animal experiments were approved by the Animal Research Ethics Committee of Cancer Hospital of the University of Chinese Academy of Sciences (Approval Number: 2020–06). Ten male BALB/c nude mice (6–8 weeks old, average weight: 24.5 g ± 2.20) were purchased from Zhejiang Province Experimental Animal Center (Hangzhou, China). Mice were maintained under standard housing conditions (23°C, 40% humidity, 12 h light cycles, and free access to food and water). The mice were randomly divided into two groups (NC group and MCM3AP-AS1 overexpression group, n = 5 in per group). A549 cells (2 × 107 cells/per mouse) were injected into each mouse via tail vein. Three weeks later, the mice were euthanized with 100% oxygen/5% isoflurane, and the bilateral thoracotomy was used to confirm the death. Lung tissues were removed after lavage, fixed in 10% neutral formalin for 36 hours, and embedded in paraffin. Then, hematoxylin/eosin staining was performed for pathological examination of lung metastatic nodules of the mice. The number of pulmonary metastatic nodules in each section was counted in 5 randomly selected visual fields under the microscope (Olympus, Tokyo, China).
2.13. Statistical analysis
All the experiments were conducted in triplicate. The data were processed by GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA), plotted and represented as mean ± standard deviation. To make the comparison between two groups, One-Sample Kolmogorov–Smirnov test was used to examine whether the data are normally distributed. For the data which were normally distributed, independent sample t test was used. For the data which were skewed distributed, paired sample Wilcoxon signed rank test was used. One-way ANOVA test was performed to make the comparison among three or more groups. If there was a significant difference, Newman–Keuls analysis was performed to make the comparison between two groups. Pearson correlation analysis was used to determine the correlation. P < 0.05 signified statistical significance.
3. Results
In this study, we performed a series of experiments to investigate the biological function of MCM3AP-AS1 in NSCLC progression, and explore the regulatory mechanism of MCM3AP-AS1/miR-195-5p/E2F3 axis, which were aimed to help clarify the mechanism of NSCLC progression.
3.1. MCM3AP-AS1 and E2F3 expressions were up-regulated, and miR-195-5p expression was down-regulated in NSCLC tissue specimens
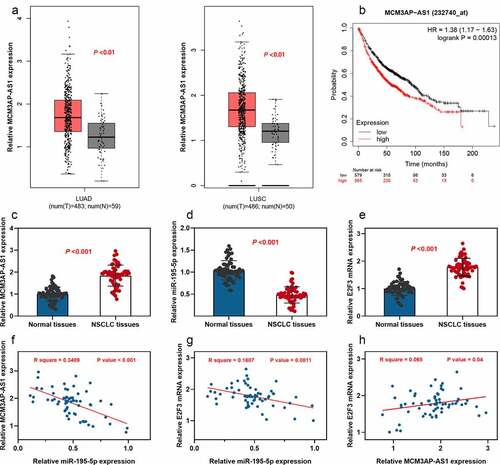
First of all, gene expression analysis and survival analysis were conducted using the GEPIA database. As shown, MCM3AP-AS1 expression was up-regulated in both lung adenocarcinoma tissues (LUAD) and lung squamous carcinoma (LUSC) tissues; additionally, MCM3AP-AS1 overexpression was significantly linked to a shorter overall survival time of NSCLC patients (–b). Then, the expression patterns of MCM3AP-AS1, miR-195-5p and E2F3 in paired specimens collected from 63 patients with NSCLC were examined by qRT-PCR, respectively. We found significant up-regulation of both MCM3AP-AS1 and E2F3 expressions and downregulation of miR-195-5p expression in NSCLC tissue specimens, in comparison with these in paired adjacent normal tissues (–e). Pearson’s correlation analysis further indicated that MCM3AP-AS1 and E2F3 were negatively correlated with miR-195-5p; conversely, MCM3AP-AS1 expression was positively correlated with E2F3 expression in NSCLC samples (–h). StarBase database also suggested that MCM3AP-AS1 expression and E2F3 expression were negatively correlated with miR-195-5p expression in NSCLC samples; conversely, MCM3AP-AS1 expression was positively correlated with E2F3 expression (Supplementary Figure 1a-c). Moreover, statistically, high MCM3AP-AS1 expression was closely associated with the larger tumor size, low differentiation and higher TNM stage of the NSCLC patients (). These results suggested that MCM3AP-AS1 was an oncogenic factor for NSCLC and might have regulatory functions on miR-195-5p and E2F3.
Table 2. Correlation between clinicopathological features and expression of MCM3AP-AS1 in NSCLC
Figure 1. Expression of MCM3AP-AS1, miR-195-5p and E2F3 in NSCLC
3.2. MCM3AP-AS1 repressed miR-195-5p expression in NSCLC cells
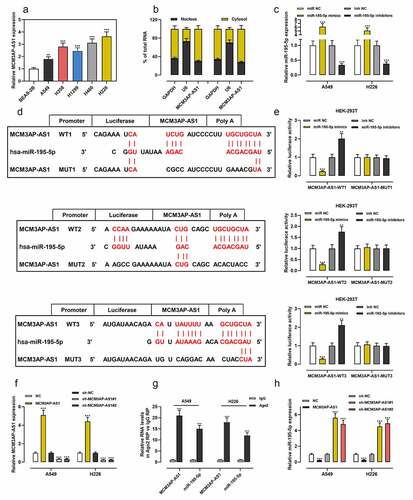
Then, we found that MCM3AP-AS1 was overexpressed in NSCLC cell lines (). Importantly, lncRNAs usually function as competitive endogenous RNA (ceRNA) by binding to miRNAs and can inhibit the expressions of miRNAs. We demonstrated that MCM3AP-AS1 was enriched in the cytoplasm but not in the nucleus of NSCLC cells (). MiR-195-5p expression in cells was much higher after the transfection of miR-195-5p mimics than the transfection with miR-NC; miR-195-5p expression was much lower after the transfection with miR-195-5p inhibitors than the transfection with inhibitors-NC (). Bioinformatics analysis predicted that there were three potential binding sites for miR-195-5p in the sequence of MCM3AP-AS1 (). Dual-luciferase reporter experiment revealed that miR-195-5p mimics weakened the luciferase activity of the MCM3AP-AS1-WT reporter vectors, while miR-195-5p inhibitors increased it, but miR-195-5p mimics or inhibitors had no significant effects on the MCM3AP-AS1-MUT reporter vectors (). Next, MCM3AP-AS1 overexpression plasmid, sh-MCM3AP-AS1#1 and sh-MCM3AP-AS1#2 were transfected into NSCLC cells, and qRT-PCR verified the success of transfection (). The RIP experiment demonstrated that MCM3AP-AS1 and miR-195-5p were remarkably enriched in Ago2-immunoprecipitates compared with the control group (). Moreover, MCM3AP-AS1 overexpression decreased miR-195-5p expression, while MCM3AP-AS1 knockdown worked oppositely in NSCLC cells ().
Figure 2. MCM3AP-AS1 was specifically regulated by miR-195-5p
3.3. The effect of MCM3AP-AS1/miR-195-5p axis on the proliferation of NSCLC cells
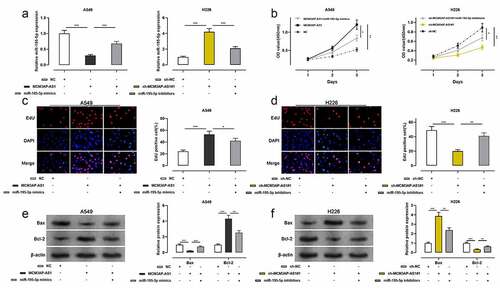
To clarify the function of MCM3AP-AS1 in NSCLC progression, A549 cells were co-transfected with MCM3AP-AS1 overexpression plasmid and miR-195-5p mimics; H226 cells were co-transfected with sh-MCM3AP-AS1#1 and miR-195-5p inhibitors, and qRT-PCR showed that the transfection was successful (). The proliferation of these two cell lines was assessed by CCK-8 and EdU assays, and the data presented that the proliferation of NSCLC cells was markedly increased after the transfection with MCM3AP-AS1 overexpression plasmid, while miR-195-5p mimics abolished this effect; the proliferation of NSCLC cells was remarkably decreased after the transfection with sh-MCM3AP-AS1#1, while co-transfection of miR-195-5p inhibitors reversed this effect (–d). Western blot assay suggested that after MCM3AP-AS1 overexpression, Bcl-2 protein level was increased and BAX protein level was decreased, while miR-195-5p mimics counteracted such effect; after knockdown MCM3AP-AS1, Bcl-2 protein level was declined and Bax protein level was elevated, while miR-195-5p inhibitors reversed this effect (–f). These results suggested that MCM3AP-AS1 promoted the growth and repressed the apoptosis of NSCLC cells, which was mediated by its inhibitory function on miR-195-5p.
Figure 3. Effect of MCM3AP-AS1 and miR-195-5p on A549 and H226 cells proliferation and apoptosis
3.4. The effect of MCM3AP-AS1/miR-195-5p axis on the metastasis of NSCLC cells
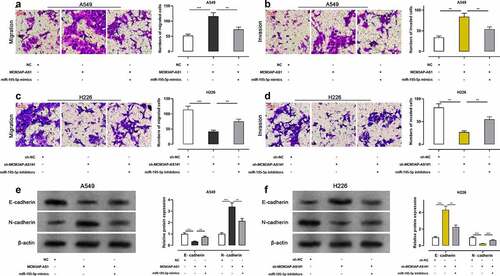
The migration and invasion of these two cell lines were assessed using Transwell assay, and the results showed that the number of migrated and invaded cells was significantly raised after NSCLC cells were transfected with MCM3AP-AS1 overexpression plasmid, while miR-195-5p mimics abrogated such effect (–b); the number of migrated and invaded cells was markedly reduced after the transfection with sh-MCM3AP-AS1#1 while miR-195-5p inhibitors reversed such effect (–d). After MCM3AP-AS1 overexpression, E-cadherin protein level was decreased and N-cadherin protein level was increased, while miR-195-5p mimics reversed this effect; after knockdown MCM3AP-AS1, E-cadherin protein level was elevated and N-cadherin protein level was declined, while miR-195-5p inhibitors reversed such effect (–f). Lung metastasis experiments in nude mice indicated that overexpression of MCM3AP-AS1 promoted lung metastasis in vivo (Supplementary Figure 1d). These results manifested that MCM3AP-AS1 facilitated the metastasis of NSCLC, which was mediated by its inhibitory function on miR-195-5p.
Figure 4. Effect of MCM3AP-AS1 on NSCLC cell migration and invasion
3.5. MCM3AP-AS1 regulated E2F3 expression by decoying miR-195-5p
TargetScan database indicated that miR-195-5p could probably target the 3ʹ-UTR of E2F3 (), and this prediction was validated by dual-luciferase reporter assay. The data illustrated that miR-195-5p mimics reduced the luciferase activity of the E2F3-WT reporter vector, while miR-195-5p inhibitors functioned oppositely; miR-195-5p mimics and inhibitors had no significant effects on E2F3-MUT reporter vector (–c). qRT-PCR and Western blot showed that overexpression of MCM3AP-AS1 promoted E2F3 expression, while miR-195-5p mimics counteracted the function of MCM3AP-AS1 in NSCLC cells; conversely, MCM3AP-AS1 knockdown inhibited E2F3 expression, while miR-195-5p inhibitors reversed such effect (–e). These results suggested that MCM3AP-AS1, as a ceRNA for miR-195-5p, could induce the expression of E2F3 by repressing miR-195-5p expression.
Figure 5. MCM3AP-AS1 up-regulated E2F3 expression by adsorbing miR-195-5p
4. Discussion
LncRNA can regulate the expressions of downstream genes at the different levels, including chromatin modification, transcription or post-transcription, thus participating in various biological processes, including cell proliferation, migration and apoptosis [Citation24,Citation25]. The role of lncRNA in cancer biology is reported in a lot of previous research. For example, in NSCLC, lncRNA KCNQ1OT1 and AWPPH expressions are significantly up-modulated in tumor tissues, and their high expressions were correlated to adverse prognosis; KCNQ1OT1 and AWPPH can enhance the proliferation, migration and invasion of NSCLC cells, respectively [Citation26,Citation27]. Reportedly, MCM3AP, which serves as an essential modulator in DNA replication by acetylating micro-chromosome maintenance protein 3 (MCM3), impedes cell cycle progression and modulates gene expression in tumors [Citation28]. MCM3AP-AS1 is the lncRNA antisense of MCM3AP gene. MCM3AP-AS1 is overexpressed in hepatocellular carcinoma tissues and MCM3AP-AS1 knockdown inhibits the malignant phenotypes of hepatocellular carcinoma cells; mechanistically, MCM3AP-AS1 promotes epidermal growth factor receptor (EGFR) expression by the adsorption of miR-455, which in turn enhances the metastasis of hepatocellular carcinoma [Citation29]. Another study reveals that MCM3AP-AS1 regulates the progression of hepatocellular carcinoma via miR-194-5p/FOXA1 axis [Citation30]. In NSCLC, MCM3AP-AS1 knockdown impedes cell proliferation and migration, and YY1 transcription factor (YY1) mediates the transcription of MCM3AP-AS1 in NSCLC [Citation14]. In this study, it was verified that MCM3AP-AS1 expression was up-modulated in NSCLC, and its overexpression was associated with unfavorable prognosis of patients and MCM3AP-AS1 facilitated the proliferation and metastasis of NSCLC cells. Our data prove that MCM3AP-AS1 is a promising biomarker and therapy target for NSCLC, which is consistent with the previous reports [Citation14].
MiRNAs also participate in regulating diverse biological process. Accumulating studies elucidate that multiple miRNAs are aberrantly expressed in diverse tumors, including NSCLC [Citation31–35]. Reportedly, miR-195-5p expression is down-modulated in esophageal carcinoma and miR-195-5p represses the proliferation and metastasis of cancer cells via targeting Fos-related antigen 1 (FOSL1) [Citation15]. In NSCLC, miR-195-5p expression is also down-regulated, and in vitro experiments unmask that miR-195-5p impedes the proliferation of NSCLC cells [Citation16]. Additionally, down-regulation of miR-195-5p expression in NSCLC is closely associated with increased TNM staging, increased tumor size and lymph node metastasis; functionally, miR-195-5p suppresses cell proliferation and induces cell cycle arrest and apoptosis [Citation17]. LncRNA can adsorb miRNA as ceRNA and thus participate in tumorigenesis and development [Citation36,Citation37]. For instance, in glioma and HCC, miR-195-5p is sponged by LINC00473 and lncRNA SNHG1, respectively [Citation38,Citation39]. The current study confirmed that miR-195-5p was a downstream target of MCM3AP-AS1 and MCM3AP-AS1 negatively regulated its expression; functional experiments suggested that MCM3AP-AS1/miR-195-5p axis regulated the proliferation and metastasis of NSCLC cells. To the best of our knowledge, our study is the first to validate the interaction between MCM3AP-AS1 and miR-195-5p in cancer biology.
E2F3, which is an important cell cycle regulator and figures prominently in regulating cell proliferation, apoptosis and differentiation, belongs to the E2F transcriptional regulatory family and is closely related to tumorigenesis [Citation40–42]. Reportedly, E2F3 expression is up-regulated in tumors, such as osteosarcoma and breast cancer, and enhances the proliferation and colony formation of tumor cells [Citation42,Citation43]. In NSCLC, E2F3 improves the malignancy of cancer cells by increasing the expressions of cyclinD1, cyclinD2, and CDK4 while inhibiting p21 and p57 expressions [Citation44,Citation45]. The present work verified the binding sites between miR-195-5p and E2F3 3ʹUTR and proved that MCM3AP-AS1 could up-regulate E2F3 expression by adsorbing miR-195-5p, which explained the mechanism of E2F3 dysfunction in NSCLC. Importantly, our data also suggest that MCM3AP-AS1, miR-195-5p and E2F3 form a novel ceRNA network to take part in regulating NSCLC progression.
5. Conclusion
MCM3AP-AS1 plays a promotive role in NSCLC progression via modulating the miR-195-5p/E2F3 axis, indicating that MCM3AP-AS1 is a new target for gene therapy in NSCLC. However, this work has several limitations. First, the value of MCM3AP-AS1 as a biomarker to predict the prognosis of NSCLC patients’ needs to be further verified by a larger cohort of patients from different medical centers. Second, whether MCM3AP-AS1 can promote other malignant phenotypes of NSCLC cells, such as drug resistance and radioresistance, needs further investigation. Furthermore, there are other potential downstream miRNAs of MCM3AP-AS1 remained to be screened and validated in the future.
Research highlights
MCM3AP-AS1 is up-regulated in NSCLC tissues;
MCM3AP-AS1 promotes the malignant biological behaviors of NSCLC cells such as proliferation, migration and invasion.
The functions of MCM3AP-AS1 are partly dependent on miR-195-5p/E2F3 axis in NSCLC.
Acknowlegement
We thank Beijing TP Med Science & Technology Co., Ltd. for its linguistic assistance during the preparation of this manuscript.
Authors contribution
Conceived and designed the experiments: Kaiyi Tao, Youhua Jiang;
Performed the experiments: Dijian Shen, Jianqiang Li;
Statistical analysis: Dijian Shen, Jianqiang Li and Kaiyi Tao;
Wrote the paper: Dijian Shen, Jianqiang Li.
All authors read and approved the final manuscript.
Ethics statement
Our study was approved by the Ethics Review Board of Cancer Hospital of the University of Chinese Academy of Sciences.
Supplemental Material
Download ()Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Shen DJ, Jiang YH, Li JQ, et al. A-binding protein RBM47 inhibits non-small cell lung carcinoma metastasis through modulation of AXIN1 mRNA stability and Wnt/β-catentin signaling. Surg Oncol. 2020 Sep;34:31–39.
- Gelatti ACZ, Drilon A, Santini FC. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer. 2019;137:113–122.
- Wang H, Yu X, Fan Y, et al. Multiple treatment modalities for brain metastasis in patients with EGFR-mutant non-small-cell lung cancer. Oncol Targets Ther. 2018;11:2149–2155.
- Jiang YH, Xu XL, Ruan HH, et al. The impact of functional LIG4 polymorphism on platinum-based chemotherapy response and survival in non-small cell lung cancer. Med Oncol. 2014;31(5):959.
- Kanda S, Ohe Y, Goto Y, et al. Five-year safety and efficacy data from a phase Ib study of nivolumab and chemotherapy in advanced non-small-cell lung cancer. Cancer Sci. 2020;111(6):1933–1942.
- Dong Y, Wan G, Yan P, et al. Long noncoding RNA LINC00324 promotes retinoblastoma progression by acting as a competing endogenous RNA for microRNA-769-5p, thereby increasing STAT3 expression. Aging (Albany NY). 2020;12(9):7729–7746.
- Sun CB, Wang HY, Han XQ, et al. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J Gastrointest Oncol. 2020;12(4):394–404.
- Cao F, Wang Z, Feng Y, et al. lncRNA TPTEP1 competitively sponges miR‑328‑5p to inhibit the proliferation of non‑small cell lung cancer cells. Oncol Rep. 2020;43(5):1606–1618.
- Ye Y, Gu J, Liu P, et al. Long non-coding RNA SPRY4-IT1 reverses cisplatin resistance by downregulating MPZL-1 via suppressing EMT in NSCLC. Oncol Targets Ther. 2020;13:2783–2793.
- Chen T, Li J, Zhou MH, et al. 6 stimulates lncRNA ZEB2-AS1 to aggravate the progression of non-small cell lung cancer through activating STAT1. Eur Rev Med Pharmacol Sci. 2020;24(7):3734–3740.
- Li X, Lv J, Liu S. MCM3AP-AS1 KD inhibits proliferation, invasion, and migration of PCa cells via DNMT1/DNMT3 (A/B) methylation-mediated upregulation of NPY1R. Mol Ther Nucleic Acids. 2020;20:265–278.
- Wang H, Xu T, Wu L, et al. Molecular mechanisms of MCM3AP-AS1 targeted the regulation of miR-708-5p on cell proliferation and apoptosis in gastric cancer cells. Eur Rev Med Pharmacol Sci. 2020;24(5):2452–2461.
- Lan L, Liang Z, Zhao Y, et al. LncRNA MCM3AP-AS1 inhibits cell proliferation in cervical squamous cell carcinoma by down-regulating miRNA-93. Biosci Rep. 2020;40:2.
- Li X, Yu M, Yang C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J Cell Biochem. 2020;121(3):2258–2267.
- Shen S, Li K, Liu Y, et al. Silencing lncRNA AGAP2-AS1 upregulates miR-195-5p to repress migration and invasion of EC cells via the decrease of FOSL1 expression. Mol Ther Nucleic Acids. 2020;20:331–344.
- Luo J, Pan J, Jin Y, et al. MiR-195-5p inhibits proliferation and induces apoptosis of non-small cell lung cancer cells by targeting CEP55. Oncol Targets Ther. 2019;12:11465–11474.
- Zheng J, Xu T, Chen F, et al. MiRNA-195-5p functions as a tumor suppressor and a predictive of poor prognosis in non-small cell lung cancer by directly targeting CIAPIN1. Pathol Oncol Res. 2019;25(3):1181–1190.
- Szász AM, Lánczky A, Á N, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017 Jul 3;45(W1):W98–W102. Oncotarget. 2016 Aug 2;7(31):49322–49333.
- Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016 Aug 2;7(31):49322–49333.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014 Jan;42( Database issue):D92–7.
- Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016 Jan 4;44(D1):D231–8.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 Aug;12(4):e05005.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001 Dec;25(4):402–408.
- Shang A, Wang W, Gu C, et al. Long non-coding RNA CCAT1 promotes colorectal cancer progression by regulating miR-181a-5p expression. Aging (Albany NY). 2020;12(9):8301–8320.
- Guo J, Ding Y, Yang H, et al. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp Mol Pathol. 2020;104448. DOI:10.1016/j.yexmp.2020.104448
- Wang Y, Zhang L, Yang J, et al. LncRNA KCNQ1OT1 promotes cell proliferation, migration and invasion via regulating miR-129-5p/JAG1 axis in non-small cell lung cancer. Cancer Cell Int. 2020;20:144.
- Wu D, Qin BY, Qi XG, et al. LncRNA AWPPH accelerates the progression of non-small cell lung cancer by sponging miRNA-204 to upregulate CDK6. Eur Rev Med Pharmacol Sci. 2020;24(8):4281–4287.
- Yang M, Sun S, Guo Y, et al. Long non-coding RNA MCM3AP-AS1 promotes growth and migration through modulating FOXK1 by sponging miR-138-5p in pancreatic cancer. Mol Med. 2019;25(1):55.
- Zhang H, Luo C, Zhang G. LncRNA MCM3AP-AS1 regulates epidermal growth factor receptor and autophagy to promote hepatocellular carcinoma metastasis by interacting with miR-455. DNA Cell Biol. 2019;38(8):857–864.
- Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18(1):28.
- Wei L, Jiang J. Targeting the miR-6734-3p/ZEB2 axis hampers development of non-small cell lung cancer (NSCLC) and increases susceptibility of cancer cells to cisplatin treatment. Bioengineered. 2021 Dec;12(1):2499–2510.
- Liang Z, Xu J, Ma Z, et al. MiR-187 suppresses non-small-cell lung cancer cell proliferation by targeting FGF9. Bioengineered. 2020 Dec;11(1):70–80.
- Liu B, Sun X. miR-25 promotes invasion of human non-small cell lung cancer via CDH1. Bioengineered. 2019 Dec;10(1):271–281.
- Huang HQ, Chen G, Xiong DD, et al. Down-regulation of microRNA-125b-2-3p is a risk factor for a poor prognosis in hepatocellular carcinoma. Bioengineered. 2021 Dec;12(1):1627–1641.
- Zhao G, Zhang Y, Zhao Z, et al. MiR-153 reduces stem cell-like phenotype and tumor growth of lung adenocarcinoma by targeting Jagged1. Stem Cell Res Ther. 2020;11(1):170.
- Wu Y, Bi QJ, Han R, et al. Long noncoding RNA KCNQ1OT1 is correlated with human breast cancer cell development through inverse regulation of hsa-miR-107. Biochem Cell Biol. 2020;98(3):338–344.
- Zhang S, Wang B, Xiao H, et al. LncRNA HOTAIR enhances breast cancer radioresistance through facilitating HSPA1A expression via sequestering miR-449b-5p. Thorac Cancer. 2020;11(7):1801–1816.
- Wang X, Li XD, Fu Z, et al. Long non‑coding RNA LINC00473/miR‑195‑5p promotes glioma progression via YAP1‑TEAD1‑Hippo signaling. Int J Oncol. 2020;56(2):508–521.
- Huang D, Wei Y, Zhu J, et al. Long non-coding RNA SNHG1 functions as a competitive endogenous RNA to regulate PDCD4 expression by sponging miR-195-5p in hepatocellular carcinoma. Gene 2019;714:143994.
- Gao W, Zhou X, Lin R. miR-378a-5p and miR-630 induce lens epithelial cell apoptosis in cataract via suppression of E2F3. Braz J Med Biol Res. 2020;53(5):e9608.
- Li H, Pan R, Lu Q, et al. MicroRNA‑145‑5p inhibits osteosarcoma cell proliferation by targeting E2F transcription factor 3. Int J Mol Med. 2020;45(5):1317–1326.
- Deng YJ, Ren EH, Yuan WH, et al. GRB10 and E2F3 as diagnostic markers of osteoarthritis and their correlation with immune infiltration. Diagnostics (Basel). 2020;10(3):171.
- Han R, Zhao J, Lu L. MicroRNA‑34a expression affects breast cancer invasion in vitro and patient survival via downregulation of E2F1 and E2F3 expression. Oncol Rep. 2020;43(6):2062–2072.
- Zhang J, Li Y, Dong M, et al. Long non-coding RNA NEAT1 regulates E2F3 expression by competitively binding to miR-377 in non-small cell lung cancer. Oncol Lett. 2017;14(4):4983–4988.
- Liu N, Liu Z, Zhang W, et al. MicroRNA‑433 reduces cell proliferation and invasion in non‑small cell lung cancer via directly targeting E2F transcription factor 3. Mol Med Rep. 2018;18(1):1155–1164.