ABSTRACT
Polycystic ovary syndrome (PCOS) is an endocrine disease that affects the health of many women. Circular RNAs (circRNAs) are associated with the occurrence and progression of PCOS. This study aimed to explore the function of circ_RANBP9 in PCOS. First, the circ_RANBP9 level was found to be increased in the plasma of patients with PCOS and ovarian granulosa cells (GCs) using Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR). In GCs, loss of circ_RANBP9 decelerated proliferation and accelerated apoptosis of KGN and COV434 cells, as determined by MTT assay, colony formation assay, and flow cytometry. Furthermore, bioinformatics analysis showed that circ_RANBP9 and XIAP can be targeted by the microRNA, miR-136-5p. Luciferase reporter assay and RNA pull-down assay further verified the interaction between miR-136-5p and circ_RANBP9 or XIAP. Importantly, knockdown of circ_RANBP9 suppressed proliferation and promoted apoptosis of KGN and COV434 cells, whereas inhibition of miR-136-5p reversed these effects. Additionally, XIAP abolished the repression of proliferation and acceleration of apoptosis induced by miR-136-5p. The promotion of apoptosis was accompanied by upregulation of caspase-3 and Bax, and downregulation of Bcl-2, as estimated by western blotting. In conclusion, silencing of circ_RANBP9 inhibited GC proliferation and facilitated apoptosis by mediating the miR-136-5p/XIAP pathway. These findings provide a new theoretical basis for screening and treatment of PCOS.
Introduction
Polycystic ovary syndrome (PCOS) is a widespread endocrine disease in women of childbearing age, and is characterized by hirsutism, anovulation, and polycystic ovaries [Citation1]. According to epidemiological studies, up to 10% of women develop polycystic ovaries, worldwide, leading to abnormal reproductive functions, metabolic disorders, and an increased risk of diabetes and cardiovascular disease [Citation2]. Because of the complexity of its pathogenesis, including heredity, environment, living habits, and diet, early screening is a challenging task [Citation3]. PCOS is the cause of anovulatory infertility in 70% of women and about 30% of couples [Citation4,Citation5]. However, till date, no cure is available for this syndrome. Thus, novel targets for early diagnosis and treatment are required to alleviate the progression of PCOS.
Circular RNAs (circRNAs) are covalently closed continuous loops, the majority of which are conserved and stable in different species [Citation6]. Accumulating evidence has revealed that circRNAs mediate gene expression through the regulation of transcription and translation by sponging RNA-binding proteins and/or microRNAs (miRNAs) [Citation7]. circRNAs are involved in the pathogenesis of various diseases, such as cardiovascular diseases, neurologic diseases, and cancers in a tissue-specific manner [Citation8–10]. Based on RNA sequencing (RNA-seq) data, abundant circRNAs were upregulated or downregulated in PCOS-affected women [Citation11,Citation12]. Dysregulation of circRNAs affects the incidence and progression of PCOS, leading to abnormal steroid production, adipocyte dysfunction, and alteration of ovarian cell proliferation and/or apoptosis [Citation13]. circ_RANBP9 (hsa_circ_0001577), a non-coding RNA on chromosome 6, is upregulated in PCOS patients [Citation11]. However, there are no studies on the functions of circ_RANBP9 in PCOS.
Thus, this study aimed to investigate the role of circ_RANBP9 in PCOS and explore its underlying mechanism. We found that a loss of circ_RANBP9 inhibited proliferation and facilitated apoptosis of ovarian granulosa cells (GCs). Furthermore, the miR-136-5p/XIAP axis was speculated be involved in the molecular mechanisms underlying the role of circ_RANBP9 in PCOS. Hence, this study may provide novel evidence for circ_RANBP9 as a therapy target of PCOS.
Materials and methods
Patients
A total of 90 participants, including 45 patients with PCOS and 45 healthy controls, were diagnosed in the Xiamen Key Laboratory of Genetic Testing (Xiamen, China). PCOS was diagnosed according to the Rotterdam standard. All the patients had not been previously diagnosed with PCOS (i.e., before attending this clinic) and had not received any treatment. Women with normal menstrual cycles and ovarian function served as controls. Plasma was isolated from blood (3 ml) of the participants under fasting conditions and stored in liquid nitrogen. All the participants provided written informed consent. This study was approved by the Ethics Committee of the Xiamen Key Laboratory of Genetic Testing (No. 201,801,005).
Cell collection and culture
Human GCs (KGN and COV434) and human ovarian surface epithelial cells (IOSE80) were purchased from Chuanqiu Biotechnology (Shanghai, China). HEK293T cells were obtained from Biovector NTCC (Beijing, China). All the cells were maintained at 37°C under 5% CO2 in DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (The reagents mentioned above were purchased from Hyclone, South Logan, UT, USA).
Cell transfection
KGN and COV434 cells in the exponential growth phase were used for transfection when the cell confluence reached 75%. Empty vector, circ_RANBP9 expressing vector, siRNA (si)-negative control (si-nc: 5ʹ-CACUGAUUUCAAAUGGUGCUAUU-3ʹ), si-circ_RANBP9 1# (5ʹ-TAAACTTGGGTATAACTGT-3ʹ), si-circ_RANBP9 2# (5ʹ-TTGGGTATAACTGTTGTGT-3ʹ), miR-136-5p mimic (5ʹ-ACUCCAUUUGUUUUGAUGAUGGA-3ʹ)/inhibitor (5ʹ-UCCAUCAUCAAAACAAAUGGAGU-3ʹ), mimic/inhibitor nc (5ʹ-UCACAACCUCCUAGAAAGAGUAGA-3ʹ, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ), adenovirus (Ad)-nc, and Ad-XIAP were transfected into KGN and COV434 cells. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for transient transfection of the cells. Next, the transfected cells were incubated for 48 h until the transfection efficiency was determined by Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR).
MTT assay
MTT assay was performed as previous described [Citation14]. KGN and COV434 cells were seeded in 96-well plates. After incubation for 12, 24, 48, and 72 h, the cells were washed with PBS and incubated with 200 μl MTT solution for 2 h. Next, 150 μl of DMSO was added to the wells after discarding the supernatants. After 10 min, absorbance at 490 nm was measured using a microplate reader (BD Biosciences, San Jose, CA, USA).
Colony formation assay
Cell proliferation was also assessed by colony formation assay [Citation15]. Cells, post-transfection, were seeded in 6 cm cell culture dishes (Thermo Fisher Scientific, Waltham, MA, USA) and incubated at 37°C for 14 days. After discarding the medium, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The cells were visualized under a microscope, and the numbers of colonies formed were counted in five random fields (>100 cells/each random field).
Flow cytometry
Flow cytometry was conducted as previous described [Citation16]. Cell apoptosis was assessed using the Annexin V PE/7-AAD apoptosis detection kit (Solarbio, Beijing, China). Briefly, the transfected cells were washed with PBS, and resuspended in 1ⅹ binding buffer to adjust the cell concentration to 2ⅹ106 cells/ml. The cell suspension (100 μl) was incubated with 5 μl Annexin V/PE for 5 min in the dark. Next, 5 μl of 7-AAD solution at a concentration of 20 μg/ml was added, and the cells were washed with 400 μl PBS. Cell apoptosis rates were determined using flow cytometry on CytoFlex flow cytometer (Beckman Coulter, Miami, FL, USA).
Luciferase reporter assay
3ʹUTRs of circ_RANBP9 and XIAP containing miR-136-5p binding sites were inserted into pmirGLO vectors (Promega, Madison, WI, USA). Plasmids with the target-site mutations were constructed (MUT circ_RANBP9 and MUT XIAP). HEK293T cells were seeded in 24-well plates. The cells were co-transfected with wild type (WT) plasmids or the corresponding MUT plasmids and miR-136-5p mimic or mimic NC using Lipofectamine 2000 (Invitrogen). Luciferase activity was analyzed using the Dual Luciferase Reporter Gene Assay Kit (Solarbio, Beijing, China) following the manufacturer’s instructions.
RNA pull-down assay
Cells were transfected with biotinylated miR-136-5p (biotin-miR-136-5p) and matched biotinylated NC (biotin-NC). After 48 h, the cells were washed with PBS and incubated in a lysis buffer (Ambion, Austin, TX, USA) for 10 min. The lysates (50 μl) thus obtained were incubated with M‐280 streptavidin magnetic beads (Sigma–Aldrich, St. Louis, MO, USA). The Input group is lysates (50 μl) not incubated with without magnetic beads. Cells were incubated at 4°C for 3 h and washed with a washing buffer. Total RNA was isolated, and the levels of circ_RANBP9 or XIAP were measured using RT-qPCR.
RT-qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen) to detect the relative expression of circ_RANBP9, miR-136-5p, and XIAP. RNA (1 μg) was used for reverse transcription using PrimeScript™ RT Master Mix (Perfect Real Time) (Takara, Tokyo, Japan) and Mir-X miRNA First-Strand Synthesis Kit (Clontech, Mountain View, CA, USA). qPCR for circ_RANBP9 and XIAP was performed using TB Green® Premix Ex Taq™ (Tli RNaseH Plus) (Takara), while qPCR for miR-136-5p was performed using Mir-X miRNA qRT-PCR TB Green® Kit (Clontech). All the reaction were carried out according to the manufacturers’ instructions. GAPDH was used as an internal control for circ_RANBP9 and XIAP, while U6 was used for normalization of miR-136-5p. A fold change in relative expression was calculated using the 2−ΔΔCt method. The primers used in this study were synthesized by GenScript (Nanjing, China). Specific primers sequences were as follows: circ_RANBP9 (F: 5ʹ-GGGCTTCAAACACCAGGAGA-3ʹ; R: 5ʹ-GTGCTTCCTTTGCCTGATGC-3ʹ), miR-136-5p (F: 5ʹ-ACTCCATTTGTTTTGATGATGGA-3ʹ; R: 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ), XIAP (F: 5ʹ-CCGTGCGGTGCTTTAGTTGT-3ʹ; R: 5ʹ-TTCCTCGGGTATATGGTGTCTGAT-3ʹ), U6 (5ʹ-CTCGCTTCGGCAGCACA-3ʹ; 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ) and GAPDH (F: 5ʹ-GAAAGCCTGCCGGTGACTAA-3ʹ; R: 5ʹ-GCGCCCAATACGACCAAATC-3ʹ).
Western blot
Total protein was extracted using a protein extraction kit (KeyGEN BioTECH, Nanjing, China) to measure the expression of apoptosis-related factors. Protein concentrations were measured using the BCA kit (Sigma–Aldrich). Next, the proteins (30 μg) were separated using 10% SDS-PAGE and then transferred onto PVDF membranes (Merck Millipore, Billerica, MA, USA). After blocking with 5% skimmed milk, the membranes were incubated with primary antibodies at 4°C overnight, followed by incubation with secondary antibodies at room temperature for 1 h. Protein bands were developed using the ECL detection kit (KeyGEN BioTECH) and images were obtained. GAPDH was used as an internal control. The antibodies (Cell Signaling Technology, Danvers, MA, USA) used in this study were shown as follows: anti-XIAP (#2042, 1:1000), Caspase-3 (#9662, 1:1000), Bax (#2774, 1:1000), Bcl-2 (#3498, 1:1000) GAPDH (#2118, 1:1000), and HRP linked anti-Rabbit (#7074, 1:3000).
Data analysis
Each experiment was replaced three times. All data in this study were analyzed using GraphPad Prism (Version 6; La Jolla, CA, USA). Measurement data are presented as mean ± SD. Comparisons among multiple groups were evaluated using one-way ANOVA, and Student’s t-test was used for comparison between two groups. P < 0.05 suggests a significant difference.
Results
Here, we aimed to explore the role of circ_RANBP9 in PCOS. We conducted proliferation and apoptosis assays to evaluate the biological behaviors. We found a loss of circ_RANBP9 inhibited proliferation and facilitated apoptosis of GCs via regulating miR-136-5p/XIAP axis. Therefore, our data was the first to investigate the role of circ_RANBP9 and provide novel evidence for circ_RANBP9 as a therapy target of PCOS.
Upregulation of circ_RANBP9 in patients with PCOS and GCs
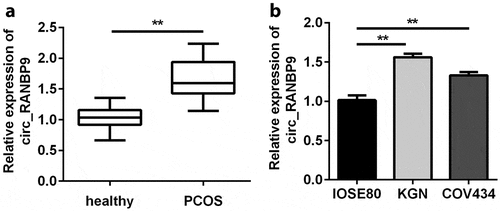
First, plasma samples obtained from 45 PCOS patients and 45 healthy controls were used to detect circ_RANBP9 levels. The expression of circ_RANBP9 in the plasma of PCOS patients was significantly higher than that in controls (P < 0.01; ). These results are consistent with those obtained for GCs. As shown in , circ_RANBP9 was significantly elevated in KGN and COV434 cells than in IOSE80 cells (P < 0.01).
Figure 1. Circ_RANBP9 levels were elevated in patients with polycystic ovary syndrome (PCOS) and in ovarian granulosa cells (GCs). Different levels of circ_RANBP9 were compared in (a) plasma of patients with PCOS (n = 45) and healthy controls (n = 45), as well as (b) normal ovarian epithelium (IOSE80) cells and GCs (KGN, COV434). **P < 0.01
Loss of circ_RANBP9 suppressed proliferation and accelerated apoptosis of GCs
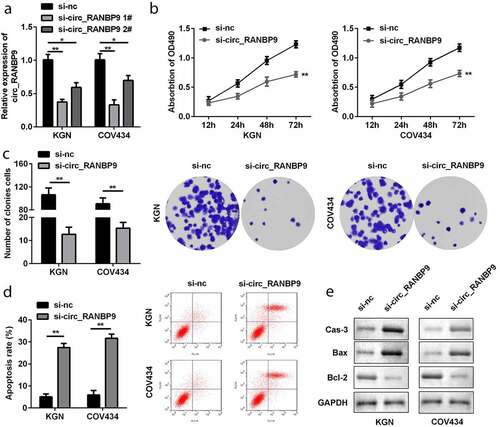
To explore the function of circ_RANBP9, its level was modulated in GCs by transfection with si-nc and si-circ_RANBP9. Downregulation of circ_RANBP9 was observed in the si-circ_RANBP9 1# (P < 0.01) and si-circ_RANBP9 2# groups (P < 0.05), particular in si-circ_RANBP9 1# group (). si-circ_RANBP9 1# was used in the following experiment. Functionally, knockdown of circRANBP9 significantly suppressed the proliferation of GCs (P < 0.01; and c). By contrast, apoptosis of GCs was significantly promoted in the si-circ_RANBP9 than in the si-nc group (P < 0.01; ). Moreover, knockdown of circ_RANBP9 enhanced caspase-3 and Bax levels, but repressed Bcl-2 levels ().
Figure 2. Effects of circ_RANBP9 on proliferation and apoptosis of GCs. (a) circ_RANBP9 levels were tested by RT-qPCR. (b) MTT assay was conducted to determine the proliferation of GCs. (c) Colony formation assay was performed to detect the proliferation of GCs. (d) Flow cytometry was carried out to measure the apoptosis rates of GCs. (e) Protein levels of caspase-3, Bax, and Bcl-2 via western blot analyses were determined post-transfection. **P < 0.01
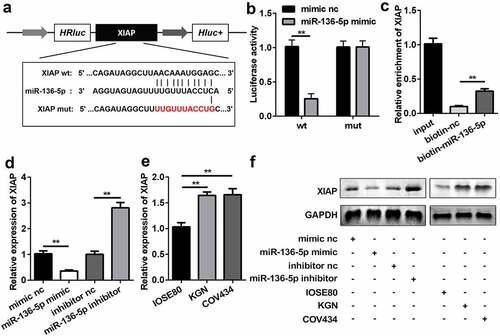
Circ_RANBP9 is a sponge of miR-136-5p
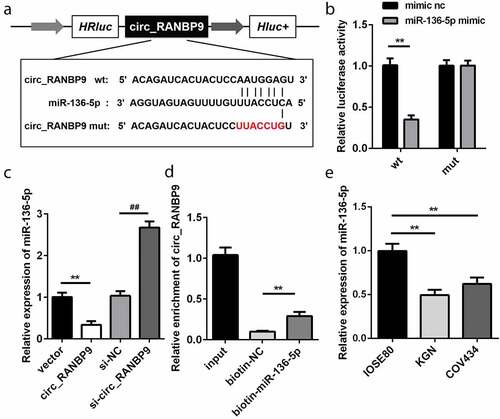
Based on bioinformatics analysis, the WT 3ʹ-UTR of circ_RANBP9 rather than MUT 3ʹ-UTR of circ_RANBP9 was able to partly bind to miR-136-5p (). As compared to the transfection of mimic nc, that of miR-136-5p mimic decreased luciferase activity in HEK293T cells expressing the WT 3ʹ-UTR of circ_RANBP9 (P < 0.01). However, luciferase activity did not influence the cells that expressed MUT 3ʹ-UTR of circ_RANBP9 (). miR-136-5p levels were significantly decreased by circ_RANBP9 (P < 0.01) and were significantly increased by knockdown of circ_RANBP9 (P < 0.01; ). The interaction between circ_RANBP9 and miR-136-5p was further confirmed by an RNA pull-down assay (P < 0.01; ). The level of miR-136-5p was significantly reduced in the GCs of KGN and COV434 cells than in those of IOSE80 cells (P < 0.01; ).
Figure 3. miR-136-5p was negatively regulated by circ_RANBP9. (a) Direct binding sites of miR-136-5p in WT 3ʹ-UTR of circ_RANBP9 and MUT 3ʹ-UTR of circ_RANBP9 were designed. (b) WT circ_RANBP9 or MUT circ_RANBP9 recombinant plasmids and miR-136-5p mimic or mimic nc were co-transfected and luciferase reporter assay was performed. (c) miR-136-5p levels determined using RT-qPCR were detected in GC overexpression or knockdown of circ_RANBP9. (d) Interaction between miR-136-5p and circ_RANBP9 was confirmed by RNA pull-down. (e) Expression of miR-136-5p in normal ovarian epithelium (IOSE80) cells and GCs (KGN, COV434) was detected by RT-qPCR. **P < 0.01. ##P < 0.01
Loss of circ_RANBP9 targeting miR-136-5p suppressed proliferation and accelerated apoptosis of GCs
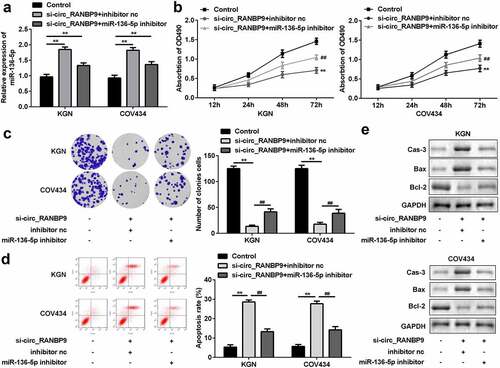
To investigate the underlying molecular mechanisms, the level of miR-136-5p was estimated in GCs transfected with si-circ_RANBP9 and miR-136-5p inhibitor. The expression of miR-136-5p was upregulated by the knockdown of circ_RANBP9 and was rescued by the inhibitor (P < 0.01; ). Silencing of circ_RANBP9 inhibited cell proliferation, while the miR-136-5p inhibitor reversed these effects (P < 0.01; and c). By contrast, cell apoptosis was accelerated by circ_RANBP9 knockdown, which was abolished by miR-136-5p inhibition (P < 0.01; ). Regarding apoptosis-related factors, si-circ_RANBP9 significantly enhanced caspase-3 and Bax levels but decreased Bcl-2 levels, while miR-136-5p inhibitor abolished the effects induced by si-circ_RANBP9 ().
Figure 4. Inhibition of miR-136-5p abolished the effects on proliferation and apoptosis induced by knockdown of circ_RANBP9. (a) Expression of miR-136-5p was tested in transfected cells by RT-qPCR. Cell proliferation was assessed post-transfection by (b) MTT assay and (c) colony formation assay. (d) Flow cytometry was performed to detect apoptosis rates of GCs. (e) Protein levels of caspase-3, Bax, and Bcl-2 at was determined by western blotting. **P < 0.01. ##P < 0.01
XIAP is a target of miR-136-5p
The results of bioinformatics analysis showed that XIAP had possible binding sites for miR-136-5p (). HEK293T cells co-transfected with miR-136-5p mimic and WT 3ʹ-UTR of XIAP significantly reduced luciferase activity (P < 0.01), which was not affected in the MUT group (). Additionally, the RNA pull-down assay further verified the interaction between XIAP and miR-136-5p (P < 0.01; ). Overexpression of miR-136-5p significantly repressed the expression of XIAP, whereas inhibition of miR-136-5p enhanced the expression of XIAP (P < 0.01; and f). The levels of XIAP were higher in KGN and COV434 cells than in IOSE80 cells ( and f).
Figure 5. miR-136-5p directly targeted downstream XIAP. (a) Direct binding sites between miR-136-5p and WT 3ʹ-UTR of XIAP are shown. MUT 3ʹ-UTR of circ_RANBP9 was also designed. (b) Luciferase activity was tested in HEK293T cells transfected by WT XIAP or MUT XIAP recombinant plasmids and miR-136-5p mimic or mimic nc. (c) Interaction between XIAP and miR-136-5p was assessed using RNA pull-down assay. (d) XIAP expression was determined by RT-qPCR in overexpression or inhibition of miR-136-5p cells. (e) Comparison of XIAP using RT-qPCR in IOSE80 cells and GCs (KGN, COV434). (f) The expression of XIAP was measured using western blotting. **P < 0.01. ##P < 0.01
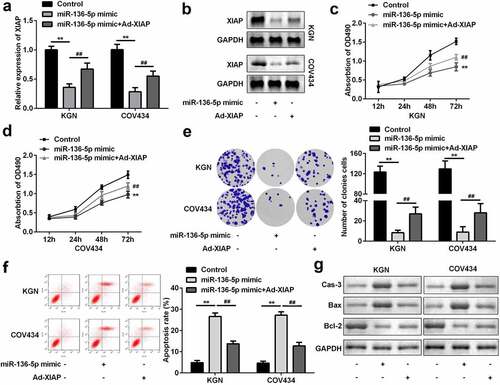
miR-136-5p targets XIAP to influence proliferation and apoptosis of GCs
To further explore the mechanism, miR-136-5p mimic and Ad-XIAP were transfected into GCs. The level of XIAP was significantly downregulated by overexpression of miR-136-5p, which was reversed by overexpression of XIAP (P < 0.01; and b). Functionally, overexpression of miR-136-5p significantly inhibited the proliferation of GCs; however, upregulation of XIAP abolished this inhibition (P < 0.01; –e). Moreover, upregulation of XIAP significantly suppressed the effects of miR-136-5p and suppressed apoptosis of GCs (P < 0.01; ). As illustrated in , the upregulation of caspase-3 and Bax, as well as the downregulation of Bcl-2 were induced by miR-136-5p, while XIAP rescued these effects.
Figure 6. XIAP reversed the effects on proliferation and apoptosis induced by miR-136-5p. (a) XIAP levels were measured by RT-qPCR. (b) XIAP levels were detected by western blotting. Cell proliferation by (c) MTT assay and (d) colony formation assay performed post-transfection is shown. (e) Cell apoptosis was determined by flow cytometry. (f) Protein levels of caspase-3, Bax, and Bcl-2 were determined using western blotting. **P < 0.01. ##P < 0.01
Discussion
Growing evidence indicates that circRNAs are involved in the incidence and development of PCOS [Citation13]. However, the pathogenesis of PCOS is still not fully understood. Excessive proliferation of granulosa cells is one of the main causes of ovarian dysfunction, which contributes to follicular atresia and further induces PCOS [Citation17]. It is well known that circRNAs are collectively involved in the initiation and development of PCOS. For example, circPSMC3 suppresses granulosa tumor cell proliferation and facilitates their apoptosis in PCOS [Citation18]. Additionally, downregulation of circPUM1 enhances proliferation and inhibits apoptosis [Citation19]. Besides, circASPH knockdown represses proliferation and accelerates apoptosis of KGN cells [Citation20]. However, there are no reports on the role of circ_RANBP9 (hsa_circ_0001577) in PCOS. circ_RANBP9, a circRNA derived from the RANBP9 gene, codes for the scaffolding protein RANBP9, which exists in the cytoplasm and nucleus and is associated with cellular processes [Citation21]. A previous study revealed that hsa_circ_0001577 is a candidate circRNA for colorectal cancer [Citation22]. Additionally, circ_RANBP9 levels were found to be increased in granulosa cells of patients with PCOS [Citation11]. In the current study, we found that circ_RANBP9 levels were elevated in the plasma of patients with PCOS and granulosa cells KGN and COV434. Knockdown of circ_RANBP9 decelerates cell proliferation and promotes apoptosis. Therefore, knockdown of circ_RANBP9 may be a promising therapy for the treatment of PCOS. However, the underlying mechanisms of action require further study.
circRNAs act as competing endogenous RNAs (ceRNAs) and regulate biological processes by sponging miRNAs [Citation7]. In this study, miR-136-5p was the target of circ_RANBP9. miR-136-5p is involved in the progression of human cancers, such as thyroid carcinoma [Citation23], hepatocellular carcinoma [Citation24], and renal cell carcinoma [Citation25]. In gynecological diseases, miR-136-5p is regulated by long noncoding RNA (lncRNA) FAM83H-AS1 to suppress cell proliferation, invasion, and migration of breast cancer cells [Citation26]. Similarly, miR-136-5p is also involved in the proliferation, migration, and invasion of cervical cancer cells [Citation27]. In PCOS, miR-136 levels are decreased, which reverse the effects of circ_0118530 on cell survival rate, apoptosis, migration, and oxidative stress in KGN cells [Citation28]. In our study, miR-136-5p was found to be downregulated in PCOS. Moreover, circ_RANBP9 sponged miR-136-5p to modulate the proliferation and apoptosis of GCs. These findings suggest that loss of circ_RANBP9 inhibits proliferation and facilitates apoptosis of GCs by sponging miR-136-5p.
XIAP is a member of the inhibitor of apoptosis protein (IAP) family and has an inhibitory effect on the activation of caspases 3, 7, and 9 [Citation29]. XIAP is also involved in the development of cells and signaling pathways [Citation30]. XIAP is widely expressed in the ovary, especially in thecal cells and granulosa cells [Citation31]. XIAP has been reported to be upregulated in PCOS [Citation30]. However, inhibition of XIAP and restoration of the PPARγ pathway could suppress proliferation and induce apoptosis of KGN cells [Citation32]. In the current study, XIAP was upregulated in GCs; this result was consistent with previous research [Citation33] and XIAP was confirmed to be a target of miR-136-5p. XIAP abolished the inhibition of proliferation and enhanced apoptosis induced by miR-136-5p. Taken together, these results suggest that miR-136-5p targets XIAP to repress GC cell proliferation and accelerate apoptosis.
During the process of apoptosis, Bax and Bcl-2 are two members of the Bcl-2 family possessing pro-apoptotic and anti-apoptotic effects, respectively [Citation34]. Caspase-3 is also activated during apoptosis [Citation35]. Therefore, the facilitation of apoptosis was accompanied by upregulation of caspase-3 and Bax; however, Bcl-2 levels were repressed in our study.
Conclusion
The expression of circ_RANBP9 was upregulated in patients with PCOS and in GCs. miR-136-5p was downregulated, and XIAP was upregulated in GCs. Furthermore, knockdown of circ_RANBP9 suppressed proliferation and facilitated apoptosis of GCs through the miR-136-5p/XIAP axis. This might provide a novel theoretical basis for the screening and treatment of PCOS.
Research highlights
circ_RANBP9 is overexpressed in polycystic ovary syndrome.
Knockdown of circ_RANBP9 inhibits the proliferation and promotes apoptosis of GCs.
circ_RANBP9 sponges miR-136-5p to regulate XIAP.
Knockdown of circ_RANBP9 alleviates PCOS progression by miR-136-5p/XIAP axis.
Disclosure statement
The authors declared that they have no competing interest.
References
- Meier RK. Polycystic Ovary Syndrome. Nurs Clin North Am. 2018;53(3):407–420.
- Goodarzi MO, Dumesic DA, Chazenbalk G, et al. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7(4):219–231.
- Azziz R. Introduction: Determinants of polycystic ovary syndrome. Fertil Steril. 2016;106(1):4–5.
- Neven ACH, Laven J, Teede HJ, et al. A summary on polycystic ovary syndrome: diagnostic criteria, prevalence, clinical manifestations, and management according to the latest international guidelines. Semin Reprod Med. 2018;36(1):5–12.
- Barthelmess EK, Naz RK. Polycystic ovary syndrome: current status and future perspective. Front Biosci (Elite Ed). 2014;6:104–119.
- Qu S, Yang X, Li X, et al. RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Altesha MA, Ni T, Khan A, et al. RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588–5600.
- Shao Y, Chen Y. Roles of circular RNAs in neurologic disease. Front Mol Neurosci. 2016;9:25.
- Lei B, Tian Z, Fan W, et al. RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301.
- Zhang C, Liu J, Lai M, et al. RNA expression profiling of granulosa cells in women of reproductive age with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;300(2):431–440.
- Ma Z, Zhao H, Zhang Y, et al. Novel circular RNA expression in the cumulus cells of patients with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;299(6):1715–1725.
- Mu L, Sun X, Tu M, et al. RNAs in polycystic ovary syndrome: a systematic review and meta-analysis. Reprod Biol Endocrinol. 2021;19(1):10.
- Han B, Ge Y, Cui J, et al. Down-regulation of lncRNA DNAJC3-AS1 inhibits colon cancer via regulating miR-214-3p/LIVIN axis. Bioengineered. 2020;11(1):524–535.
- Yin D, Lu X. Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered. 2021;12(1):225–239.
- Lyu K, Xu Y, Yue H, et al. Long noncoding RNA GAS5 Acts as a tumor suppressor in laryngeal squamous cell carcinoma via miR-21. Cancer Manag Res. 2019;11:8487–8498.
- Yang H, Xie Y, Yang D, et al. Oxidative stress-induced apoptosis in granulosa cells involves JNK, p53 and Puma. Oncotarget. 2017;8(15):25310–25322.
- Liu J, Ding J, Qu B, et al. alleviates the symptoms of PCOS by sponging miR-296-3p and regulating PTEN expression. J Cell Mol Med. 2020;24(18):11001–11011.
- Deng L, Chen Q, Xie J, et al. circPUM1 promotes polycystic ovary syndrome progression by sponging to miR-760. Gene. 2020;754:144903.
- Wu G, Xia J, Yang Z, et al. CircASPH promotes KGN cells proliferation through miR-375/MAP2K6 axis in polycystic ovary syndrome. J Cell Mol Med. 2020. DOI:10.1111/jcmm.16231
- Dhakal S, Sapkota K, Huang F, et al. Cloning, expression and purification of the low-complexity region of RanBP9 protein. Protein Expr Purif. 2020;172:105630.
- Li M, Liu M, Bin Y, et al. Prediction of circRNA-disease associations based on inductive matrix completion. BMC Med Genomics. 2020;13(Suppl 5):42.
- Gao RZ, Que Q, Lin P, et al. Clinical roles of miR-136-5p and its target metadherin in thyroid carcinoma. Am J Transl Res. 2019;11(11):6754–6774.
- Ding H, Ye ZH, Wen DY, et al. Downregulation of miR-136-5p in hepatocellular carcinoma and its clinicopathological significance. Mol Med Rep. 2017;16(4):5393–5405.
- Chen P, Zhao L, Pan X, et al. Tumor suppressor microRNA-136-5p regulates the cellular function of renal cell carcinoma. Oncol Lett. 2018;15(4):5995–6002.
- Han C, Fu Y, Zeng N, et al. FAM83H-AS1 promotes triple-negative breast cancer progression by regulating the miR-136-5p/metadherin axis. Aging (Albany NY). 2020;12(4):3594–3616.
- Zhao J, Yang T, LncRNA LL. FOXP4-AS1 is involved in cervical cancer progression via regulating miR-136-5p/CBX4 axis. Onco Targets Ther. 2020;13:2347–2355.
- Jia C, Wang S, Yin C, et al. Loss of hsa_circ_0118530 inhibits human granulosa-like tumor cell line KGN cell injury by sponging miR-136. Gene. 2020;744:144591.
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7(10):988–994.
- Latour S, Aguilar C. XIAP deficiency syndrome in humans. Semin Cell Dev Biol. 2015;39:115–123.
- Phillipps HR, Hurst PR. XIAP: a potential determinant of ovarian follicular fate. Reproduction. 2012;144(2):165–176.
- Leung DTH, Rainczuk A, Nguyen T, et al. XIAP and PPARgamma in granulosa cell tumors alters metabolic signaling. J Proteome Res. 2019;18(4):1691–1702.
- Mao Z, Li T, Zhao H, et al. Identification of epigenetic interactions between microRNA and DNA methylation associated with polycystic ovarian syndrome. J Hum Genet. 2021;66(2):123–137.
- Bagci EZ, Vodovotz Y, Billiar TR, et al. Bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys J. 2006;90(5):1546–1559.
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326(Pt 1)(Pt 1)):1–16.
