ABSTRACT
Nonalcoholic fatty liver disease (NAFLD) is characterized by high morbidity. Although long noncoding RNAs (lncRNAs) are known to have a role in NAFLD pathogenesis, the identified lncRNA types are limited. In this study, NAFLD models were established in vitro and in vivo using free fatty acid-treated LO2 cells and high-fat diet-fed mice, respectively. Microarray data were downloaded from the Gene Expression Omnibus database, and AC012668 was selected for further analysis. Cell viability and apoptosis were measured using Cell Counting Kit 8 and flow cytometry assays. RNA expression was detected using reverse transcription-quantitative polymerase chain reaction. Triglyceride (TG) content and lipid deposition were detected using enzyme-linked immunosorbent assay and Oil-Red O staining. Western blotting was used to visualize protein expression. Starbase and TargetScan were used to predict the target miRNA and gene, and the predictions were verified through RNA pull-down and luciferase reporter assays. AC012668 expression levels were significantly suppressed in NAFLD models, whereas AC012668 overexpression inhibited lipogenesis-related gene (SCD1, SREBP1, FAS) expression and TG/lipid accumulation in vitro. Subsequently, miR-380-5p was predicted and verified to target AC012668, and its expression was notably increased in the NAFLD cell model. Moreover, transfection of miR-380-5p antagonized the effects of AC012668 on lipid formation and accumulation. LRP2 was confirmed to be the target gene of miR-380-5p and was downregulated in the NAFLD cell model. Silencing LRP2 reversed the effects of the miR-380-5p inhibitor on lipid formation and accumulation. AC012668 inhibited NAFLD progression via the miR-380-5p/LRP2 axis. These findings may provide a novel strategy against NAFLD.
Introduction
Fatty liver, one of the most common liver disorders worldwide, is frequently caused by excessive intake of carbohydrates and lipids [Citation1]. According to its pathogenesis, fatty liver is classified into two categories, alcoholic fatty liver disease and nonalcoholic fatty liver disease (NAFLD), of which NAFLD is more common [Citation2]. NAFLD is a disease spectrum that progresses from simple hepatic steatosis to inflammatory responses, developing into nonalcoholic steatohepatitis (NASH) and hepatic fibrosis and ultimately leading to loss of liver function [Citation3]. Hepatic steatosis refers to a triglyceride (TG) level in the liver exceeding the 95% percentile in healthy individuals or TG lipid droplets filling > 5% of the liver cytoplasm [Citation4]. Simple hepatic steatosis is a self-limiting disease. NASH is considered present once hepatocellular damage, inflammation, or fibrosis occurs [Citation5]. Approximately 10% to 29% of patients with NASH will progress to cirrhosis within 10 years [Citation6]. At present, about 1 billion people worldwide have NAFLD [Citation7]. However, the treatment of NAFLD remains unsatisfactory and therapeutic drugs based on the pathogenesis of the disease are lacking [Citation6]. Therefore, the development of a novel and effective treatment for NAFLD is an urgent requirement to improve the life quality of patients.
Noncoding RNAs (ncRNAs) are crucial regulators of molecules that modulate biological processes [Citation8]. Long ncRNAs (lncRNAs) are a type of ncRNAs with a length of > 200 nucleotides [Citation9]. lncRNAs are required for many epigenetic processes, such as dose compensation, genomic imprinting, DNA methylation, histone modification, and chromatin remodeling [Citation10]. Dysregulated lncRNAs are closely involved in the development of NAFLD [Citation11]. For instance, high expression of the lncRNA GAS5 is closely associated with the evolution from NAFLD to advanced liver fibrosis [Citation12]. HULC knockdown inhibits the development of NAFLD [Citation13]. Moreover, lncRNAs act as competing endogenous RNAs (ceRNAs) to regulate biological processes by sponging microRNAs (miRNAs). NEAT1 deteriorates NAFLD by regulating the miR-140/AMPK axis [Citation14]. Gm12664-001 sponges miR-295-5p to induce the activation of CAV1, which alleviates NAFLD [Citation15]. The lcnRNA AC012668 is located in chromosome 2. However, the role of AC012668 has not been previously elucidated. Low-density lipoprotein-related protein 2 (LRP2) is primarily expressed in absorptive epithelial tissues [Citation16]. LRP2 has a pivotal function in metabolic diseases [Citation17]. To date, many lncRNA-miRNA pathways have been demonstrated to modulate NAFLD development [Citation18,Citation19]. Our group identified a novel lncRNA, AC012668, that is downregulated in NAFLD and functions as a sponge of miR-380-5p to promote the expression of LRP2.
Thereby, this study aimed to explore the role of AC012668 in NAFLD via establishing the model in vivo and in vitro. We hypothesized that AC012668 may suppress the progression of NAFLD via the miR-380-5p/LRP2 axis.
Materials and methods
NAFLD mouse model
Twenty 8-week-old male specific pathogen-free grade C57BL/6 J mice were purchased from Shanghai Southern Model Center. The mice were housed in a dry and ventilated pathogen-free barrier facility with 60% relative humidity, 12 h/day lighting, and 25°C temperature. The animals were randomly divided into two groups, 10 mice fed a regular diet and 10 mice fed a high-fat diet (HFD) for 12 weeks, to establish NAFLD models. The HFD (D12492, Beijing, China) consisted of protein, fat, and carbohydrates at 20%, 60%, and 20% of the total calories, respectively. The control group received a regular diet consisting of protein, fat, and carbohydrates at 20%, 10%, and 70% of the total calories, respectively. After 12 weeks of feeding, the mice were sacrificed through intraperitoneal injection of sodium pentobarbital (150 mg/kg) and the livers were collected for the subsequent experiments.
This study supervised by the Ethics Committee of Beijing Friendship Hospital, Capital Medical University Experimental Animal Center.
Cell culture and transfection
Normal human hepatocyte lines LO2 were purchased from Biowit Biotechnology Inc. (cat no. C0009). The cells were maintained in Dulbecco’s modified Eagle’s medium containing penicillin, streptomycin (Macklin), and 10% fetal bovine serum (Ausbian) at 37°C. After 24 h, the cells were cultured with 1 mM free fatty acid (FFA; containing oleic acid and palmitic acid at a 2:1 volume ratio).
The cells were transfected with AC120668/AC012668 small interfering RNA (si-AC012668), miR-380-5p mimic/inhibitor, and si-LRP2 (NovaBio Co. Inc.) in Lipofectamine® 2000 reagent (Invitrogen). The negative controls were transfected with Lipofectamine 2000 alone.
Microarray analysis
The microarray dataset GSE107231 was analyzed using Agilent-067406 Human CBC lncRNA + mRNA microarray V4.0. |logFC| > 2 and corrected P < 0.05 were used as standards. A total of 39 differentially expressed genes were screened, of which 18 were upregulated genes and 21 were downregulated genes. AC012668 was selected for further analysis.
Cell Counting Kit 8 (CCK8) assay
Thr CCK8 assay was conducted according to a previous study [Citation20]. Cells were collected and resuspended at a density of 1 × 105 cells/mL. Thereafter, the cell suspension was inoculated into 96-well plates at 100 μL/well. The cells were treated by adding 10 μL CCK8 reagent to each well and cultured at 37°C for 4 h. The absorbance values (450 nm) were obtained using a microplate reader (51119000, Thermo Fisher Inc.).
Flow cytometric apoptotic assay
Apoptotic cells were stained using an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit and detected using Attune NxT Flow Cytometer and its supporting software (Thermo Fisher Inc.). The Annexin V-FITC reagent (5 μL) was added to the cells, and the apoptosis rates were determined using flow cytometry as described by Zan et al. [Citation21].
Oil-Red O staining assay
Oil-Red O staining assay was performed according to a previous study [Citation22]. The LO2 cells were rinsed twice with phosphate-buffered saline (Meilunbio Biotechnology Co., Ltd) and placed on a coverslip after discarding the medium. The cells were fixed in 10% formaldehyde for 30 min. Oil-Red O (Solarbio) was added dropwise until the coverslip was completely covered. After 5 min of staining, the cells were washed with 60% isopropanol (Solarbio) and distilled water. Thereafter, the cells were counterstained with hematoxylin (Solarbio) for 2 min and observed under an optical microscope (SteREO Discovery.V20, Carl Zeiss Inc.).
Enzyme-linked immunosorbent assay (ELISA)
The mouse livers were homogenized and centrifuged at 5000 rpm for 5 min, and the supernatant was collected for later use. The TG levels in the mouse livers were measured using an ELISA kit following the manufacturer’s protocol (SEKH-0380, Solarbio). The obtained values were normalized to the total protein levels. The hepatic TG level was expressed as μg/g protein. The total protein levels were measured using a bicinchoninic acid (BCA) protein assay kit (PC0020, Solarbio).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNAs were collected from cells and tissues. RT-qPCR was performed using One Step SuperRT-PCR Mix Kit (T2240, Solarbio) on a Mastercycler® nexus (6330000072, Eppendorf Inc.). All primers used in the present study were designed and synthesized by Genewiz Inc. GAPDH and U6 served as loading controls. The thermocycling conditions were as follows: denaturation at 94°C for 60 s, annealing at 37°C for 60 s for 30 cycles, and extension at 72°C for 120 s. The results were analyzed using the 2−ΔΔCt method [Citation23]. The sequences of the primers were as follows: AC012668 forward (F) 5'-ATCAGAATCACCTGGCGGTC-3', reverse (R) 5'-TGTACTAGCGGCATCAGCAG-3'; SCD1 F 5''GCTGATCCTCATAATTCCCGA‐3', R 5';TTAAGCACCACAGCATATCGC‐3'; SREBP1 F 5'-ACAGTGACTTCCCTGGCCTAT-3', R 5'-GCATGGACGGGTACATCTTCAA-3'; FAS F 5'-AAATGAAAGCCAACTGCATCGAC-3', R 5'-ATTGGACCCTCGCTGAGCAC-3'; miR-380-5p F 5'-CTCGCTTCGGCAGCACA-3', R 5'-CAGTGCGTGTCGTGGAGT-3'; LRP2 F 5'-CCTTGCCAAACCCTCTGAAAAT-3'; R 5'-CACAAGGTTTGCGGTGTCTTTA-3'; and GAPDH F 5'-GGGAGCCAAAAGGGTCATCA-3', R 5'-TGATGGCATGGACTGTGGTC-3'.
Western blotting assay
Western blotting assay was conducted according to previous study [Citation24]. The total protein was collected and its level was determined using a BCA kit. The protein (40 μg) was separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels for 1.5 h at 120 V, and the separated protein fractions were transferred onto polyvinylidine diflouride membranes (Millipore) for 2 h at 200 mA. The membranes were blocked with 5% fat-free milk for 1 h and incubated with primary antibodies, including anti-SCD1 (PAB15990, 1:1000; Abnova Biotech Inc.), anti-SREBP1 (3961–100, 1:1000; BioVision Inc.), anti-FAS (ANT-205, 1:500; Prospec Technologies Inc.), and anti-GAPDH (3777 R, 1:3000; BioVision), and with a secondary antibody (6916; 1:1000; BioVision). Finally, protein expression was analyzed using an electrochemiluminescence system.
Luciferase reporter assay
Luciferase reporter assay was performed as described by Unal [Citation25]. The wild and mutant type of AC012668 and LRP2 luciferase reporter vectors were designed and synthesized by Guangzhou RiboBio Co., Ltd. The wild-/mutant-type vectors and miR-380-5p mimic/control (RiboBio Co., Ltd.) were cotransfected into the LO2 cells, and the cells were incubated for 24 h. Thereafter, the cells were lysed to detect the luciferase activity using a TransDetect® Double-Luciferase Reporter Assay Kit (FR201-01, TransGen Biotech Co., Ltd.) 48 h after transfection.
RNA pull-down assay
RNA pull-down assay was performed using an RNA pull-down kit (Bes5102, Bersin Biotechnology Co., Ltd.) according to a previous study [Citation26]. Abiocenter (Beijing) Biotechnology Co., Ltd synthesized the biotinylated miR-380-5p probe and its control probe. Magnetic beads were resuspended in 50 μL RNA immunoprecipitation wash buffer and incubated with the biotin-labeled probes (50 pmol) at 4°C overnight. The cells were lysed with radioimmunoprecipitation assay buffer and RNase for 1 h to release the total RNA. Finally, the beads were washed six times with the lysis buffer. After the separation, the same RT-qPCR process as described above was used to quantify the relative expression of LRP2 or AC012668.
Statistical analysis
Data were analyzed using SPSS19.0 statistical software and expressed as mean ± standard deviation values. Student’s t test was used to assess the difference between two groups and analysis of variance (Duncan’s multiple range test) was applied for analyzing the data among multiple groups. P < 0.05 was deemed to indicate statistical significance.
Results
Thereby, this study aimed to explore the effects of lncRNA AC012668 on the TG accumulation and lipogenesis-related gene expression in NAFLD via establishing the model in vivo and in vitro. We demonstrated that lncRNA AC012668 may suppress the progression of NAFLD via the miR-380-5p/LRP2 axis.
lncRNA AC012668 expression level was decreased in HFD mice
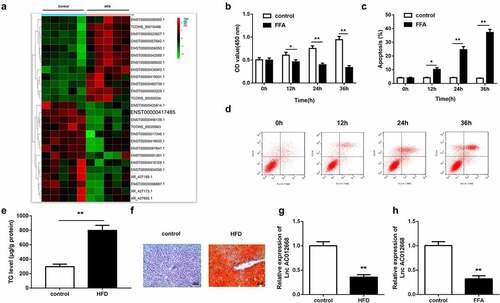
Through microarray analysis, AC012668 was predicted to be downregulated in NAFLD (). To further verify the role of AC012668 in NAFLD, we generated FFA-treated LO2 cells and an HFD mouse model to simulate NAFLD in vitro and in vivo, respectively. FFA exposure significantly suppressed cell viability () but significantly increased the apoptosis rate ( and d), indicating the successful establishment of the NAFLD cell model. The results of both Oil-Red O staining and ELISA indicated that HFD significantly increased the TG content in the mouse livers ( and f). RT-qPCR revealed that the AC012668 expression level was downregulated in the in vivo and in vitro NAFLD models ( and h).
Figure 1. NAFLD models established to investigate changes in AC012668 expression. (a) Heat map of differentially expressed lncRNAs in NAFLD. (b) Cell viability and (c and d) apoptosis of LO2 cells treated with FFA. (e and f) Oil-Red O staining image and TG level in the liver tissues of HFD mice. (g) AC012668 expression levels in the liver tissues of HFD mice. (h) Expression of AC012668 in FFA-treated cells. *P < 0.05 vs. control; **P < 0.01 vs. control
Overexpression of lncRNA AC012668 inhibited TG accumulation and lipogenesis-related gene expression in LO2 cells
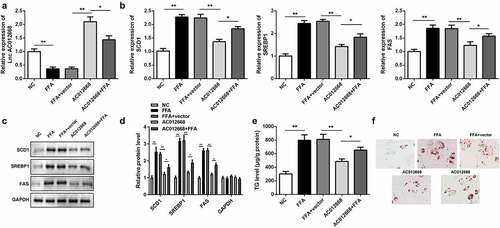
Given the observed downregulation of AC012668 in NAFLD, we further investigated the potential role of AC012668 in vitro by transfecting cells with its overexpression plasmid. The AC012668 overexpression plasmid significantly increased the expression of AC012668 (). Additionally, AC120668 suppressed the expression of lipogenesis-related genes, including SCD1, SREBP1, and FAS, in the FFA-treated cells in mRNA () and protein levels ( and d). Furthermore, the TG content was prominently reduced () and lipid droplet deposition was ameliorated in the AC012668 group ().
Figure 2. AC012668 suppresses TG/lipid accumulation and lipogenesis in LO2 cells. (a) AC012668 expression level in FFA-treated LO2 cells transfected with the AC012668 overexpression plasmid. (b) mRNA and (c and d) protein expression levels of SCD1, SREBP1, and FAS in FFA-treated LO2 cells. (e) TG level and (f) Oil-Red O staining in FFA-treated LO2 cells. *P < 0.05 vs. pcDNA3.1 or AC012668.
lncRNA AC012668 targeted miR-380-5p
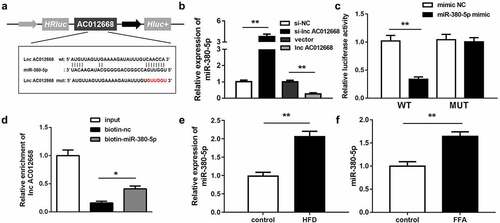
AC012668 was predicted to target miR-380-5p in the analysis using Starbase (http://starbase.sysu.edu.cn/) (). The miR-380-5p expression level was significantly increased by si-AC012668 and inversely inhibited by AC012668 (). Cotransfection with the miR-380-5p mimic and AC012668 (3′-untranslated region wild-type) markedly decreased the luciferase activity (). The RNA pull-down assay results further confirmed the interaction between miR-380-5p and AC012668 (). In addition, FFA evidently promoted the expression of miR-380-5p in the in vivo and in vitro NAFLD models ( and f).
Figure 3. miR-380-5p directly targets AC012668. (a) Wild and mutant types of AC012668 reporters labeled with luciferase. (b) Expression levels of miR-380-5p in AC012668-enhanced/-inhibited LO2 cells. (c) Relative luciferase activities in wild-type and mutant AC012668 groups compared with the mimic negative control group. (d) Relative enrichment of AC012668 in the biotinylated miR-380-5p group. (e) AC012668 expression levels in the liver tissues of HFD mice. (f) Expression level of miR-380-5p in LO2 cells treated with 1 mM FFA. *P < 0.05 vs. biotin-NC; **P < 0.01 vs. si-NC, vector, mimic NC, or control
miR-380-5p-facilitated lipogenesis antagonized lncRNA AC012668
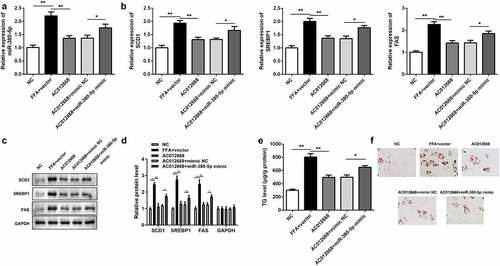
The effects of miR-380-5p on lipogenesis were also investigated. Cotransfection with the miR-380-5p mimic alleviated the effects of AC012668 on the expression of miR-380-5p compared with the AC012668 + negative control mimic group (). The inhibitory effects of AC120668 on the expression of SCD1, SREBP1, and FAS were all partially reversed via the miR-380-5p mimic in the mRNA () and protein levels ( and d). Moreover, the TG content () and deposition of lipid droplets () were increased by miR-380-5p.
Figure 4. miR-380-5p promotes TG/lipid accumulation and lipogenesis in LO2 cells transfected with AC012668. (a) AC012668 expression detected using reverse transcription-quantitative polymerase chain reaction. (b) mRNA and (c and d) protein levels of SCD1, SREBP1, and FAS. (e) TG level and (f) lipid deposition. *P < 0.05 vs. AC012668 + mimic NC; **P < 0.01 vs. vector
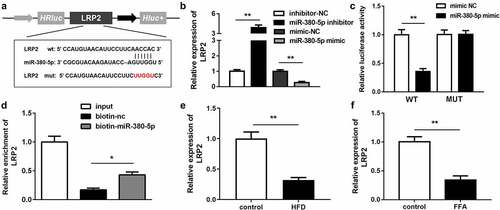
LRP2 was the downstream gene of miR-380-5p
LRP2 was predicted to be the downstream target gene of miR-380-5p in the analysis using TargetScan (http://www.targetscan.org/mamm_31/) (). The LRP2 expression level was significantly upregulated in the miR-380-5p inhibition group and downregulated in the miR-380-5p mimic group (). The RNA pull-down and luciferase reporter assay results further confirmed the interaction between miR-380-5p and LRP2 ( and d). In addition, the LRP2 expression level was decreased in the in vivo and in vitro NAFLD models ( and f).
Figure 5. LRP2 is a target of miR-380-5p. (a) Wild and mutant types of LRP2 reporters labeled with luciferase. (b) mRNA expression of LRP2 in FFA-treated LO2 cells transfected with the miR-380-5p mimic or inhibitor. (c) Relative luciferase activities in wild-type and mutant LRP2 groups compared with the mimic negative control group. (d) Relative enrichment of LRP2 in the biotinylated miR-380-5p group. (e) AC012668 expression levels in the liver tissues of HFD mice. (f) Expression level of LRP2 in FFA-treated LO2 cells. *P < 0.05 vs. biotin-NC; **P < 0.01 vs. inhibitor NC, mimic NC, or control
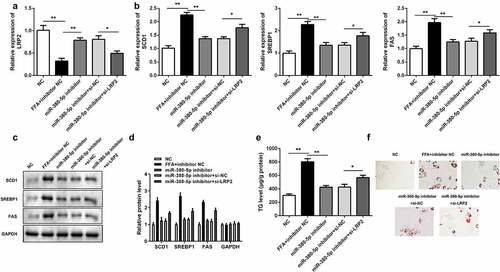
Silencing LRP2 neutralized the effects of miR-380-5p inhibitor on lipogenesis
si-LRP2 reversed the upregulation of LRP2 induced by the miR-380-5p inhibitor in LO2 cells (). Knockdown of LRP2 antagonized the effects of miR-380-5p and significantly increased expression levels of lipogenesis-related genes (SCD1, SREBP1, FAS) in the mRNA () and protein ( and d) levels. Furthermore, LRP2 knockdown increased the TG level () and the amount of lipid deposition ( and f).
Figure 6. Effects of LRP2 knockdown on TG/lipid accumulation and lipogenesis in miR-380-5p-inhibited LO2 cells. (a) miR-380-5p expression level in LO2 cells cotransfected with the miR-380-5p inhibitor and si-LRP2. (b) mRNA and (c and d) protein levels of SCD1, SREBP1, and FAS in the miR-380-5p inhibitor- and si-LRP2-cotransfected cells. (e) TG level and (f) lipid deposition in the miR-380-5p inhibitor- and si-LRP2-cotransfected cells. *P < 0.05 vs. control or miR-380-5p mimic + si-NC
Discussion
NAFLD is the leading cause of chronic hepatopathy [Citation27]. However, few targeted drugs have been clinically applied [Citation28]. In the current study, we screened for downregulated lncRNAs in NAFLD. AC012668 was first identified and selected for subsequent experiments. The expression of AC012668 was inhibited in vitro and in vivo. In addition, FFA-induced lipogenesis and lipid deposition were alleviated by AC012668. AC012668 upregulated LRP2 by sponging miR-380-5p. Surprisingly, overexpression of miR-380-5p or silencing of LRP2 reversed the effects of AC012668 overexpression and promoted lipogenesis. These findings demonstrated the role of an AC012668/miR-380-5p/LRP2 pathway in the pathogenesis of NAFLD.
Numerous studies have demonstrated that lncRNAs participate in the progression of NAFLD [Citation29–31]. For instance, the lncRNA NEAT1 was upregulated in NAFLD models and promoted lipid accumulation by targeting the miR-146a-5p/ROCK1 axis [Citation14]. Huang et al. [Citation32] observed that overexpression of MEG3 inhibited the process of lipogenesis via the miR-21/LRP6 axis. Sponging of miR-742-3p by Gm15622 enhanced the expression of the transcriptional regulator SREBP1c and promoted lipid accumulation in the NAFLD models [Citation33]. Herein, we first documented that AC012668 was downregulated in NAFLD in vivo and in vitro. Overexpression of AC012668 reduced the expression of lipogenesis-related genes and the TG level. These inhibitory effects on lipogenesis suppressed the progression of NAFLD, consistent with the findings of previous studies [Citation14,Citation32,Citation33].
lncRNAs function as ceRNAs to regulate gene expression through binding to miRNAs [Citation11]. In this study, miR-380-5p was a target of AC012668. miR-380-5p was first reported to be highly expressed in the majority of primary neuroblastomas and to function as a proto-oncogene in a mouse mammary transplant model [Citation34]. A microarray analysis study revealed that miR-380-5p was associated with metastasis and poor prognosis in breast cancer [Citation35]. In this study, overexpression of miR-380-5p attenuated the effects of AC012668 and promoted lipogenesis in LO2 cells, suggesting that miR-380-5p overexpression is closely related to the progression of NAFLD. Meanwhile, transfection of the miR-380-5p mimic exacerbated lipogenesis and lipid deposition, attenuating the effects of AC012668.
Previous studies mainly focused on the possible role of miR-380-5p in tumors [Citation34,Citation35]. However, its potential role in lipid metabolism has not been reported. In this study, miR-380-5p played a positive role in lipogenesis and enhanced the development of NAFLD from lipid metabolism disorders. These findings may add to the existing knowledge about miR-380-5p. Suppressing the expression of miR-380-5p may be an alternative method to suppress the progression of NAFLD.
LRP2, also known as megalin, is a multiligand receptor expressed by macrophages in the liver (Kupffer cells) [Citation36]. In a previous study, peritubular capillary damage and HFD-induced glomerular alterations were ameliorated in LRP2-knockout mice [Citation17]. Another study reported that LRP2 antagonized the attenuated effects of palmitate on clusterin-mediated insulin signaling and APOA1 expression in human adipocytes and reduced hepatic gluconeogenesis in mice [Citation37]. In this study, LRP2 was found to be a target gene of miR-380-5p and downregulation of LRP2 promoted lipid formation. Collectively, these results suggest the role of an AC012668/miR-380-5p/LRP2 signaling pathway in the pathogenesis of NAFLD.
Conclusion
In summary, the lncRNA AC012668 was downregulated in NAFLD. Overexpression of AC012668 suppressed lipid formation and protected against NAFLD by regulating the miR-380-5p/LRP2 axis. These results may provide a promising strategy against NAFLD.
Research highlights
miR-380-5p was verified to target AC012668
AC012668 expression was suppressed in NAFLD
LRP2 was confirmed to be the target gene of miR-380-5p
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Cariou B, Byrne CD, Loomba R, et al. Nonalcoholic fatty liver disease as a metabolic disease in humans: a literature review. Diabetes Obes Metab. 2021;23:1069–1083.
- Ullah R, Rauf N, Nabi G, et al. Role of nutrition in the pathogenesis and prevention of non-alcoholic fatty liver disease: recent updates. Int J Biol Sci. 2019;15(2):265–276.
- Sumida Y, Shima T, Mitsumoto Y, et al. Epidemiology: pathogenesis, and diagnostic strategy of diabetic liver disease in Japan. Int J Mol Sci. 2020;21(12):4337.
- Mahdi L, Kahn A, Dhamija R, et al. Hepatic steatosis resulting from LMNA-associated familial lipodystrophy. ACG Case Rep J. 2020;7(4):e00375.
- Boeckmans J, Natale A, Rombaut M, et al. Anti-NASH drug development hitches a lift on PPAR agonism. Cells. 2019;9(1):37.
- Khomich O, Ivanov AV, Bartosch B. Metabolic hallmarks of hepatic stellate cells in liver fibrosis. Cells. 2019;9(1):24.
- Youssry S, Kamel MA. Effect of folate supplementation on immunological and autophagy markers in experimental nonalcoholic fatty liver disease. Eur Cytokine Netw. 2019;30(4):135–143.
- Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166.
- Wu Y, Zhang F, Li X, et al. Systematic analysis of lncRNA expression profiles and atherosclerosis-associated lncRNA-mRNA network revealing functional lncRNAs in carotid atherosclerotic rabbit models. Funct Integr Genomics. 2020;20(1):103–115.
- Furio-Tari P, Tarazona S, Gabaldón T, et al. spongeScan: a web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44(W1):W176–80.
- Rohilla S, Awasthi A, Kaur S, et al. Evolutionary conservation of long non-coding RNAs in non-alcoholic fatty liver disease. Life Sci. 2021;264:118560.
- Han MH, Lee JH, Kim G, et al. Expression of the long noncoding RNA GAS5 correlates with liver fibrosis in patients with nonalcoholic fatty liver disease. Genes (Basel). 2020;11(5):545.
- Shen X, Guo H, Xu J, et al. Inhibition of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis by inhibiting the MAPK signaling pathway in rats with nonalcoholic fatty liver disease. J Cell Physiol. 2019;234(10):18169–18179.
- Sun Y, Song Y, Liu C, et al. LncRNA NEAT1-MicroRNA-140 axis exacerbates nonalcoholic fatty liver through interrupting AMPK/SREBP-1 signaling. Biochem Biophys Res Commun. 2019;516(2):584–590.
- Zhang Q, Wang J, Li H, et al. LncRNA Gm12664-001 ameliorates nonalcoholic fatty liver through modulating miR-295-5p and CAV1 expression. Nutr Metab (Lond). 2020;17:13.
- Ye F, Wang Y, Wu C, et al. Angiotensinogen and megalin interactions contribute to atherosclerosis-brief report. Arterioscler Thromb Vasc Biol. 2019;39(2):150–155.
- Kuwahara S, Hosojima M, Kaneko R, et al. Megalin-mediated tubuloglomerular alterations in high-fat diet-induced kidney disease. J Am Soc Nephrol. 2016;27(7):1996–2008.
- Liu J, Tang T, Wang GD, Liu B. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARgamma axis in non-alcoholic fatty liver disease. Biosci Rep. 2019;39(7):BSR20181722.
- Huang F, Liu H, Lei Z, et al. Long noncoding RNA CCAT1 inhibits miR-613 to promote nonalcoholic fatty liver disease via increasing LXRα transcription. J Cell Physiol. 2020;235(12):9819–9833.
- Wang S, Li P, Jiang G, et al. Long non-coding RNA LOC285194 inhibits proliferation and migration but promoted apoptosis in vascular smooth muscle cells via targeting miR-211/PUMA and TGF-β1/S100A4 signal. Bioengineered. 2020;11(1):718–728.
- Zan L, Chen Q, Zhang L, et al. Epigallocatechin gallate (EGCG) suppresses growth and tumorigenicity in breast cancer cells by downregulation of miR-25. Bioengineered. 2019;10(1):374–382.
- Andrés-Manzano MJ, Andrés V, Dorado B. Oil red O and hematoxylin and eosin staining for quantification of atherosclerosis burden in mouse aorta and aortic root. Methods Mol Biol. 2015;1339:85–99.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Hu X, Chen J, Huang H, et al. Syndecan-4 promotes vascular beds formation in tissue engineered liver via thrombospondin 1. Bioengineered. 2020;11(1):1313–1324.
- Unal H. Luciferase reporter assay for unlocking ligand-mediated signaling of GPCRs. Methods Cell Biol. 2019;149:19–30.
- Zhou D, Lin X, Wang P, et al. Circular RNA circ_0001162 promotes cell proliferation and invasion of glioma via the miR-936/ERBB4 axis. Bioengineered. 2021;12(1):2106–2118.
- Shabangu CS, Huang J-F, Hsiao -H-H, et al. Liquid biopsy for the diagnosis of viral hepatitis, fatty liver steatosis, and alcoholic liver diseases. Int J Mol Sci. 2020;21(10):3732.
- Pennisi G, Celsa C, Giammanco A, et al. The burden of hepatocellular carcinoma in non-alcoholic fatty liver disease: screening issue and future perspectives. Int J Mol Sci. 2019;20(22):5613.
- Zhang M, Chi X, Qu N, et al. Long noncoding RNA lncARSR promotes hepatic lipogenesis via Akt/SREBP-1c pathway and contributes to the pathogenesis of nonalcoholic steatohepatitis. Biochem Biophys Res Commun. 2018;499(1):66–70.
- Zhang B, Li H, Li D, et al. Long noncoding RNA Mirt2 upregulates USP10 expression to suppress hepatic steatosis by sponging miR-34a-5p. Gene. 2019;700:139–148.
- Wu H, Zhong Z, Wang A, et al. LncRNA FTX represses the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via regulating the M1/M2 polarization of Kupffer cells. Cancer Cell Int. 2020;20:266.
- Huang P, Huang F-Z, Liu H-Z, et al. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR-21 with LRP6. Metabolism. 2019;94:1–8.
- Ma M, Duan R, Shen L, et al. The lncRNA Gm15622 stimulates SREBP-1c expression and hepatic lipid accumulation by sponging the miR-742-3p in mice. J Lipid Res. 2020;61(7):1052–1064.
- Swarbrick A, Woods SL, Shaw A, et al. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16(10):1134–1140.
- Nygren MK, Tekle C, Ingebrigtsen VA, et al. Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br J Cancer. 2014;110(8):2072–2080.
- Pieper-Furst U, Lammert F. Low-density lipoprotein receptors in liver: old acquaintances and a newcomer. Biochim Biophys Acta. 2013;1831(7):1191–1198.
- Bradley D, Blaszczak A, Yin Z, et al. Clusterin impairs hepatic insulin sensitivity and adipocyte clusterin associates with cardiometabolic risk. Diabetes Care. 2019;42(3):466–475.
