ABSTRACT
The lncRNAs have been made certain to take part in the development of most cancers in multiple ways. Here, our purpose is to making observation of the biological role and function of lncRNA CDKN2B-AS1 in human breast cancer. Twenty-eight pairs of breast cancer tissue and adjacent normal tissue from breast cancer patients were used to investigate the expression of CDKN2B-AS1 by qRT-PCR. And a lentivirus-shRNA guided CDKN2B-AS1 were to reduce its expression. The function of CDKN2B-AS1 was analyzed using a series of in vitro assays. Meanwhile, the xenograft model was used to further explicate the role of CDKN2B-AS1 in breast cancer. As for the results, there is a relative rich expression of CDKN2B-AS1 in breast cancer tissues compared with the corresponding adjacent normal tissues. Compared with the human breast epithelial cell line, the abundant expression of CDKN2B-AS1 in breast cancer cells were revealed as well. Then, knockdown CDKN2B-AS1 inhibited the malignant biological behaviors of MCF7 and T47D cells. In mechanism, CDKN2B-AS1 sponged the miR-122-5p to regulate STK39 expression. Furthermore, the inhibition effect with sh-CDKN2B-AS1 on breast cancer cells was alleviated by miR-122-5p inhibitor. Last, an in vivo model also confirmed that knockdown CDKN2B-AS1 retarded the growth of breast cancer. Our data concluded that knockdown of CDKN2B-AS1 suppresses the progression of breast cancer by miR-122-5p/STK39 axis.
1. Introduction
Breast cancer (BC), as one of the highest degree of malignant cancers, seriously endangers the public health [Citation1]. Only in 2020, among all newly diagnosed women cancer patients, the breast cancer alone accounts nearly for 30% [Citation1–3]. The continuously increasing incidence of breast cancer caused a huge burden for public health [Citation3,Citation4]. Breast cancer is clinically classified into various subtypes, such as the luminal, human epidermal growth factor receptor 2 (HER2)-enriched, and triple-negative (TN) subtype [Citation5]. A constant stream of breast cancer patients are putting the huge pressure on clinical treatment for breast cancer [Citation4,Citation6]. Nowadays, the therapeutic strategy for breast cancer is mainly surgery supplemented with chemotherapy [Citation6,Citation7]. Revealing the underlying mechanism about tumorigenesis and progression of breast is beneficial to clinical treatment. Thus, seeking a novel and effective therapeutic target in breast cancer is an urgent demand for breast cancer patients.
Recently, increasing researches have pointed that non-coding RNAs equally exert their biological function [Citation8,Citation9]. The long non-coding RNAs (lncRNAs) which participate in the progression in multiple ways have been proven in varieties of cancers [Citation9,Citation10]. Up to date, more and more studies have realized that lncRNAs could be an oncogene or a tumor suppressor in development of human tumors [Citation9,Citation11,Citation12]. Under normal circumstances, lncRNAs perform as competing endogenous RNA (ceRNA) to adsorb miRNA as sponges [Citation13,Citation14]. The aberrant expression of lncRNAs would induce dysregulated miRNA expression, eventually impact on the cellular signaling pathways to affect cancer progression [Citation14,Citation15].
The lncRNA CDKN2B Antisense RNA 1 (CDKN2B-AS1) is located within the CDKN2B-CDKN2A (Cyclin Dependent Kinase Inhibitor 2B, Cyclin Dependent Kinase Inhibitor 2A) gene cluster at chromosome 9p21 [Citation16]. It was reported that lncRNA CDKN2B-AS1 is associated with a number of pathologies, such as cardiovascular disease, Alzheimer’s disease and type-2 diabetes [Citation17–19]. In breast cancer, several studies indicated that lncRNA CDKN2B-AS1 could be an independent bio-markers for diagnosis due to it has an aberrant expression pattern [Citation20–22]. However, rare researches investigated how lncRNA CDKN2B-AS1 exerts its effects on breast cancer till date. The underlying mechanism about lncRNA functions in breast cancer remains enigmatic. Therefore, revealing the functional characteristic of lncRNA CDKN2B-AS1 in breast cancer is urgent and significant.
MicroRNAs (miRNAs, ~20 nt) belong to a class of non-coding RNAs that are highly conserved in evolution [Citation23]. With the development of the tumors, the expression profile of miRNAs alteration occurs [Citation23,Citation24]. It has been verified that this alteration could promote and repress the progression of most cancers [Citation25–27]. In general, miRNAs binds to the matched target 3ʹ-UTR of mRNA to restrict the target gene expression through forming a RNA-induced silencing complex (RISC) [Citation28]. Besides, some studies also indicated that the combined mRNA would be spliced to degrade [Citation28,Citation29]. Either way, miRNAs regulate the expressions of genes on decreasing the targets protein level [Citation28]. MiR-122-5p as an RNA gene is related to various diseases, including majorities of cancers like hepatocellular carcinoma (HCC), non-small cell lung cancer (NSCLC) and so on [Citation30–32]. Also in breast cancer, miR-122-5p was involved in the progression or chemo-sensitizes of breast cancer [Citation33].
Nevertheless, the miRNAs function often rely on the target downward protein [Citation28]. Serine/Threonine Kinase 39 (STK39) encodes a serine/threonine kinase affecting cellular stress response pathway [Citation34]. This kinase is closest implicated in oncogenesis, such as HCC, osteosarcoma and so on [Citation34,Citation35]. Previous researches have revealed that STK39 excited the mitogen-activated protein kinase (MAPK) pathway in tumorigenesis [Citation36]. As is well known, MAPK signaling usually play a vital factor to participate in varieties of cancers [Citation37]. Especially in breast cancer, MAPK signaling pathway regulates the cancer progression tightly [Citation38]. Meanwhile, many studies reported that STK39 accelerated the development of breast cancer [Citation39]. Hence, STK39 could regulate the development of breast cancer and reducing STK39 expression receded the malignancy of breast cancer.
To sum up, the following hypothesis is proposed that lncRNA CDKN2B-AS1 took part in regulating the progression of breast cancer. Herein, our aim and goal are to reveal the role and the mechanism of lncRNA CDKN2B-AS1 in human breast cancer. Through a systematical investigation of lncRNA CDKN2B-AS1, it would provide for clinical treatment of breast cancer with a potential therapeutic target and a novel diagnostic bio-marker.
2. Materials and methods
2.1 Tissue samples
The tumor tissue samples and matched adjacent normal tissues were obtained from breast cancer patients (28 females; age: 45–65 years’ old) at General Hospital of Ningxia Medical University between 2019 and 2020 with written informed consent. The clinical characteristics of patients with breast cancer are shown in . This study was reviewed and approved by the Ethics Review Committee of General Hospital of Ningxia Medical University and was performed in accordance with Declaration of Helsinki.
Table 1. The relationships between CDKN2B-AS1 expression and clinicopathological characteristics of breast cancer patients
2.2 Cells culture
Human normal breast basal epithelial cell, MCF10A, and human breast cancer cell lines, MCF7, T47D and MDA-MB-231 were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Medium selection for these cells was base DMEM medium with 10% FBS, 100 IU/mL penicillin, and 100 μg /mL streptomycin (Gibco, the US). All cells were incubated at 37°C, 5% CO2.
2.3 Cell transfections
MCF7 and T47D cells were infected with the lentivirus (sh CDKN2B-AS1 and sh NC, MOI = 25) to stably knockdown CDKN2B-AS1 in MCF7 and T47D cells. The lentivirus and infection regent HitransG A were obtained from Genechem (Shanghai, China). Through incubation with 2 mg/ml puromycin for 48 h, sh CDKN2B-AS1 and sh NC breast cancer cells were harvested by trypsin digestion and centrifugation. When cells reach at 50% confluence, cells were transfected with a miR-122-5p mimic (100 pmol), a miR-122-5p inhibitor (100 pmol) and their corresponding negative control were carried out as required in assigned experiment by using Lipofectamine™3000 (ThermoFisher, USA) to conduct follow-up experiments. All the miRNA mimic and inhibitor were synthesized from Invitrogen (Invitrogen, Shanghai, China).
2.4 Cell viabilities
The cell viabilities were assessed by the CCK-8 (Beyotime Institute of Biotechnology, Shanghai, China) solution and absorbance was measured at 450 nm through a Microplate Reader (Bio-Rad, USA). Briefly, planted 4000 transfected MCF7 cells or T47D cells in 96-well plates per well on day 0. After 6 hours and on Day1, Day2 and Day3, respectively, the absorbance was measured to determine the cell viability by adding 10 μl CCK-8 solution for another 4 hours at 37°C and 5% CO2.
2.5 Colony formation assays
About 600 cells were evenly planted into a 6-well plate for 10 days, then washed with PBS once, fixed with formaldehyde solution and stained with 0.1% crystal violet solution in turn. More than 50 cells were considered to a colony. The numbers of colonies were counted under an inverted microscope (Leica Microsystems, German).
2.6 Apoptotic cells
The apoptotic cells rates were analyzed by flow cytometry (FACScan, Beckman Coulter, USA) with an Annexin V-FITC/PI (Invitrogen, USA) assay. Briefly, all suspended and adherent cells were collected, and then resuspended and stained with 5 μl of Annexin V-FITC and 10 μl of PI for 15 min in dark. Finally, the apoptotic rate of different treatment groups was analyzed by flow cytometry.
2.7 Cell cycle
Cells were washed with PBS twice and fixed with 70% ethanol at 4°C overnight. After centrifugation, cells were washed and resuspended in 500 μL BD Pharmingen PI/RNase staining buffer (BD Biosciences). Then cells were incubated for 15 minutes at room temperature and analyzed on the same flow cytometer.
2.8 qRT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen). cDNAs were produced with the RNA templates by using a reverse transcription kit (Invitrogen). qRT-PCR analysis was performed on a CFX96 Thermal Cycler DiceTM real-time PCR system with SYBR Premix Ex Taq II (TaKaRa, Dalian, China). GAPDH and U6 were used as reference gene, respectively. The primers and shRNA in this article are in . The relative expressions were calculated by using 2−ΔΔCt method.
Table 2. The primers and shRNA in this article are as follow
2.9 Luciferase reporter assays
All plasmids were made from Genepharma, China. We first insert the containing underlying binding site sequence of CDKN2B-AS1 Wild Type (WT) or CDKN2B-AS1 mutant type (MUT) and into a luciferase report gene vectors (pRL-TK, Promega) to establish the two types plasmid. Subsequently, miR-122-5p mimic or miR NC was co-transfected into MCF7 and T47D cells with reporter plasmids for 48 h. The relative luciferase activity was detected by Dual-Luciferase Reporter Assay System (Promega). About 50 ng renilla plasmid was used as an internal reference. The same approach applies to the STK39 Wild Type (WT) or STK39 mutant type (MUT) plasmid.
2.10 Western blotting
The protein is extracted by cell lysis (RIPA Lysis Buffer, Beyotime, Shanghai, China)) and denatured with loading buffer to load into 10% SDS-PAGE gels. Subsequently, the samples were separated and transferred to PVDF membrane (Bio-Rad, USA). Then, the primary antibody, anti-STK39 (1:1000, orb100341, biorbyt) and anti-GAPDH (1:1000, ab8245, abcam), was incubated after blocking with blocking buffer overnight. On the second day, HRP conjugated secondary antibodies were incubated after washing. Finally, the proteins were examined through chemiluminescence. GAPDH was a reference.
2.11 Xenograft mouse model
Ten female mice (8 weeks old) were divided into two groups (sh NC and sh CDKN2B-AS1) and amount of 5 × 106 sh CDKN2B-AS1 and sh NC MCF7 cells were inoculated in the axils of each mouse. Tumor volume was measured every 3 days according to the following formula: volume = 1/2 × length × width2. Animal protocols, housing, and care were performed with the approval of the Ethics Committee of General Hospital of Ningxia Medical University and conducted according to the guidelines set forth in the National Institutes of Health’s (NIH) ‘Guide for the Care and Use of Laboratory Animals’ (8th edition).
2.12 Immunohistochemistry (IHC)
The dissected tumors were paraffin-embedded and cut into 4-μm sections. Through an antigenic repair process, the sections were incubated with antibodies, anti-Ki67 antibody (1:200, ab15580, abcam) or anti-STK39 (1:200, orb100341, biorbyt), at 4°C overnight. Subsequently, the sections were washed the primary antibodies away to incubate biotinylated secondary antibodies for 2 h at room temperature. IHC images were taken using an Olympus microscope after visualizing with diaminobenzidine substrate (Sigma-Aldrich, St Louis, MO, USA).
2.13 Statistical analysis
All statistical analyses were conducted using the SPSS statistical package (16.0, SPSS Inc. Chicago, IL). Unpaired student’s t test was used to compare the means of two groups of data. The data were shown as mean ± SD. P-values were calculated by ANOVA, with P < 0.05 considered as significant.
3. Results
In our study, we proposed that lncRNA CDKN2B-AS1 took part in regulating the progression of breast cancer. Thus, our aim and goal was to reveal the role and the mechanism of lncRNA CDKN2B-AS1 in human breast cancer. In this study, we first found that the different expression pattern of lncRNA CDKN2B-AS1 and miR-122-5p in breast cancers by detecting the collected clinical cancer samples. Subsequently, knockdown the inherently highly expressed lncRNA CDKN2B-AS1 with sh-RNA in order to investigate the function of CDKN2B-AS1. Through a series of functional and mechanism experiments, we concluded that lncRNA CDKN2B-AS1 acts as a miR-122-5p sponge to regulate the STK39 expression, and promotes breast cancer progression.
3.1 The aberrant expressions of lncRNA CDKN2B-AS1 in breast cancer tissues and cells.
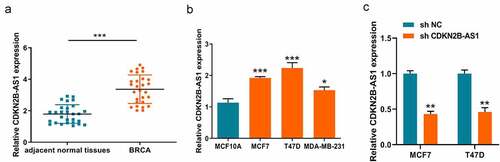
Finding out the expression levels of CDKN2B-AS1 gave a hint on the role in development of breast cancer. Therefore, we initially detected the CDKN2B-AS1 expression in the 28 collected breast cancer tissues from breast patients. As shown in , CDKN2B-AS1 present a higher expression level in breast cancer tissues compared with the adjacent normal tissues. Due to CDKN2B-AS1 is upregulated in breast cancer tissues, we then detected the expression in breast cancer cells. Similarly, the level of CDKN2B-AS1 in breast cancer cells is consistent with the previous result in breast tissues. All the three breast cancer cells had the relative rich expression in comparison to the human normal breast basal epithelial cell (MCF10A). In , the expressions of CDKN2B-AS1 in MCF7 and T47D have more significant difference with the expression of MCF10A (***P < 0.001) compared with the difference between MDA-MB-231 and MCF10A (*P < 0.05). Therefore, we chose this two breast cancer cells with the shRNA system to conduct follow-up experiments. Knockdown of CDKN2B-AS1 with an shRNA lowered the CDKN2B-AS1 level in MCF7 and T47D cells (). All these results indicated the dysregulated expressions of CDKN2B-AS1 might participate in progression of breast cancer.
Figure 1. The aberrant expressions of lncRNA CDKN2B-AS1 in breast cancer tissues and cells. (a). The level of CDKN2B-AS1 in breast cancer tissues and adjacent normal tissues was examined by RT-qPCR; (b). The expression levels of CDKN2B-AS1 were detected by RT-qPCR in MCF10A and breast cancer cells; (c). The expression of CDKN2B-AS1 were detected by qRT-PCR after knockdown of CDKN2B-AS1 with shRNA. Data are presented as mean ± SD. n = 6, *P < 0.05, **P < 0.01, ***P < 0.001
3.2 Knockdown of CDKN2B-AS1 repressed the MCF7 and T47D cell proliferation and induced the apoptosis.
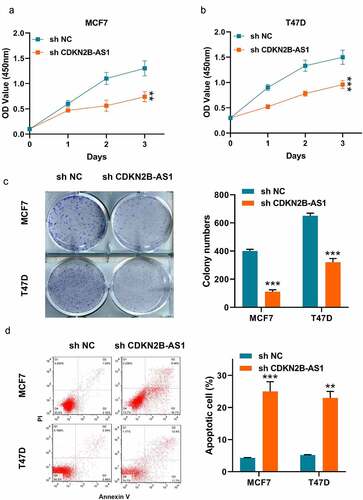
Then, we examined the effect of sh-CDKN2B-AS1 on the proliferation of the two cells. As shown in , the CCK-8 results showed that inhibition of CDKN2B-AS1 decreased the cell viability of MCF7 and T47D cells. Meanwhile, the clonality of the two cells were repressed by knockdown of CDKN2B-AS1 (). In addition, we detected the apoptosis of the two cancer cells as well. It was illustrated that lowering the expression of CDKN2B-AS1 would lead to induce the apoptosis (). From the above results, we could summarize that knockdown of CDKN2B-AS1 affected the MCF7 and T47D cellular functions.
Figure 2. Knockdown of CDKN2B-AS1 inhibited the breast cancer cell proliferation and induced apoptosis. (a) and (b). The relative cell viability was detected by CCK-8 assay in MCF7 and T47D cells; (c). The cell proliferation was evaluated with the colony formation assay in MCF7 and T47D cells; (d). The cell apoptosis was evaluated by flow cytometry in MCF7 and T47D cells; Data are presented as mean ± SD. n = 6, **P < 0.01, ***P < 0.001
3.3 MiR-122-5p is a target of lncRNA CDKN2B-AS1.
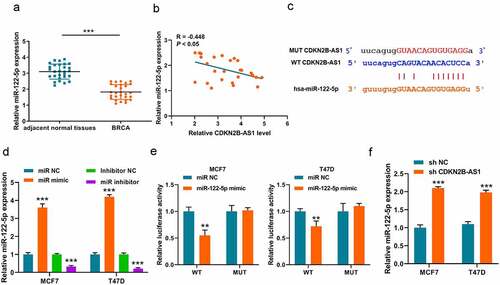
Upon most occasions, lncRNAs perform as ceRNA to adsorb miRNA as sponges. Thus, we first analyzed the underlying downstream miRNAs of lncRNA CDKN2B-AS1 on Online database (http://starbase.sysu.edu.cn/). Among the numerous of results, miR-122-5p is a potential target of CDKN2B-AS1. Thus, we also detected the expression of miR-122-5p in breast cancer tissues and a downward trend was discovered in breast cancer (). Through analyzing correlation between the expression pattern of lncRNA CDKN2B-AS1 and miR-122-5p in breast cancer, we found that there was a negative correlation (R = – 0.448, P < 0.05) in . Subsequently, to verified this interaction, we construct the luciferase reporter plasmid system () and the miR-122-5p mimic, miR-122-5p inhibitor and respective negative control were transfected into MCF7 and T47D cells for elevating or repressing the miR-122-5p level (). The luciferase system results indicated that miR-122-5p could lower the luciferase activity of WT plasmid in MCF7 and T47D cells, but made no difference on MUT plasmid (). Finally, we verified that the expressions of miR-122-5p were elevated in sh-CDKN2B-AS1 MCF7 and T47D cells (). All these results indicated that knockdown of CDKN2B-AS1 could elevate the expression of miR-122-5p.
Figure 3. MiR-122-5p is a target of CDKN2B-AS1. (a). The expression levels of miR-122-5p in breast cancer tissues and adjacent normal tissues were detected by RT-qPCR; (b). The correlation between CDKN2B-AS1 and miR-122-5p was analyzed in breast cancer tissues (R = −0.448, P < 0.05); (c). The binding site between CDKN2B-AS1 and miR-122-5p; (B). Luciferase activity was examined in MCF7 and T47D cells; (d). The levels of miR-122-5p were detected by qRT-PCR in MCF7 and T47D cells; (e). Luciferase activity was examined in MCF7 and T47D cells; (f). The levels of miR-122-5p in MCF7 and T47D cells. Data are presented as mean ± SD. n = 5, **P < 0.01, ***P < 0,001
3.4 STK39 is a target of miR-122-5p.
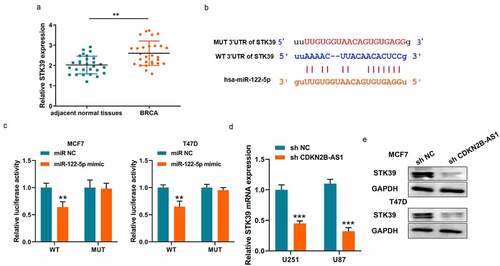
Similarly, we also analyzed the underlying downstream targets of miR-122-5p on Online database. STK39 is a potential target of miR-122-5p and an upward trend was discovered in breast cancer (). The binding site and corresponding luciferase reporter plasmid were also verified the interaction between miR-122-5p and STK39 (). And it also illustrated that miR-122-5p only affected WT fluorescence intensity and doesn’t affect MUT intensity (). Meanwhile, the mRNA and protein level of STK39 were both downregulated when CDKN2B-AS1 expression was restricted in MCF7 and T47D cells (). Thus, CDKN2B-AS1 regulate the expressions of miR-122-5p and STK39 as a ceRNA sponge.
Figure 4. STK39 is a target of miR-122-5p. (a). The expression levels of STK39 in breast cancer tissues and adjacent normal tissues were detected by RT-qPCR; (b). The binding site between miR-122-5p and STK39; (c). Luciferase activity was examined in MCF7 and T47D cells; (B). Luciferase activity was examined in MCF7 and T47D cells; (d). The mRNA levels of STK39 in MCF7 and T47D cells; (e). The protein levels of STK39 in MCF7 and T47D cells. Data are presented as mean ± SD. n = 5, **P < 0.01, ***P < 0,001
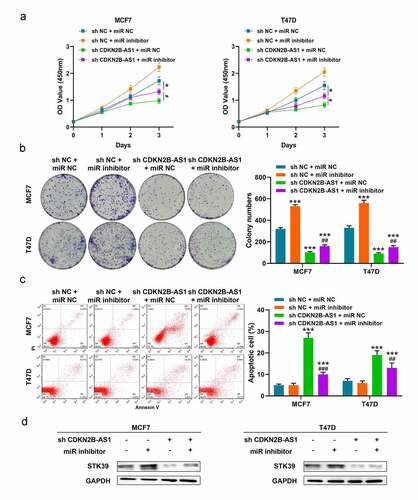
3.5 LncRNA CDKN2B-AS1 affected the biological behaviors of MCF7 and T47D cells through miR-122-5p/STK39 axis
Based on previous results, we speculated that lncRNA CDKN2B-AS1 functions via miR-122-5p and STK39 and restricting the expression of miR-122-5p with miR inhibitor could alleviate the impact of sh-CDKN2B-AS1 in MCF7 and T47D cells. In , the inhibition of sh-CDKN2B-AS1 on cell viability and colony formation were relieved with miR-122-5p inhibitor in MCF7 and T47D cells. Likewise, miR-122-5p inhibitor also weakened the apoptosis occurrence when CDKN2B-AS1 were knockdown in MCF7 and T47D cells (). In addition, the protein level of STK39 was elevated after transfecting with miR-122-5p inhibitor in sh-CDKN2B-AS1 MCF7 and T47D cells (). Thus, we considered that CDKN2B-AS1 regulated tumor progression of MCF7 and T47D cells through miR-122-5p/STK39 axis.
Figure 5. LncRNA CDKN2B-AS1 affected the functions of MCF7 and T47D cells through miR-122-5p/STK39 axis. (a). The relative cell viability was detected by CCK-8 assay; (b). The cell proliferation was evaluated with the colony formation assay; (c). The cell apoptosis was evaluated by flow cytometry; (d). The protein levels of STK39 in MCF7 and T47D cells. Data are presented as mean ± SD. n = 5, * indicated the difference compared with the group of sh NC + miR NC; # indicated the difference compared with the group of sh CDKN2B-AS1 + miR NC; *P < 0.05, ***P < 0.001, #P < 0.05, ##P < 0.01, ###P < 0.001
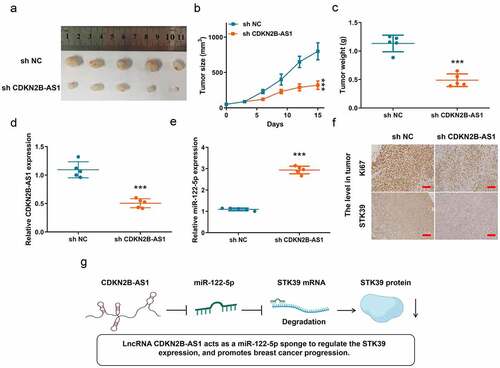
3.6 The biological role of CDKN2B-AS1 was further clarified in the xenograft breast cancer model.
To further clarify the biological role of lncRNA CDKN2B-AS1 in breast cancer, we established a xenograft tumor model on BALB/C nude mice with sh-CDKN2B-AS1 MCF7 and sh-NC MCF7 cells. The tumors were collected form the mice on day 15 after inoculation (). In , inhibition of lncRNA CDKN2B-AS1 significantly suppressed the tumor growth and decreased tumor weight. Meanwhile, in sh-CDKN2B-AS1 tumors, the expressions of CDKN2B-AS1 and miR-122-5p were repressed and increased, respectively. For detecting the level of Ki67 and STK39, sh-CDKN2B-AS1 lowered both the expression in tumor (). Hence, knockdown of CDKN2B-AS1 could retard the progression of breast cancer. To sum up, we found that CDKN2B-AS1 acts as a miR-122-5p sponge to regulate the STK39 expression, and promotes breast cancer progression ().
Figure 6. In vivo experiment confirmed that knockdown of CDKN2B-AS1 inhibited the progression of breast cancer. (a). The image of tumors; (b). The tumor volume; (c). The tumor weight; (d) and (e). The levels of CDKN2B-AS1 and miR-122-5p in tumors; (f). The Ki67 level in tumor and the STK39 level in tumor, Scale bar: 50 µm; (g). The schematic diagram of mechanism. n = 5, ***P < 0.001
4. Discussions
In our study, we first revealed that CDKN2B-AS1 acted as a miR-122-5p sponge to regulate the STK39 expression, and promoted breast cancer progression. Our data confirmed that reducing the lncRNA CDKN2B-AS1 expression restrained the proliferation of MCF7 and T47D cells, and increased apoptotic rate of the two breast cancer cells. Moreover, we further uncovered that miR-122-5p mediated the effect of sh-CDKN2B-AS1 through altering STK39 expression. In addition, a xenograft model with MCF7 cells confirmed knockdown of CDKN2B-AS1 retarded the breast cancer progression. To sum up, these results support the conclusion that CDKN2B-AS1 participates in the development of breast cancer cells.
Based on our data, lncRNA CDKN2B-AS1 played an oncogenic factor in breast cancer. It is consistent with the previous other studies about its role in most cancers, which indicated that CDKN2B-AS1 exerted a pro-cancer effect [Citation40–42]. For examples, in renal clear cell carcinoma, lncRNA CDKN2B-AS1 affected the malignancy through epigenetically activating NUF2 (NUF2 Component of NDC80 Kinetochore Complex) transcription [Citation16]. Besides, CDKN2B-AS1 had a stimulative function in osteosarcoma and facilitates lung cancer development through their respective ways of regulation [Citation43]. From the views of most reports, their conclusions about the role of lncRNAs CDKN2B-AS1 agreed with our findings and further verified that our results are reliable as well. In summary, in breast cancer, it should be considered lncRNA CDKN2B-AS1 to act as an oncogenic factor.
Similarly, the role of miR-122-5p in breast cancer were further revealed that they took part in the development of breast cancer. Based on our results, depressing the level of miR-122-5p could alleviate the inhibition of knockdown CDKN2B-AS1. In addition, it increased the malignancy of MCF7 and T47D tumor cells with miR-122-5p inhibitor alone. Therefore, we considered that miR-122-5p played a tumor suppressor in breast cancer. However, more evidence is needed to support this view despite there have been articles reported miR-122-5p functions as anti-oncogene in other cancers. Meanwhile, we subsequently verified the STK39 was identified as a target of the miR-122-5p as well. Previous studies have indicated that STK39 accelerates the development of most cancer, such as hepatocellular carcinoma, osteosarcoma and NSCLC [Citation34,Citation35,Citation44,Citation45]. More importantly, several reports have revealed that STK39 promoted progression of breast cancer [Citation39]. For instance, in breast cancer, Li C reported that STK39 regulated by miR-299-5p promoted cell metastasis [Citation39]. These researches uncovered the biological roles of miR-122-5p and STK39 in malignancy are accordance with our results.
Notwithstanding, there are still a few limitations in this work. Apart from the roles of the factors we revealed, more unknown biological functions are still need to be investigated about CDKN2B-AS1 and miR-122-5p. They may play a synergistic role in breast cancer through multi-level regulation. However, this work just uncovered the interaction between the CDKN2B-AS1 and miR-122-5p. Meanwhile, whether CDKN2B-AS1 regualting other molecules such as enzymes or kinases, which participate in tumor progression, is also worth studying.
5. Conclusion
In conclusion, these data support our hypothesis that lncRNA CDKN2B-AS1 acts as a miR-122-5p sponge to regulate the STK39 expression, and promotes breast cancer progression.
Abbreviations
BC: breast cancer; lncRNAs: long non-coding RNAs; ceRNA: competing endogenous RNA; CDKN2B-AS1: CDKN2B Antisense RNA 1; miRNAs: MicroRNAs; RISC: RNA-induced silencing complex; HCC: hepatocellular carcinoma; NSCLC: non-small cell lung cancer; STK39: Serine/Threonine Kinase 39; MAPK: mitogen-activated protein kinase; IHC: immunohistochemistry; NUF2: NUF2 Component Of NDC80 Kinetochore Complex; HER2: human epidermal growth factor receptor 2; TN: triple-negative.
Ethics and Consent Statement
This study was performed with the approval of the Research Ethics Committee of General Hospital of Ningxia Medical University. In addition, written informed consent forms were signed by all the patients who participated in this research. All animal experiments were performed with the approval of the animal ethics committee of General Hospital of Ningxia Medical University and conducted according to the guidelines set forth in the National Institutes of Health’s (NIH) ‘Guide for the Care and Use of Laboratory Animals’ (8th edition).
Author contributions
Q.L. and S.Q. conceived and designed the experiments. All authors performed the experiments and analyzed the data. Q.L. contributed reagents and materials. Q.L. and S.Q. wrote the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download ()Disclosure statement
There are no financial interests to be disclosed.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Siegel RL, et al. Cancer statistics, 2021. Ca-a Cancer Journal for Clinicians. 2021;71(1):7–33.
- Loibl S, Poortmans P, Morrow M. Breast cancer (vol 397, pg 1750, 2021). Lancet. 2021;397(10286): 1710-1710.
- Husseini A, Abu-Rmeileh NME, Mikki N. Cardiovascular disease, diabetes mellitus, and cancer in the occupied Palestinian territory. (vol 373, pg 1041, 2009). Lancet. 2009;373(9677): 1764-1764.
- Collignon J, et al. Prognostic value of androgen receptor expression in triple negative breast carcinomas: personal experience and comments on a review about “Triple-negative breast cancer: treatment challenges and solutions” by Collignon et al reply. Breast Cancer-Targets and Therapy. 2016;8: 159-159.
- Carey LA, et al. Race, breast cancer subtypes, and survival in the carolina breast cancer study. Jama-Journal of the American Medical Association. 2006;295(21):2492–2502.
- Gradishar WJ, et al. NCCN guidelines (R) insights breast cancer, version 1.2017 featured updates to the NCCN guidelines. J National Compr Cancer Network. 2017;15(4):433-+.
- Jatoi I, Kemp Z. Surgery for breast cancer prevention. Jama-Journal of the American Medical Association. 2021;325(17): 1804-1804. DOI:10.1001/jama.2021.1647
- Liu SJ, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–460.
- Statello L, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118.
- Ma JY, et al. Metabolism-related long non-coding RNAs (lncRNAs) as potential biomarkers for predicting risk of recurrence in breast cancer patients. Bioengineered. 2021;12(1):3726–3736.
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551.
- Ning ML, et al. LncRNA AFAP-AS1 promotes anaplastic thyroid cancer progression by sponging miR-155-5p through ETS1/ERK pathway. Bioengineered. 2021;12(1):1543–1554.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Volovat SR, et al. MiRNA and LncRNA as potential biomarkers in triple-negative breast cancer: a review. Front Oncol. 2020;10. DOI:10.3389/fonc.2020.526850
- Lv YF, et al. LncRNA SNHG6/miR-125b-5p/BMPR1B axis: a new therapeutic target for triple-negative breast cancer. Front Oncol. 2021;11. DOI:10.3389/fonc.2021.678474
- Xie XN, et al. LncRNA CDKN2B-AS1 stabilized by IGF2BP3 drives the malignancy of renal clear cell carcinoma through epigenetically activating NUF2 transcription. Cell Death Dis. 2021;12(2.
- Huang K, et al. Effects of CDKN2B-AS1 polymorphisms on the susceptibility to coronary heart disease. Mol Genet Genomic Med. 2019;7(11.
- Timmers PRHJ, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. Elife. 2019;8.
- Toraih EA, et al. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Archive for Clinical and Experimental Ophthalmology. 2019;257(9):1897–1913.
- Luo ZB, et al. A competing endogenous RNA network reveals novel lncRNA, miRNA and mRNA biomarkers with diagnostic and prognostic value for early breast cancer. Technol Cancer Res Treat. 2020;19:153303382098329.
- Song CY, et al. CDKN2B-AS1: An indispensable long non-coding RNA in multiple diseases. Curr Pharm Des. 2020;26(41):5335–5346.
- Kattan SW, et al. Association of cyclin-dependent kinase inhibitor 2B antisense RNA 1 gene expression and rs2383207 variant with breast cancer risk and survival. Cell Mol Biol Lett. 2021;26(1.
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36.
- Chuffa LGD, et al. A meta-analysis of microRNA networks regulated by melatonin in cancer: portrait of potential candidates for breast cancer treatment. J Pineal Res. 2020;69(4.
- Fabris L, et al. The potential of MicroRNAs as prostate cancer biomarkers. Eur Urol. 2016;70(2):312–322.
- Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149(5):1177-+.
- Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143(1):35–U543.
- Pu MF, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76(3):441–451.
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A. 2003;100(17):9779–9784.
- Liang YJ, et al. lncRNA-SOX2OT promotes hepatocellular carcinoma invasion and metastasis through miR-122-5p-mediated activation of PKM2. Oncogenesis. 2020;9(5.
- Gao L, et al. Up-regulation of FSTL3, regulated by lncRNA DSCAM-AS1/miR-122-5p axis, promotes proliferation and migration of non-small cell lung cancer cells. Onco Targets Ther. 2020;13:2725–2738.
- Ma WG, et al. The LINC01410/miR-122-5p/NDRG3 axis is involved in the proliferation and migration of osteosarcoma cells. Iubmb Life. 2021;73(4):705–717.
- Zhang W, et al. Resveratrol chemosensitizes adriamycin-resistant breast cancer cells by modulating miR-122-5p. J Cell Biochem. 2019;120(9):16283–16292.
- Zhang CF, et al. STK39 is a novel kinase contributing to the progression of hepatocellular carcinoma by the PLK1/ERK signaling pathway. Theranostics. 2021;11(5):2108–2122.
- Huang T, et al. STK39, overexpressed in osteosarcoma, regulates osteosarcoma cell invasion and proliferation. Oncol Lett. 2017;14(4):4599–4604.
- Shen CH, et al. SPAK-p38 MAPK signal pathway modulates claudin-18 and barrier function of alveolar epithelium after hyperoxic exposure. BMC Pulm Med. 2021;21(1.
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9(8):537–549.
- Wen SY, et al. Cancer-associated fibroblast (CAF)-derived IL32 promotes breast cancer cell invasion and metastasis via integrin beta 3-p38 MAPK signalling. Cancer Lett. 2019;442:320–332.
- Li CX, et al. MicroRNA-299-5p inhibits cell metastasis in breast cancer by directly targeting serine/threonine kinase 39. Oncol Rep. 2020;43(4):1221–1233.
- Wang GL, Xu GH, Wang WG. Long noncoding RNA CDKN2B-AS1 facilitates lung cancer development through regulating miR-378b/NR2C2. Onco Targets Ther. 2020;13:10641–10649.
- Dasgupta P, et al. LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis. 2020;11(8.
- Cai YX, et al. Low-coverage sequencing of urine sediment DNA for detection of copy number aberrations in bladder cancer. Cancer Manag Res. 2021;13:1943–1953.
- Abula A, et al. The stimulative function of long noncoding RNA CDKN2B-AS1 in osteosarcoma by targeting the microRNA-122/CCNG1 axis. J Recept Signal Transduct. 2020;1–9. doi:10.1080/10799893.2020.1850784
- Li Z, et al. Role of high expression levels of STK39 in the growth, migration and invasion of non-small cell type lung cancer cells. Oncotarget. 2016;7(38):61366–61377.
- Shen XY, et al. LncRNA CDKN2B-AS1 promotes cell viability, migration, and invasion of hepatocellular carcinoma via sponging miR-424-5p. Cancer Manag Res. 2020;12:6807–6819.
