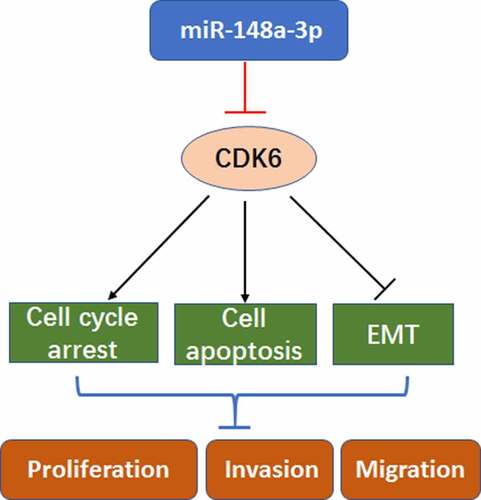
ABSTRACT
To study the regulation of miR-148a-3p on CDK6 and its mechanism in the progress of acute myeloid leukemia (AML), differential miRNAs were analyzed by bioinformatics, and the miR-148a-3p levels in AML cell lines were detected. Results showed that miR-148a-3p played a crucial role in AML, and the level was lower in AML cells, especially in J111 and KG-1a cells. In J111 and KG-1a cells, the up-regulation of miR-148a-3p mimics blocked the cell growth by arresting cell cycle at G2/M and enhancing cell apoptosis. Transwell and EMT markers detection indicated that miR-148a-3p reduced the cell migration and invasion. Afterward, through bioinformatics analysis, it showed that the CDK6 is one of the direct target genes of miR-148a-3p. DLR assay confirmed the target regulation. CDK6 overexpression reversed the effects of miR-148a-3p on AML cells. Collectively, miR-148a-3p inhibited the process of AML cells through disturbing the CDK-6 expression, implying that the trageting miR-148a-3p might be regarded as effective therapy of AML.
Introduction
Acute myeloid leukemia (AML) is an acute leukemia with complex etiology and high heterogeneity, accounting for approximately 34% of all leukemia cases [Citation1]. It is a clonal expansion of immature myeloid stem cells with abnormal proliferation and differentiation [Citation2]. Current treatments for AML include cytarabine and anthracycline combined with hematopoietic stem cell transplantation technology [Citation3], as well as targeted drug therapies such as FLT3-ITD inhibitors [Citation4]. Although some subtypes of AML have been improved, the prognosis is still poor for most patients, with a five-year survival rate lower than 50%[Citation5]. As the molecular biological characteristics are significant prognostic markers of AML survival, it is important to explore new regulatory mechanisms for AML development.
Till now, it has been reported by many researches that miRNA is related to the development of various tumors [Citation6]. MiRNA is an 18–25nt regulatory molecule, and it always complementarily binds to the 3’UTR of the target gene mRNA, thus negatively regulates gene expression[Citation7]. MiR-148a has been reported playing a key role in cancer cells [Citation8]. In osteosarcoma, miR-148a indicated a poor prognosis for osteosarcoma patients [Citation9]. miR-148a can inhibit colitis and colitis-associated tumor [Citation10]. Furthermore, miR-148a-3p could inhibit bovine myoblast cell proliferation and induce cell apoptosis via KLF6 [Citation11]. Nevertheless, miR-148a-3p regulatory mechanism in AML has rarely been studied.
Cyclin-dependent kinase 6 (CDK6) is a member of the cyclin-dependent kinases (CDKs) family. During the G1 phase, CDK6 binds Cyclin D to initiate the cell cycle progression. At the same time, the complex involves in the transcription of genes for cell proliferation [Citation12]. Studies have shown that ZFP36L1-CDK6 mediates the monocyte/macrophage differentiation regulatory pathway in AML patients [Citation13]. Prostratin induces cell proliferation inhibition, cell cycle arrest and differentiation by down-regulating CDK6 [Citation14], and the knockdown of CDK6 significantly inhibits the occurrences of NUP98 fusion protein-driven AML [Citation15]. The above studies have shown that CDK6 is quite crucial for the occurrence and development of AML, but miR-148a-3p's regulation on CDK6 needs to be further clarified.
Here, our study showed that the up-regulation of miR-148a-3p inhibits cell proliferation, invasion and migration in AML cells. Bioinformatics prediction combined with dual-luciferase reporter (DLR) assay verified the relation of CDK6 and miR-148a-3p, and up-regulation of CDK6 could reverse the effect. Our study provided new insights into the mechanisms in AML and upregulating miR-148a-3p has potential effect in AML treatment.
Methods
Bioinformatics analysis
The miRNA expression matrix and mRNA expression matrix related to AML were downloaded through the GEO database. The data set was named GSE142700, and the mRNA platform was GPL19956, including a total of 48 cases – 24 control cases (GSM4238029-GSM4238052) and 24 AML patients (GSM4237957-GSM4237980). Besides, the miRNA chip platform was GPL26945, also including a total of 48 cases – 24 control cases (GSM712488-GSM712507) and 24 AML patients (GSM4238005-GSM4238028). Limma package was used to analyze the difference between miRNA data and mRNA data in GSE142700 (|log2FC|>2, adj.P.Val<0.05) to obtain differential miRNAs and differentially expressed genes (DEGs). Then, FunRich (version 3.13) was applied to predict the differential miRNA target genes, and the target genes were intersected with DEGs. According to the negative regulation relationship between miRNA and the target gene, the eligible relationship pairs were screened, and Cytoscape 3.8.1 was used to display the regulatory network.
Cell culture
AML cell lines THP-1, NB-4, HL-60, KG-1a and mesenchymal stem cell HS5 were obtained from ATCC (USA), and J111 cell was obtained from the National Collection of Authenticated Cell Culture (China). NB-4, KG-1a, and HL-60 cells were inoculated into the DMEM medium (Gibco, USA) containing 20% FBS, 1% penicillin and streptomycin. Besides, THP-1, J111 and HS5 cells were inoculated into the DMEM medium containing 10% FBS, and then cultured in the 5% CO2, 37°C incubator (ThermoFisher, USA).
qRT-PCR
Cells were mixed with 1 mL Trizol (Invitrogen, USA), 200 μL chloroform and 200 μL isopropanol successively. Then, the precipitate was dissolved by 50 μL DEPC water. Mir-X miRNA First-Strand Synthesis Kit (Takara, USA) and Reverse transcription kit (Beyotime, China) were used to synthesize cDNA for miRNA and total RNA. Then SYBR Green Master Mix Kit (Thermo Fisher, USA) was applied for qRT-PCR analysis. GAPDH and U6 were regarded as endogenous control genes for CDK6 and miRNA respectively. mRNA expression levels were analyzed by 2−∆∆Ct method. The primer sequence was listed as follows:
Cell transfection
To manipulate the miR-148a-3p levels in J111 and KG-1a cells, miR-148a-3p mimics (overexpression of miR-148a-3p), miR-148a-3p inhibitor (knockdown of miR-148a-3p), and negative control (miR-NC and inhibitor NC) were constructed (Sangon Biotech, Shanghai, China). To overexpress CDK6, the pcDNA-CDK6 and the control vector (Sangon Biotech) were applied. Cell transfection was executed with Highgene (Wuhan Aibotec Biotechnology Co., Ltd., China). The process was strictly following the instruction. After the transfection, the cells were continully cultured for 48 h.
CCK-8 assay
Referring to the protocol of CCK-8 Kit (Dojindo, Japan), 1 × 104/ml transfected AML cells were added in a 96-well plate. Then, 10 μL CCK-8 solution and 100 μL DMEM medium (with 10% or 20% FBS) were added. The absorbance at 450 nm was detected by the microplate reader.
Cell clone formation experiment
Colony formation experiment was assayed in methylcellulose medium (StemCell Technologies, USA). Cells wereincubated for 10 days after blending with methycellulose, and colonies were counted with a microscope.
Apoptosis assay
1 × 106/ml transfected J111 and KG-1a cells were incubated for 24 h at 37°C. AML cells were centrifuged at 1000 rpm for 2 min and then the supernatant was discarded. Next, 500 μL binding buffer, 5 μL PI solution and 5 μL annexin V-FITC solution (Liankebio, China) were added in turn, and they were all mixed up gently. The plate was placed in a dark environment for another 30 min, and the apoptosis was measured by flow cytometry.
Cell cycle assay
Cells (5 × 106) were collected for each sample and washed with PBS twice. After fixed with ethanol, the cells were cultured with 10 μg/ml RNase for 1 hour. Then, propidium iodide (PI, 100 μg/ml) was used for staining the cells at 25°C for 30 min in the dark environment. Flow cytometry (BD Bioscience, San Jose, CA) was used to analyze the cell cycle distribution.
Western blot assays
Total protein was extracted and quantified by the BCA Kit (Beyotime, China). After SDS-PAGE electrophoresis, the target proteins were transferred to PVDF membrane for 2 h. After blocking, the first antibody was added and incubated on shaker for 3 h, and the second antibody was added for 1.5 h at 37°C. The membrane was washed with TBST solution for three times. Using GAPDH as an internal reference, the expressions of apoptosis-related proteins and migration-related proteins were calculated.
Cell invasion and migration
Cell invasion and migration were studied by using Transwell assay. For Migration assay, 1 × 104 cells were added to the upper chamber in medium supplemented with 1% FBS. In the lower chamber, medium supplemented with 10% FBS was added. For the invasion assay, the lower surface was covered with matrigel. After 48 h, the cells in the upper or the lower chamber were collected and the cell number were counted under the microscope.
DLR assay
Both wild type (wt) CDK6 or mutation (mt) CDK6 reporter plasmid and miRNA-NC or miR-148a-3p mimics were co-transfected into J111 and KG-1a for 48 h. The luciferase activity of firefly and marine kidney were detected according to the instruction (Yeasen, China).
Statistical analysis
The data were shown as mean ± standard deviation, and analyzed with GraphPad Prism 8. T-test was applied to analyze the differences between the two groups. The difference was statistically significant with P < 0.05.
Results
miR-148a-3p level was decreased in AML cells
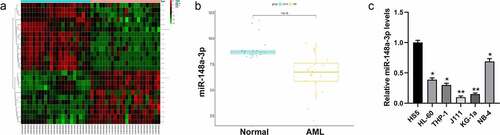
The Limma package was used to analyze the differences between miRNA sample data in the GSE142700 data set to obtain differential miRNAs (). Further analysis by FunRich (version 3.13) revealed that miR-148a-3p was essential to the progress of AML ( B). Studies indicated that miR-148a-3p decreased in bladder cancer, gastric cancer, as well as laryngeal cancer [Citation16–18], and it can be regarded as tumor suppressor. We detected the expression of miR-148a-3p in AML cells (HL-60, THP-1, J111, KG-1a, and NB-4) and bone marrow mesenchymal stem cells (HS5) by qRT-PCR and it revealed that miR-148a-3p expression in AML cells was remarkably lower than in the HS5 cells (). The cell lines J111 and KG-1a with relatively lower expressions of miR-148a-3p were used for subsequent experiments.
Figure 1. Screening the differential expression of miRNAs in AML. A: Differential analysis of miRNAs with red for up-regulated miRNAs and green for down-regulated miRNAs; B: Analysis of miR-148a-3p level in the GSE142700 data set; C: qRT-PCR was used to measure miR-148a-3p levels in human leukemia cell lines and bone marrow mesenchymal stem cells, and HS5 cells were considered as control. *P < 0.05, **P < 0.01
Up-regulation of miR-148a-3p supressed the growth of AML cells
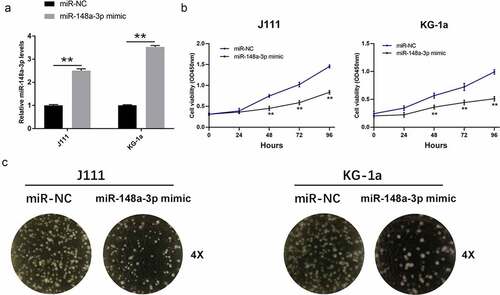
To study the effects of miR-148a-3p on the proliferation of AML cells, we transfected miR-148a-3p mimics into J111 and KG-1a cells. It turned out that mimics up-regulated the level of miR-148a-3p in AML cells (). miR-148a-3p mimics up-regulation inhibited the growth of J111 and KG-1a cells obviously (). To study the effects of miR-148a-3p in AML progress, the cell clone formation experiment was performed. The results showed that miR-148a-3p up-regulation attenuated the cell clonal ability ().
Figure 2. Effect of miR-148a-3p on the proliferation of AML cells. A: J111 and KG-1a cells were transfected with miR-NC or miR-148a-3p mimics for 48 h, and miR-148a-3p level was measured by qRT-PCR; B: CCK-8 assay detected cell proliferation; C: Cell clone formation experiment was conducted to measure cell proliferation; Compared with miR-NC, *P < 0.05, **P < 0.01
miR-148a-3p induced cell apoptosis and inhibited the cell cycle progression
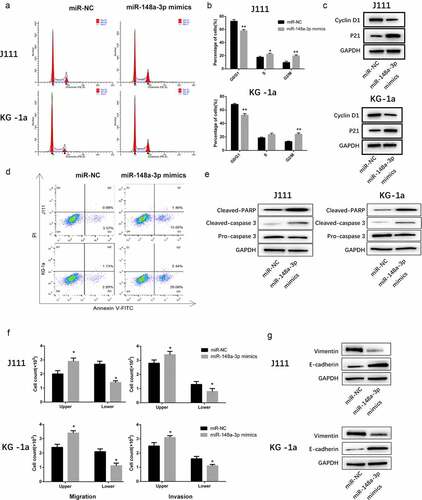
As shown in , miR-148a-3p arrested the cell cycle at G2/M phase. Meanwhile, the expression of Cyclin D1 decreased and the upregulation of p21 also validated the results. (). Furthermore, upregulating miR-148a-3p significantly enhanced the cell apoptosis (). Meanwhile, the expression of Cleaved-PARP and Cleaved-caspase 3 increased significantly ().
Figure 3. The effect of miR-148a-3p on the cell cycle progression, apoptosis, invasion and migration of AML cells. A: J111 and KG-1a cells were transfected with miR-NC or miR-148a-3p mimics for 48 h, flow cytometry was used to analyze the cell cycle. B: The percentage of each cell cycle phase were statistic with chart. C: Cyclin D1, p21 and GAPDH were detected with Western blot. D: Flow cytometry were used to analyze the cell apoptosis. E: Cleaved PARP, Cleaved caspase-3, pre-caspase-3 and GAPDH were detected with Western blot. F: Transwell was applied to detect the cell invasion and migration. G: Vimentin, E-cadherin and GAPDH were measured with Western blot. Compared with miR-NC, *P < 0.05, **P < 0.01
Up-regulation of miR-148a-3p inhibited the invasion and migration of AML cells
Transwell assay showed that the upregulated miR-148a-3p remarkably inhibited cell invasion and migration (). Western Blot results indicated that Vimentin expressions decreased while E-cadherin expressions increased (). All of those indicated that the up-regulation of miR-148a-3p weakened cell invasion and migration.
CDK6 is the direct target gene of miR-148a-3p
In order to predict the target gene of miR-148a-3p, the Limma package was used to analyze the mRNA sample data set (|log2FC|>2, adj.P.Val<0.05) to obtain 23 differential mRNAs, among which 12 were up-regulated and 11 were down-regulated (). Then, we used FunRich (version 3.13) to predict differential target genes of miRNA, and we took the intersection of the DEGs and predicted target genes (). According to the negative regulation between miRNA and the target gene, the eligible relationship pairs were screened, and the regulatory network was displayed by Cytoscape 3.8.1 (). DLR experiment demonstrated that miR-148a-3p mimics markedly decreased wt CDK6 luciferase activity, while mt CDK6 luciferase activity remained unchanged (). After miR-148a-3p mimics were transfected into J111 and KG-1a, both CDK6 mRNA and protein expressions were reduced (); On contrast, the expressions increased in AML cells when treated with miR-148a-3p inhibitor (). Those demonstrated that miR-148a-3p regulated CDK6.
Figure 4. Analysis of the direct target gene of miR-148a-3p. A: Differential analysis of mRNAs with red for up-regulated mRNAs and green for down-regulated mRNAs; B: Analyzed miR-148a-3p potential target genes; C: Screened the target genes that negatively regulated by miR-148a-3p. Red for up-regulation, green for down-regulation, triangle for miRNA, and circle for mRNA; D: wt CDK6 or mt CDK6 reporter plasmid and miR-NC or miR-148a-3p mimics were co-transfected with J111 and KG-1a cells for 48 h, and DLR experiment was conducted to measure the luciferase activity; E: miR-148a-3p mimics or miR-NC was co-transfected with J111 and KG-1a cells for 48 h, qRT-PCR and Western blot measured CDK6 mRNA and protein expressions; F: miR-148a-3p inhibitor or inhibitor-NC was transfected into J111 and KG-1a cells for 48 h, and CDK6 mRNA and protein expressions were detected by qRT-PCR and Western blot. Compared with miR-NC, **P < 0.01
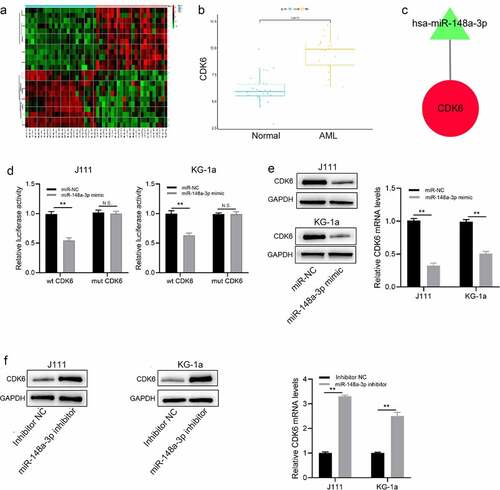
CDK6 overexpression reversed the effect of miR-148a-3p in AML cells
We successfully constructed CDK6 overexpression plasmid to study the regulation of miR-148a-3p on CDK6 for the process of AML. The CDK6 overexpression plasmid was transfected into J111 and KG-1a cells, causing up-regulation of CDK6 mRNA and protein expression (). The results of CCK8 assay demonstrated that as the viability of J111 and KG-1a cells increased, the overexpression of CDK6 reversed the effect of miR-148a-3p upregulation in AML cell (). The results of cell clone formation experiment indicated that the up-regulation of CDK6 increased the number of cell clones when compared with overexpression miR-148a-3p alone (). Flow cytometry and Western blot were applied to measure the change of cell cycles and the cell apoptosis after the CDK6 overexpression in J111 and KG-1a cells, and it showed that CDK6 overexpression neutralized the effect of miR-148a-3p mimics (, e).
Figure 5. Effect of CDK6 overexpression on the proliferation of J111 and KG-1a cells which transfected with miR-148a-3p mimics. A: The CDK6 overexpression plasmid or control plasmid was co-transfected with miR-148a-3p mimics or miR-NC in AML cells for 48 hours, then CDK6 mRNA and protein expressions were conducted by qRT-PCR and Western blot; B: CCK-8 assay was used to detect the cell viability. C: cell clone formation experiment of J111 and KG-1a cells; D: Cell cycle arrest assay and related protein expression in J111 and KG-1a cells. E: cell apoptosis ratio and related protein expression in J111 and KG-1a cells. Compared with miR-NC, *P < 0.05, **P < 0.01
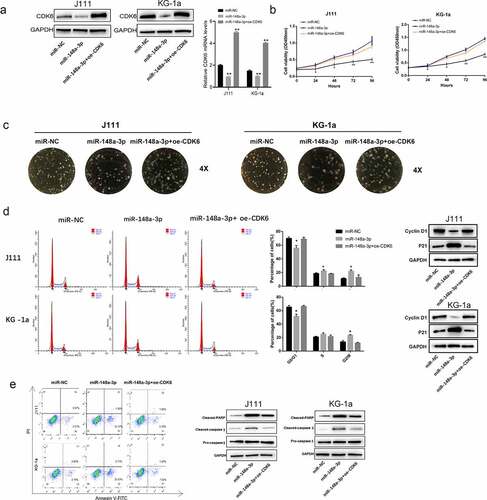
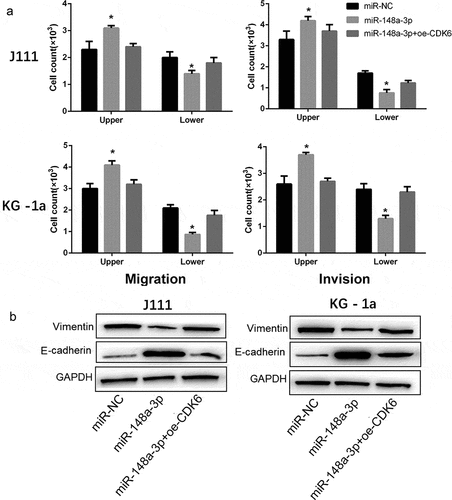
CDK6 overexpression reversed the effect of miR-148a-3p overexpression on the invasion and migration of AML cells
As shown in , CDK6 overexpression reversed the inhibitory effect of miR-148a-3p on the invasion and migration of AML cells (). And the up-regulation of CDK6 neutralized the restrain of miR-148a-3p on Vimentin, and increased the E-cadherin expression (). All results demonstrated that CDK6 overexpression could neutralize the effect of miR-148a-3p on the invasion and migration of AML cells.
Figure 6. Effect of CDK6 overexpression on the invasion and migration in AML cells which transfected with miR-148a-3p mimics. A: CDK6 overexpression plasmid or control plasmid was co-transfected with miR-148a-3p mimics or miR-NC for 48 h, and Transwell was used to measure the cell invasion and cell migration; B: Invasion and migration related protein expression was measured by Western blot. Compared with miR-NC, *P < 0.05
Discussion
As a heterogeneous tumor, gene mutations and abnormalities often caused the etiology of AML. However, the pathogenesis of AML is still unclear. In contrast to the high survival rate (more than 68%) of acute lymphoblastic leukemia (ALL) [Citation19], the diagnosis and treatment of AML has progressed slowly and relies heavily on the massive use of cytotoxic chemotherapy drugs. In recent years, targeted drugs such as FLT3-ITD inhibitors, STAT inhibitors, and IDH1/IDH2 small molecule inhibitors, have also been used effectively in the treatment of AML [Citation20]. Therefore, the search for new AML regulatory factors and target genes will not only provide new targets for the treatment of AML, but also set a new perspective for exploring the pathological mechanisms of AML.
Here, differential miRNA between AML and control cases were analyzed by bioinformatics, and miR-148a-3p was chosen to study in AML. MiR-148a-3p has anti-cancer and anti-metastasis functions, and its expression is inhibited in many diseases. For example, miR-148a-3p decreased in NSCLC patients, thus it can be implied as a diagnosis biomarker for NSCLC [Citation21]. MiR-148a-3p interacts with DNMT1 to hypermethylated the promoter region, forming a regulatory axis to regulate RUNX3 protein expression, thus inhibiting the proliferation of laryngeal squamous cell carcinoma [Citation22]. MiR-148a-3p promotes cell G1 arrest in bladder cancer, thereby suppressing cell invasion and migration [Citation16]. These results were also confirmed in this study. For example, compared with normal bone marrow mesenchymal stem cells, the levels of miR-148a-3p in AML cells (THP-1, HL-60, J111, NB-4 and KG-1a) were significantly lower, indicating that miR-148a-3p may have anti-cancer effect for AML. As shown in the CCK-8 and cell clone formation experiment, miR-148a-3p up-regulation inhibited the proliferative capacity of J111 and KG-1a cells. And miR-148a-3p overexpression promoted apoptosis and caused the cell cycle arrested at G2/M phage. Transwell assay confirmed that miR-148a-3p could also inhibit the invasion and migration of J111 and KG-1a cells.
In order to study the mechanism in which miR-148a-3p inhibits AML progression, the potential target genes were predicted by FunRich (version 3.13), and it was confirmed that miR-148a-3p targets CDK6 directly through the dual-luciferase reporter assay. CDK6, as one of the classical cell cycle kinases, is located on human chromosome 7. It was reported that CDK6 plays an important role in hematopoiesis [Citation23]. In myeloid leukemia, the abnormally high expression of CDK6 can not only prevent the myeloid differentiation of hematopoietic stem cells [Citation24], regulate the expression of FLT3, AKT, and VEGFA [Citation25,Citation26] but also cause the cycle disorders of cells and the proliferation of leukemia cells. It was found in the previous studies that the overexpression of miR-148a-3p inhibited the proliferation of AML cells. Based on this, we transfected CDK6 overexpression plasmid or control plasmid into J111 and KG-1a cells to determine whether miR-148a-3p could regulate AML development by downregulating CDK6 expression. The results indicated that CDK6 neutralized the inhibition of miR-148a-3p on AML cells.
Conclusion
In conclusion, our findings indicated that miR-148a-3p inhibited the progression of AML cells via down-regulating the CDK-6 expression. This mechanism provided a new therapy for AML treatment and hereby the overall survival rate of AML patients could also be improved as well.
Highlights
Up-regulation of miR-148a-3p inhibits the progression of AML cells.
CDK6 was the direct target gene of miR-148a-3p.
miR-148a-3p could be served as a specific biomarker for AML.
Abbreviations
Acute myeloid leukemia (AML); cyclin-dependent kinase 6 (CDK6); cyclin-dependent kinases (CDKs); dual-luciferase reporter (DLR); acute lymphoblastic leukemia (ALL); differentially expressed genes (DEGs); non-small cell lung cancer (NSCLC).
Acknowledgements
Not applicable.
Availability of data and material
The datasets used and/or; analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
The author(s) declare there are no competing interests.
Additional information
Funding
References
- Rogers HJ, Vardiman JW, Anastasi J, et al. Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv(3)(q21q26.2)/t(3;3)(q21;q26.2): a Bone Marrow Pathology Group study. Haematologica. 2014;99(5):821–829.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Estey EH. Acute myeloid leukemia: 2014 update on risk-stratification and management. Am J Hematol. 2014;89(11):1063–1081.
- Antar A, Kharfan-Dabaja MA, Mahfouz R, et al. Sorafenib maintenance appears safe and improves clinical outcomes in FLT3-ITD acute myeloid leukemia after allogeneic hematopoietic cell transplantation. Clin Lymphoma Myeloma Leuk. 2015;15(5):298–302.
- Riva L, Luzi L, Pelicci PG. Genomics of acute myeloid leukemia: the next generation. Front Oncol. 2012;2:40.
- Lee S, Jung JW, Park SB, et al. Histone deacetylase regulates high mobility group A2-targeting microRNAs in human cord blood-derived multipotent stem cell aging. Cell Mol Life Sci. 2011;68(2):325–336.
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060.
- Li Y, Deng X, Zeng X, et al. The role of mir-148a in cancer. J Cancer. 2016;7(10):1233–1241.
- Ma W, Zhang X, Chai J, et al. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014;35(12):12467–12472.
- Zhu Y, Gu L, Li Y, et al. miR-148a inhibits colitis and colitis-associated tumorigenesis in mice. Cell Death Differ. 2017;24(12):2199–2209.
- Song C, Yang J, Jiang R, et al. miR-148a-3p regulates proliferation and apoptosis of bovine muscle cells by targeting KLF6. J Cell Physiol. 2019;234(9):15742–15750.
- Meng LH, Zhang H, Hayward L, et al. Tetrandrine induces early G1 arrest in human colon carcinoma cells by down-regulating the activity and inducing the degradation of G1-S–specific cyclin-dependent kinases and by inducing p53 and p21 Cip1. Cancer Res. 2004;64(24):9086–9092.
- Chen MT, Dong L, Zhang XH, et al. ZFP36L1 promotes monocyte/macrophage differentiation by repressing CDK6. Sci Rep. 2015;5(1):16229.
- Shen X, Xiong GL, Jing Y, et al. The protein kinase C agonist prostratin induces differentiation of human myeloid leukemia cells and enhances cellular differentiation by chemotherapeutic agents. Cancer Lett. 2015;356(2):686–696.
- Schmoellerl J, Barbosa IAM, Eder T, et al. CDK6 is an essential direct target of NUP98 fusion proteins in acute myeloid leukemia. Blood. 2020;136(4):387–400.
- Wang X, Liang Z, Xu X, et al. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis. 2016;7(12):e2503.
- Song B, Du J, Song DF, et al. Dysregulation of NCAPG, KNL1, miR-148a-3p, miR-193b-3p, and miR-1179 may contribute to the progression of gastric cancer. Biol Res. 2018;51(1):44.
- Wu T, Qu L, He G, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7(10):11553–11566.
- Man LM, Morris AL, Keng M. New therapeutic strategies in acute lymphocytic leukemia. Curr Hematol Malig Rep. 2017;12(3):197–206.
- De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 2016;6(7):e441.
- Yang JS, Li BJ, Lu HW, et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36(4):3035–3042.
- Jili S, Eryong L, Lijuan L, et al. RUNX3 inhibits laryngeal squamous cell carcinoma malignancy under the regulation of miR-148a-3p/DNMT1 axis. Cell Biochem Funct. 2016;34(8):597–605.
- Malumbres M, Sotillo R, Santamaria D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118(4):493–504.
- Jena N, Sheng J, Hu JK, et al. CDK6-mediated repression of CD25 is required for induction and maintenance of Notch1-induced T-cell acute lymphoblastic leukemia. Leukemia. 2016;30(5):1033–1043.
- Uras IZ, Maurer B, Nebenfuehr S, et al. Therapeutic vulnerabilities in FLT3-Mutant AML unmasked by palbociclib. Int J Mol Sci. 2018;19(12):3987.
- Kollmann K, Heller G, Schneckenleithner C, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24(2):167–181.