ABSTRACT
Increasing evidence has indicated the implications of circular RNAs (circRNAs) in the development of colorectal cancer (CRC). In this study, we investigated the functional role and mechanism of circ_0089153 in CRC pathogenesis. The expression levels of circ_0089153, microRNA (miR)-198, and SUMO-specific peptidase 1 (SENP1) were gauged by quantitative real-time PCR (qRT-PCR) or western blot. Cell proliferation, sphere formation, tube formation, and apoptosis abilities were detected by 5-Ethynyl-2ʹ-Deoxyuridine (EdU), sphere formation, tube formation, and flow cytometry assays, respectively. The direct relationship between miR-198 and circ_0089153 or SENP1 was verified by dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. The mouse xenograft assays were performed to evaluate the role of circ_0089153 in vivo. Our data showed that circ_0089153 was overexpressed in CRC tissues and cells. Depletion of circ_0089153 repressed cell proliferation, sphere formation ability, and enhanced cell apoptosis, as well as inhibited tube formation in vitro. Moreover, circ_0089153 depletion diminished tumor growth in vivo. Mechanistically, circ_0089153 targeted miR-198, and the effects of circ_0089153 were mediated by miR-198. SENP1 was identified as a direct and functional target of miR-198. Circ_0089153 worked as a competing endogenous RNA (ceRNA) to post-transcriptionally regulate SENP1 expression by miR-198. Our findings identify circ_0089153 as a novel regulator of CRC development through the regulation of the miR-198/SENP1 axis and establish a strong rationale for developing circ_0089153 as a promising therapeutic against CRC.
Introduction
Colorectal cancer (CRC) remains a major cause of cancer death all over the world [Citation1,Citation2]. Although the treatment options, such as surgical local excision and radiochemotherapy, can control many non-metastatic tumors effectively, they have limited utility in curbing the advanced and metastatic CRC [Citation2,Citation3]. Significant players of colorectal tumorigenesis and CRC progression, including microRNAs (miRNAs) and circular RNAs (circRNAs), are under intensive exploration at present [Citation4–7]. Identifying the precise, critical actions of these regulatory RNAs will provide a new opportunity for the development of molecularly targeted therapies.
Covalently closed circRNAs are formed by a non-canonical back-splicing of pre-mRNA transcripts [Citation8]. Intense studies have documented the roles of circRNAs as post-transcriptional regulators by operating as competing endogenous RNAs (ceRNAs) for miRNAs [Citation9]. Moreover, the importance of the ceRNA regulatory networks has been highlighted in human cancers [Citation9–12], including CRC [Citation13,Citation14]. For instance, Liu et al. established that circ_100146 worked as a ceRNA for miR-149 to contribute to the development of CRC [Citation15]. Fang et al. uncovered that circ_100290 exerted a strong oncogenic activity in CRC by inducing frizzled class receptor 4 (FZD4) expression through miR-516b competition [Citation16]. Moreover, Xie et al. ascertained that circ_0000467 sponged miR-382-5p to upregulate engrailed-2 expression and thus contributed to CRC development [Citation14]. Conversely, Li and colleagues showed that circRNA integrin subunit alpha 7 (circITGA7) functioned as an anti-tumor factor in CRC via regulating ITGA7 expression by competing for binding to miR-370-3p [Citation17]. Circ_0089153, produced by head-to-tail splicing of exons of nucleoporin 214 (NUP214) mRNA, is found to be overexpressed in colon cancer tissues by using circRNA microarray analysis [Citation18]. Nonetheless, no reports demonstrated whether aberrant circ_0089153 expression is causally involved in the development of CRC.
MiRNAs have been shown to serve as tumor drivers or suppressors in CRC [Citation4]. Examples of anti-CRC miRNAs include miR-195-5p and miR-4319 [Citation19,Citation20]. Conversely, the oncomirs miR-96, miR-221, and miR-222 are overexpressed in CRC and associated with the metastatic potential [Citation21,Citation22]. MiR-198, one of the top 10 miRNAs down-regulated in CRC, has been established as a potent tumor inhibitor in CRC [Citation23,Citation24]. In a preliminary survey for targeted miRNAs of circ_0089153 using target prediction program circInteractome [Citation25], we observed a putative miR-198 binding region within circ_0089153. Thus, we set to examine whether miR-198 can work as a molecular mediator of circ_0089153 function.
SUMO-specific peptidase 1 (SENP1), a member of de-SUMOylation protease family, is frequently overexpressed in many types of cancer cells and functions as a strong oncogenic promoter in these cancers [Citation26–28]. Moreover, SENP1 has been documented to contribute to colorectal carcinogenesis [Citation29,Citation30]. In a preliminary survey for the miR-198 molecular target, we found a putative miR-198 binding site within the 3ʹuntranslated region (3ʹUTR) of SENP1.
In this study, we investigated the activity of circ_0089153 in CRC progression and examined whether the miR-198/SENP1 axis is involved in the regulation of circ_0089153, with the hope that such ceRNA crosstalk might a provide insight into the causal mechanism of CRC progression.
Materials and methods
Human subject study
In this study, we obtained 50 pairs of colorectal primary tumors and adjacent noncancerous colorectal tissues (~3 cm away from tumors) from CRC patients undergoing colorectal resection in the First Affiliated Hospital of Wenzhou Medical University. None of the patients received any conventional treatment before surgery. The clinical features of these patients were shown in . We confirmed tumor tissues by staining for cell proliferation using the cell-cycle marker Ki67 with anti-Ki67 antibody (ab15580, 1:100 dilution; Abcam, Cambridge, UK), biotinylated goat anti-rabbit IgG secondary antibody (ab64256, 1:300 dilution; Abcam), and a 3,3-diaminobenzidine (DAB) Kit (Vector Laboratories, Peterborough, UK), which was processed by immunohistochemistry using standard method [Citation31]. Human tissue samples were used to evaluate the levels of circ_0089153, miR-198, and SENP1. The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University and informed consent was provided by all subjects.
Table 1. Correlation between circ_0089153 expression and the clinical features of CRC patients (n = 50)
Cell lines
We obtained normal colonic epithelial FHC cells, HCT116, and SW480 CRC cells, and human umbilical vein endothelial cells (HUVECs) from American Type Culture Collection (ATCC, Manassas, VA, USA). Standard cell culture mediums provided by ATCC were used to propagate these cells as per the accompanying instructions. All cells were maintained in a fully humidified incubator of 5% CO2 at 37°C.
Transient transfection of cells
We subcloned human SENP1 (Accession: NM_001267595.2) coding sequence (lacking the 3ʹUTR) and a nontarget control sequence, synthesized by Genewiz (Suzhou, China), into EcoR I and Xba I sites into the pcDNA3.1(+) vector (T7 promoter, Life Technologies, Tokyo, Japan) to create an overexpression plasmid (pc-SENP1) and a control plasmid (pc-NC), respectively. We designed circ_0089153 small interfering RNA (siRNA) (si-circ_0089153, CUAAAUCCCUGGGGAGAGCCU) to knock down circ_0089153 and used a scrambled nontarget sequence (si-NC, CAAUAACGCAGCGCACACCGU) as the negative control. We employed a chemically modified miR-198 mature sequence (5ʹ-GGUCCAGAGGGGAGAUAGGUUC-3ʹ) to mimic miR-198. The sequence of miR-198 inhibitor (5ʹ- GAACCUAUCUCCCCUCUGGACC-3ʹ) was the exact antisense of the mature miR-198 sequence. All oligonucleotides, including miRNA NC control (5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ) and inhibitor NC control (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ), were synthesized by HanBio (Shanghai, China).
SW480 and HCT116 cells were plated at 1 × 105 cells/well in 24-well culture dishes before 24 h transfection with 60 nM of siRNA or/and 30 nM of miRNA mimic or/and 40 nM of miRNA inhibitor or/and 100 ng of plasmid using Lipofectamine 2000 as described by the manufacturers (Thermo Fisher Scientific, Leiden, the Netherlands). Following a 6-h incubation period, the media were replaced with a fresh growth medium, and the cells were harvested 24 h later.
RNA preparation and Ribonuclease (RNase) R assay
We prepared total RNA from cultured cells and tissue samples with the RNeasy Mini Kit as per the manufacturing protocols (Qiagen, Courtaboeuf, France). Total RNA (3 µg) of HCT116 and SW480 cells was incubated with or without 10 U of RNase R for 10 min at 37°C as recommended by the manufacturers (Geneseed, Guangzhou, China). RNA was then subjected to reverse transcription (RT) PCR and quantitative real-time PCR (qRT-PCR) as below.
qRT-PCR
cDNA was synthesized from total RNA using random hexamers with PrimeScript RT Reagent Kit (TaKaRa, Beijing, China; for circ_0089153, NUP214, or SENP1 analysis) or stem-loop RT primers and miScript II RT Kit (Qiagen; for miR-198 analysis). qRT-PCR was performed on a StepOnePlus PCR System (Applied Biosystems, Rotkreuz, Switzerland) using a SYBR Green mix (TaKaRa) and specific primers (Supplement Table S1). Threshold cycle (Ct) was normalized to β-actin or U6 expression, and fold changes in gene expression were evaluated by the 2−ΔΔCt method [Citation32].
5-Ethynyl-2ʹ-Deoxyuridine (EdU) assay
Cell proliferation was evaluated by EdU assay [Citation33] with the Cell-Light EdU Apollo488 In Vitro Kit (Ribobio, Guangzhou, China) and 4,6-diamidino-2-phenylindole (DAPI, GlpBio, Montclair, NJ, USA). Transfected SW480 and HCT116 cells were seeded into 24-well dishes at 1 × 105 cells/well and cultured overnight at 37°C. Afterward, the media were replaced by the fresh growth medium containing EdU (50 mM). The cells were then stained with Apollo488 and the cell nuclei were stained with DAPI. The proportion of EdU-positive cells relative to total nuclei was determined under a fluorescence reverse microscope (Nikon, Tokyo, Japan).
Sphere formation assay
Sphere formation ability of transfected cells was measured by sphere formation assay [Citation34]. Briefly, transfected SW480 and HCT116 cells (4 × 103 cells/well) were seeded into 6-well ultralow attachment plates (Corning, Avon, France) in serum-free medium containing 20 ng/mL of recombinant human epidermal growth factor (EGF, Sigma-Aldrich, Tokyo, Japan), B27 (1:50 dilution; Life Technologies), and 10 ng/mL of bFGF recombinant human protein (R&D Systems, Abingdon, UK). The media were changed every 3 days. After 10 days of growth, the wells with positive sphere formation were scored under a 100X microsphere (Nikon).
Tube formation assay
Tube formation assays were performed under standard protocols [Citation35]. In brief, transfected SW480 and HCT116 cells were maintained in standard growth medium at 37°C. When the cells reached approximately 85% confluence, the media were replaced by the non-serum medium. After 48 h of incubation, the medium supernatant was harvested. HUVECs (1 × 103) were suspended in 200 µL of the medium supernatant and plated in 96-well dishes pre-coated with Matrigel (BD Biosciences, Cowley, UK). Following a 12-h incubation period, images were photographed under a 100X inverted microscope. Using ImageJ software (National Institutes of Health, Bethesda, MD, USA), we quantified the number of the formed tubes in 10 randomly selected fields.
Flow cytometry for cell apoptosis
Apoptosis of transfected cells was assessed by flow cytometry using standard protocols [Citation15]. Transfected SW480 and HCT116 cells were double stained with Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI), processed as recommended by the manufacturers (BD Biosciences). Data were analyzed on a flow cytometer (Epics XL-MCL, Beckman Coulter, Krefeld, Germany) with FlowJo 10.0 software (FlowJo LLC, Ashland, OR, USA). We considered the cells that were both PI and Annexin V positive or PI negative and Annexin V positive as apoptotic cells.
Western blot
For total protein preparation, we used radio immunoprecipitation assay (RIPA) buffer with protease/phosphatase inhibitor cocktail as per the manufacturing guidance (Roche, Basel, Switzerland). Equivalent amounts of protein were resolved by electrophoresis and immunoblotted by a standard protocol [Citation15]. Antibodies against B cell lymphoma-2 (Bcl-2, PA5-27,094, 1:2,000 dilution; Life Technologies), SENP1 (ab108981, 1:3,000 dilution; Abcam), Bcl-2 associated X, apoptosis regulator (Bax, PA5-11,378, 1:2,000 dilution; Life Technologies), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, ab9485, 1:2,500 dilution; Abcam) loading buffer were used. Quantification of protein bands was done by densitometry using ImageJ software.
Bioinformatics
We searched the circInteractome prediction web (https://circinteractome.nia.nih.gov/) to predict the miRNAs that potentially bind to circ_0089153. Analysis of the miRNA-binding sites to human 3ʹUTRs was carried out using target prediction program TargetScan 7.2 (http://www.targetscan.org/vert_72/docs/help.html) with total context++ score < −0.30 and context++ score percentile > 90.
RNA immunoprecipitation (RIP) assay
RIP experiments [Citation36] were performed to evaluate the possibility of the circ_0089153-miR-198 and miR-198-SENP1 3ʹUTR interactions. We prepared the lysates of SW480 and HCT116 cells with RIPA buffer. Also, a complex of an antibody against Argonaute2 (Ago2, PA5-117,725, 1:200 dilution; Life Technologies) or isotype control IgG (ab172730, 1:500 dilution; Abcam) and protein A/G beads (Sigma-Aldrich) was prepared at 4°C for 4 h and added in the lysates, followed by the incubation at 4°C for 4 h. We harvested total RNA from the beads to quantify circ_0089153, miR-198, and SENP1 by qRT-PCR.
Dual-luciferase reporter assay
Our preliminary data showed that the copy number (absolute expression) of circ_0089153 is 484.3 for HCT116 cells and 571.3 for SW480 cells; the copy number (absolute expression) of miR-198 is 157 for HCT116 and 204.7 for SW480 cells (Supplement Figure S1). When we transfected miR-198 mimic into cells, miR-198 expression was significantly elevated (~7.6-fold for HCT116 cells and ~10-fold for SW480 cells) ()). Thus, in cells co-transfected with reporter construct and miRNA mimic, the number of miR-198 was much higher than that of circ_0089153. Although overexpression of miR-198 can engage higher association of circ_0089153, miR-198 can still bind to the predicted binding site in the reporter construct. Based on the fundamental, we carried out the dual-luciferase reporter assays as reported [Citation14]. For circ_0089153 reporters, we inserted circ_0089153 fragment harboring the predicted miR-198 pairing sites (UCUGGAC, WT-circ_0089153) or a mutant containing a mutated seed sequence (AGACCUG, MUT-circ_0089153), synthesized by Genewiz, into Sac I and Pme I sites into the pMIR-REPORTTM reporter vector (CMV promoter, Ambion, Thermo Fisher Scientific). For SENP1-3ʹUTR reporters, we cloned a segment of human SENP1 3ʹUTR (WT-SENP1-3ʹUTR) or a mutant harboring a mutated target region (GACCUG, MUT-SENP1-3ʹUTR) into Sac I and Pme I sites of the pMIR-REPORTTM firefly luciferase vector. We transfected SW480 and HCT116 cells (1 × 105) with the indicated reporter construct (200 ng), pRL-TK Renilla luciferase internal control vector (50 ng, Promega, Beijing, China), and miRNA mimic (30 nM) using Lipofectamine 2000. The cells were lysed after 48 h transfection and the ratio of firefly to Renilla luciferase was gauged using the Dual-luciferase reporter assay as recommended by the manufacturers (Promega). Normalized firefly to Renilla ratios were determined in the presence or absence of miR-198 inhibition.
Generation of stable circ_0089153 knockdown cell line
Recombinant shRNA-circ_0089153 (sh- circ_0089153) expressing lentiviruses and control lentiviral particles (sh-NC) were purchased from Geneseed and used to infect SW480 cells based on the accompanying recommendations. To obtain a stable knockdown cell line, the infected cells were cultured in media containing 2 µg/mL of puromycin (Sigma-Aldrich) for 2 weeks.
Mouse xenograft
We used 12 six-eight-week-old BALB/c female nude mice (Jiangsu ALF Biotechnology Co., LTD., Nanjing, China) for examining the tumorigenicity of sh-circ_0089153-transduced or sh-NC-infected SW480 cells (6 mice per group). For xenograft formation, we gave BALB/c nude mice a 200 µL dose of phosphate buffered saline (PBS) containing 5 × 106 transduced SW480 cells by subcutaneous injection into the left flanks. Tumor volume measurement was periodically performed (calculated volume = shortest diameter2 × longest diameter/2). Four weeks after cell inoculation, the xenografts were excised from these experimental mice. Proliferation of the tumors was assessed with paraffin-embedded tumor tissues by immunohistochemistry under the standard method [Citation31], using the anti-Ki67 antibody (ab15580, 1:100 dilution; Abcam). All animal procedures complied with protocols approved by the Animal Care and Use Committee of the First Affiliated Hospital of Wenzhou Medical University.
Statistical analysis
The means ± standard error (SEM) of at least 3 independent experiments performed in quadruplicate were reported. Unless otherwise noted, we performed statistical analysis using an unpaired Student’s t-test (two-tailed) or a two-way analysis of variance (ANOVA) with Tukey’s post hoc test or Sidak’s multiple comparison test. P < 0.05 was regarded as significant.
Results
Circ_0089153 is highly expressed in CRC
Circ_0089153 exhibits increased expression in colon cancer tissues [Citation18]. However, no reports demonstrated whether aberrant circ_0089153 expression is causally involved in the development of CRC. To investigate the involvement of circ_0089153 in CRC progression, we firstly examined its expression pattern in 50 colorectal primary tumors paired with the adjacent normal tissues from the same patients. Tumor tissues were confirmed by staining for cell proliferation using the cell-cycle marker Ki67. CRC tissues had remarkably more cells stained for Ki67 staining than the normal controls ()). Strikingly, circ_0089153 expression was elevated in CRC tissues compared with the nontumor colorectal tissues ()). Moreover, CRC cell lines (HCT116 and SW480) exhibited higher levels of circ_0089153 compared with the normal colonic FHC cells ()). Using genomic DNA (gDNA) and cDNA from HCT116 and SW480 cells as templates, circ_0089153 was amplified by divergent primers on cDNA rather than gDNA (Supplement Figure S2(a) and S2b), indicating that circ_0089153 is a bona fide circular transcript. To evaluate the RNase R resistance of circ_0089153, we adopted RNase R digestion experiments in HCT116 and SW480 cells. Circ_0089153, rather than the corresponding NUP214 linear mRNA, was resistant to RNase R ( and 1(e)), demonstrating that circ_0089153 has a higher tolerance to RNase R. Additionally, circ_0089153 expression was closely correlated with tumor TNM stage and size, rather than age and gender of patients (). These results together suggest that overexpression of circ_0089153 may play an important role in CRC development.
Figure 1. Circ_0089153 is overexpressed in CRC tissues and cell lines. (a) Representative images depicting the immunohistochemistry assay for Ki67 staining in colorectal primary tumors paired with the adjacent normal tissues from the same patients. (b) qRT-PCR analysis of circ_0089153 in 50 pairs of colorectal primary tumors and the adjacent noncancerous colorectal tissues. P-values based on an unpaired Student’s t-test (two-tailed). (c) The expression of circ_0089153 in normal colonic FHC cells, HCT116 and SW480 CRC cells by qRT-PCR analysis. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. (d and e) RNase R assay showing the RNase R resistance of circ_0089153 in HCT116 and SW480 cells. P-values based on an unpaired Student’s t-test (two-tailed). *P < 0.05
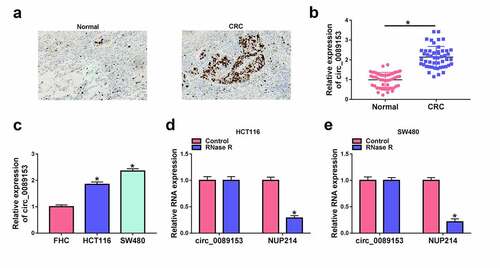
Silencing endogenous circ_0089153 regulates cell proliferation, apoptosis, sphere formation, and tube formation in vitro
Having demonstrated overexpression of circ_0089153 in CRC, we hypothesized that different expression of circ_0089153 may be implicated in CRC development by influencing cell functional properties. In order to direct elucidate the functional role of circ_0089153 in CRC, we knocked down its expression in HCT116 and SW480 cells, which express high levels of circ_0089153. The effectiveness of circ_0089153-siRNA (si-circ_0089153) in inhibiting circ_0089153 expression was validated by qRT-PCR analysis ()). EdU proliferation assays showed that silencing endogenous circ_0089153 markedly suppressed cell proliferation as compared with that in the si-NC controls ()). We then used sphere formation assays to examine the effect of circ_0089153 on cell sphere formation ability. Notably, circ_0089153 knockdown clearly reduced sphere numbers and sizes in both cell lines ()). HUVECs, a type of vascular endothelial cells that can exert important functions in cancer angiogenesis, have been widely used to perform the tube formation assay in vitro [Citation35,Citation37]. Thus, we next incubated HUVECs with the medium supernatant of HCT116 and SW480 cells transfected by si-circ_0089153 or si-NC to observe the effect of circ_0089153 on tube formation. Knocking down circ_0089153 strongly impeded tube formation of HUVECs ()). Moreover, the cells with circ_0089153 depletion exhibited enhanced apoptosis rates compared with the controls ()). Additionally, circ_0089153 silencing resulted in increased levels of pro-apoptotic protein Bax and decreased expression of anti-apoptotic protein Bcl-2 in HCT116 and SW480 cells ( and 2(g)), reinforcing that circ_0089153 depletion enhanced cell apoptosis. Taken together, these data strongly establish the notion that silencing of circ_0089153 inhibits cell proliferation, sphere formation, and promotes cell apoptosis, as well as suppresses tube formation ability.
Figure 2. Circ_0089153 affects cell proliferation, apoptosis, sphere formation, and tube formation in vitro. HCT116 and SW480 cells were transfected with si-circ_0089153 or si-NC. (a) The relative expression of circ_0089153 in transfected cells detected by qRT-PCR analysis. (b) Representative images showing a cell proliferation assay and cell proliferation by EdU assay. (c) Representative images depicting a sphere formation assay and cell sphere formation by sphere formation assay. (d) Representative images presenting the tube formation ability of HUVECs performed by incubating HUVECs with the medium supernatant of transfected cells. (e) Representative images depicting a cell apoptosis assay and flow cytometry for cell apoptosis. (f and g) Western blot showing the expression levels of Bax and Bcl-2 in transfected HCT116 and SW480 cells. *P < 0.05 based on a two-way ANOVA with Sidak’s multiple comparison test
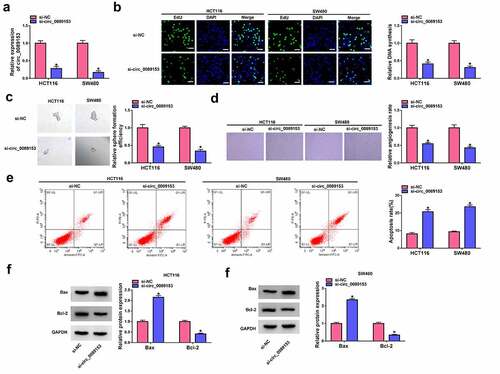
Circ_0089153 targets miR-198 by directly binding to miR-198
CircRNAs have been identified as crucial players in human tumorigenesis by acting as inhibitors of specific miRNAs. To understand the molecular mechanisms by which circ_0089153 regulates the functional properties of CRC cells, we considered its targeted miRNAs that directly bind to circ_0089153. Using the circInteractome prediction web, we found that seven predicted nucleotides (UCUGGAC) of sequence complementarity to the miR-198 target region ()) within circ_0089153. To ascertain this, we cloned circ_0089153 fragment encompassing the putative target sequence into a luciferase reporter vector (WT-circ_0089153) and transfected it into HCT116 and SW480 cells together with miR-198 mimic. The transfection efficiency of miR-198 mimic was confirmed by qRT-PCR analysis ()). Remarkably, the luciferase activity of the reporter construct appeared to be suppressed by miR-198 overexpression () and 3(d)). To determine whether the putative miR-198 complementary nucleotides are required for this effect, we generated a reporter mutation (MUT-circ_0089153) in the target region ()). The reporter mutation carrying a mutated binding sequence (AGACCUG) was refractory to repression by miR-198 overexpression () and 3(d)). We then performed RIP experiments using an antibody against Ago2, which is the core component of the RNA-induced silencing complex (RISC), where miRNAs silence gene expression [Citation38]. In comparison to the anti-IgG control, incubation of cell lysates with anti-Ago2 antibody led to a synchronous elevation in the enrichment levels of circ_0089153 and miR-198 () and 3(f)). Analysis of miR-198 expression in CRC tissues and cell lines showed that miR-198 expression was significantly suppressed in colorectal primary tumors and CRC cells compared with their counterparts () and 3(h)). Intriguingly, we observed a remarkable up-regulation in the levels of endogenous miR-198 in circ_0089153-silenced HCT116 and SW480 cells ()). Collectively, these findings demonstrate that circ_0089153 can bind to miR-198 and regulate miR-198 expression.
Figure 3. Circ_0089153 targets miR-198 to regulate miR-198 expression. (a) Sequence of miR-198, the putative miR-198 binding sequence within circ_0089153, and the mutation in the target sites. (b) qRT-PCR analysis showing the overexpression of miR-198 in HCT116 and SW480 cells transfected by miR-198 mimic or miRNA NC control. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. (c and d) Dual-luciferase reporter assays in HCT116 and SW480 cells co-transfected with circ_0089153 reporter construct (WT-circ_0089153) or the mutant reporter (MUT-circ_0089153) and miR-198 mimic or miRNA NC control. Normalized firefly to Renilla ratios were determined in the presence or absence of miR-198 inhibition. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. (e and f) RIP experiments in HCT116 and SW480 cells performed by incubating cell lysates with antibody against Ago2 or IgG control. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. The expression levels of miR-198 by qRT-PCR analysis in 50 pairs of colorectal primary tumors and the adjacent noncancerous colorectal tissues (g), normal colonic FHC cells, HCT116, and SW480 CRC cells (h), HCT116 and SW480 cells transfected by si-NC or si-circ_0089153 (i). P-values based on an unpaired Student’s t-test (two-tailed) or a two-way ANOVA with Sidak’s multiple comparison test. *P < 0.05
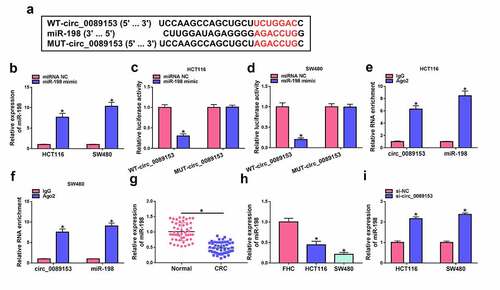
The effects of circ_0089153 on cell proliferation, apoptosis, sphere formation, and tube formation are mediated by miR-198 in vitro
Having demonstrated that circ_0089153 targets miR-198, we next determined whether the effects of circ_0089153 are due to the alteration of miR-198. To elucidate this hypothesis, we reduced miR-198 expression in circ_0089153-silenced cells with miR-198 inhibitor. The effectiveness of miR-198 inhibitor in repressing miR-198 expression was verified by qRT-PCR analysis (). Strikingly, reduced expression of miR-198 reversed si-circ_0089153-driven cell proliferation ()) and sphere formation ()) defects in HCT116 and SW480 cells. Down-regulation of miR-198 also markedly abolished circ_0089153 depletion-mediated suppression of tube formation of HUVECs ()). Furthermore, reduced expression of miR-198 strikingly counteracted circ_0089153 silencing-induced promotion of apoptosis of HCT116 and SW480 cells (). All these results suggest that the effects of circ_0089153 depletion may be due to, at least in part, the up-regulation of miR-198.
Figure 4. Circ_0089153 silencing affects cell proliferation, apoptosis, sphere formation, and tube formation in vitro by increasing miR-198. (a) qRT-PCR analysis showing the down-regulation of miR-198 expression in HCT116 and SW480 cells transfected by inhibitor NC or miR-198 inhibitor. HCT116 and SW480 cells were transfected with si-NC, si-circ_0089153, si-circ_0089153+ inhibitor NC, or si-circ_0089153+ miR-198 inhibitor and checked for cell proliferation by EdU assay (b) and sphere formation ability by sphere formation assay (c). (d) Tube formation assay for tube formation ability of HUVECs incubated with the medium supernatant of HCT116 and SW480 cells transfected by si-NC, si-circ_0089153, si-circ_0089153+ inhibitor NC, or si-circ_0089153+ miR-198 inhibitor. HCT116 and SW480 cells were transfected with si-NC, si-circ_0089153, si-circ_0089153+ inhibitor NC, or si-circ_0089153+ miR-198 inhibitor, followed by the determination of cell apoptosis by flow cytometry (e), Bax and Bcl-2 levels by western blot (f and g). P-values based on a two-way ANOVA with Sidak’s multiple comparison test. *P < 0.05
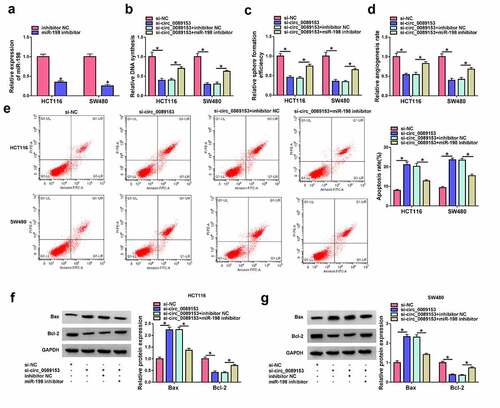
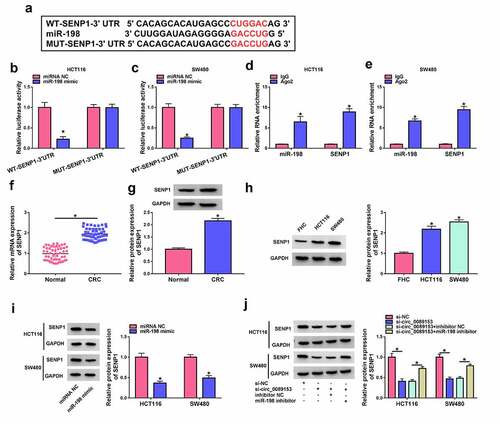
Circ_0089153 regulates SENP1 expression by operating as a ceRNA for miR-198
MiRNAs are crucial for neoplastic transformation and tumor progression because miRNAs can post-transcriptionally regulate entire sets of genes [Citation38]. To identify the downstream effector of miR-198, we searched target prediction program TargetScan 7.2. We found that a putative binding sequence for miR-198 was present in the 3ʹUTR of SENP1 ()). To test whether SENP1 is a target of miR-198, we subcloned the SENP1 3ʹUTR segment harboring the predicted miR-198 target sequence downstream of a luciferase reporter vector (WT-SENP1-3ʹUTR). In HCT116 and SW480 cells, miR-198 overexpression significantly reduced luciferase activity in this construct () and 5(c)). However, the reduction was abrogated by a SENP1 3ʹUTR mutant reporter (MUT-SENP1-3ʹUTR) that contains specific point mutations in the predicted miR-198 binding sequence () and 5(c)), indicating the validity of the binding sites for interaction. RIP experiments showed that incubation of antibody against Ago2 caused a synchronous increase in the enrichment levels of miR-198 and SENP1 () and 5(e)). The data of qRT-PCR and western blot assays revealed that SENP1 was strikingly overexpressed in colorectal primary tumors and CRC cell lines compared with the corresponding controls ((f-h)). We then analyzed SENP1 protein levels in HCT116 and SW480 cells after overexpression of miR-198. By contrast, miR-198 overexpression remarkably repressed the levels of endogenous SENP1 protein in the two CRC cell lines ()). All these data strongly suggest that SENP1 is directly targeted and inhibited by miR-198.
Figure 5. Circ_0089153 operates as a regulator of SENP1 expression by competing for binding to miR-198. (a) Sequence of miR-198, the putative binding sequence for miR-198 in the 3ʹUTR of SENP1 and the mutation in the seed region. (b and c) Dual-luciferase reporter assays in HCT116 and SW480 cells co-transfected with SENP1 3ʹUTR reporter construct (WT-SENP1-3ʹUTR) or the mutant reporter (MUT-SENP1-3ʹUTR) and miR-198 mimic or miRNA NC control. Normalized firefly to Renilla ratios were determined in the presence or absence of miR-198 inhibition. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. (d and e) RIP experiments in HCT116 and SW480 cells performed by incubating cell lysates with antibody against Ago2 or IgG control. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. qRT-PCR analysis of SENP1 mRNA and western blot of SENP1 protein level in colorectal primary tumors and the adjacent noncancerous colorectal tissues (f and g), normal colonic FHC cells, HCT116, and SW480 CRC cells (h), HCT116 and SW480 cells transfected by miR-198 mimic or miRNA NC control (i), HCT116 and SW480 cells transfected with si-NC, si-circ_0089153, si-circ_0089153+ inhibitor NC, or si-circ_0089153+ miR-198 inhibitor (j). P-values based on an unpaired Student’s t-test (two-tailed) or a two-way ANOVA with Sidak’s multiple comparison test. *P < 0.05
Based on the preceding observations that circ_0089153 and SENP1 contain a shared binding sequence (GACCUG) for miR-198, we next examined whether circ_0089153 could modulate SENP1 expression by miR-198 competition. Notably, silencing endogenous circ_0089153 led to a reduction in the levels of SENP1 protein in HCT116 and SW480 cells ()). To explore whether this effect was due to the elevation of miR-198, miR-198 inhibitor was co-transfected and assayed for SENP1 expression. Indeed, reduced expression of miR-198 abolished circ_0089153 depletion-mediated down-regulation of SENP1 protein expression ()). Together, these findings point to the role of circ_0089153 as a ceRNA for miR-198 to modulate SENP1 expression.
MiR-198-mediated suppression of SENP1 impacts cell proliferation, apoptosis, sphere formation, and tube formation in vitro
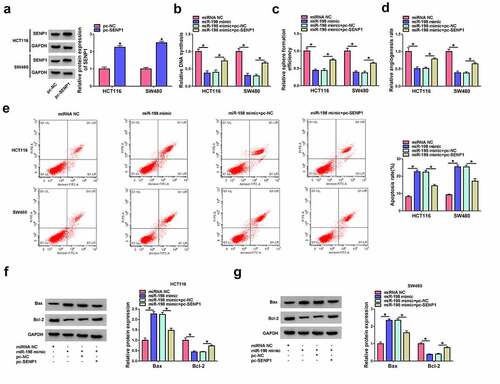
Our above data showed that SENP1 is a direct target of miR-198. In order to elucidate whether inhibition of SENP1 by miR-198 is responsible for the regulation of miR-198 on cell functional properties, we increased SENP1 expression using a SENP1 overexpression plasmid (pc-SENP1) lacking the 3ʹUTR region in HCT116 and SW480 cells after overexpression of miR-198. The effectiveness of pc-SENP1 in increasing SENP1 expression was confirmed by western blot ()). By contrast, overexpression of miR-198 strongly suppressed cell proliferation ()) and sphere formation ()) in HCT116 and SW480 cells. Moreover, overexpression of miR-198 impeded tube formation of HUVECs compared with the controls ()). Additionally, increased expression of miR-198 remarkably enhanced apoptosis of HCT116 and SW480 cells (). Furthermore, restored expression of SENP1 dramatically abolished these effects of miR-198 (). These findings together suggest that the effects of miR-198 may be at least partially due to the inhibition of SENP1.
Figure 6. MiR-198 overexpression impacts cell proliferation, apoptosis, sphere formation, and tube formation in vitro by suppressing SENP1. (a) Western blot showing the up-regulation of SENP1 protein in HCT116 and SW480 cells transfected by SENP1 overexpression plasmid (pc-SENP1) or pc-NC control plasmid. P-values based on a two-way ANOVA with Sidak’s multiple comparison test. HCT116 and SW480 cells were transfected with miRNA NC control, miR-198 mimic, miR-198 mimic+pc-NC, or miR-198 mimic+pc-SENP1, followed by the assessment of cell proliferation by EdU assay (b), and sphere formation ability by sphere formation assay (c). P-values based on a two-way ANOVA with Tukey’s post hoc test. (d) Tube formation assay for tube formation ability of HUVECs incubated with the medium supernatant of HCT116 and SW480 cells transfected with miRNA NC control, miR-198 mimic, miR-198 mimic+pc-NC, or miR-198 mimic+pc-SENP1. P-values based on a two-way ANOVA with Tukey’s post hoc test. HCT116 and SW480 cells were transfected with miRNA NC control, miR-198 mimic, miR-198 mimic+pc-NC, or miR-198 mimic+pc-SENP1 and checked for cell apoptosis by flow cytometry (e), Bax and Bcl-2 levels by western blot (f and g). P-values based on a two-way ANOVA with Tukey’s post hoc test. *P < 0.05
Silencing of circ_0089153 impedes tumor growth in vivo
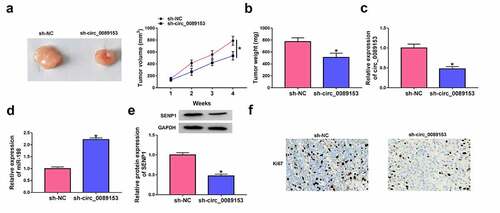
An important question was whether circ_0089153 could affect the tumorigenicity of CRC cells in vivo. To address this possibility, we knocked down circ_0089153 expression in SW480 cells using shRNA-circ_0089153 (sh-circ_0089153) expressing lentiviruses and subcutaneously inoculated sh-circ_0089153-transduced or sh-NC-infected SW480 cells into the nude mice. Sh-circ_0089153-transduced SW480 cells generated remarkably smaller tumors than the same cells infected with sh-NC controls () and 7(b). Moreover, qRT-PCR and western blot analyses showed that sh-circ_0089153-transduced SW480 tumors exhibited lower levels of circ_0089153 and SENP1 protein and higher expression of miR-198 compared with the controls (). Additionally, circ_0089153-silenced xenografts had markedly fewer cells stained for Ki67 staining than the sh-NC controls ()), reinforcing that circ_0089153 depletion diminishes tumor growth. Taken together, these results suggest that the suppression of tumor growth may be due to down-regulation of circ_0089153 and SENP1 and up-regulation of miR-198.
Figure 7. Knocking down circ_0089153 suppresses tumor growth in vivo. Subcutaneous tumors were produced by inoculating sh-circ_0089153-transduced or sh-NC-infected SW480 cells (5 × 106) into the flanks of BALB/c nude mice (6 mice per group) and harvested from the sacrificed mice at week 4 after implantation. (a) Representative images and growth curves of the tumors. (b) Average weight of the tumors at the end point. (c and d) qRT-PCR analysis of circ_0089153 and miR-198 expression in the excised xenografts. (e) Western blot showing SENP1 protein levels in the tumors. (f) Representative images showing Ki67 staining of sections of the tumors performed by immunohistochemistry. *P < 0.05 based on an unpaired Student’s t-test (two-tailed)
Discussion
CircRNAs have been implicated in the pathogenesis of human diseases, including CRC [Citation39]. Emerging experimental evidence also shows the importance of the ceRNA activity of circRNAs in colorectal tumorigenesis [Citation9,Citation13]. Identifying the precise actions of circRNAs in CRC progression has been challenging. Here we showed, for the first time, the role of circ_0089153 as a novel regulator of CRC cell functional properties depending on the regulation of the miR-198/SENP1 axis.
Our results showed that circ_0089153 is overexpressed in colorectal primary tumors and CRC cells. In agreement with our findings, dysregulation of circ_0089153 has been reported in ameloblastoma and breast cancer [Citation40,Citation41]. Moreover, Li et al. uncovered that circ_0089153 functioned as an oncogenic driver in papillary thyroid cancer by sponging miR-145 to induce E-box binding homeobox 2 (ZEB2) expression [Citation42]. Cancer stem cells (CSCs), a population of cells with stem cell-like properties, play essential roles in neoplastic transformation and tumor progression [Citation43]. Colorectal CSCs exert important effects on colorectal tumorigenesis [Citation44]. Using sphere formation assays, we first demonstrated that circ_0089153 depletion can weaken the stemness feature of CRC cells in vitro. Angiogenesis is one of the hallmarks of tumor, which contributes to tumor growth and progression [Citation45]. Anti-angiogenic therapy has been accepted as a novel therapeutic strategy for cancer [Citation46]. Using tube formation assays of HUVECs, we first uncovered that circ_0089153 silencing suppresses tumor angiogenesis in vitro.
Numerous studies have documented the anti-tumor activity of miR-198 in human cancers, such as papillary thyroid cancer, prostate cancer, and lung adenocarcinoma [Citation47–49]. Our results first ascertained that circ_0089153 directly targets miR-198, a potent tumor inhibitor in CRC [Citation23,Citation24]. Moreover, we first demonstrated the regulation of circ_0089153 in colorectal tumorigenesis through miR-198. Similarly, circ_0136666, a highly expressed circRNA in CRC, promotes CRC progression depending on the modulation of miR-198 [Citation50].
SENP1 has been identified as a functionally downstream effector of miR-133a-3p and miR-193-5p in the context of repressing CRC cell growth [Citation30,Citation51]. In this study, we first identified SENP1 as a direct and functional target of miR-198. Similarly, Wang et al. uncovered that miR-198 exerted a strong anti-tumor activity in CRC by targeting fucosyl transferase 8 (FUT8) [Citation23]. The miR-198/SENP1 and miR-198/FUT8 axes may be two paralleled or interactional networks implicated in CRC development, which is expected to be further explored in future work. More importantly, we identified circ_0089153 as a ceRNA to regulate SENP1 expression by miR-198 competition. However, the in vivo direct evidence between tumor growth suppression and the circ_0089153/miR-198/SENP1 ceRNA network is limited, which is expected to be studied in further work. A future challenge will be to determine how the novel ceRNA network regulates CRC progression in vitro and in vivo.
Additionally, our data revealed that tumor TNM stage and size, but not patient gender and age, significantly affected the expression of circ_0089153 (), which prompts that tumor TNM stage and size may influence the expression levels of miR-198 and the downstream signaling by affecting circ_0089153 expression. This novel ceRNA network may also exist in other normal tissues and cells, where the regulatory network is normally regulated and the expression of these molecules are normal. Therefore, the novel ceRNA network does not increase the susceptibility of other normal tissues and cells to cancer.
Conclusion
Collectively, we identify herein circ_0089153 as a novel regulator of CRC progression. We uncover a novel circ_0089153/miR-198/SENP1 ceRNA network in CRC development. Since circ_0089153 depletion suppresses tumor growth, the circ_0089153 inhibitors appear to be promising candidates for the development of new anti-tumor therapies.
Highlights
(1) Circ_0089153 targets miR-198 by a specific binding site.
(2) SENP1 is a direct and functional of miR-198.
(3) Circ_0089153 acts as a ceRNA to regulate colorectal cancer development by the miR-198/SENP1 axis.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Brody H. Colorectal cancer. Nature. 2015;521:S1.
- Modest DP, Pant S, Sartore-Bianchi A. Treatment sequencing in metastatic colorectal cancer. Eur J Cancer. 2019;109:70–83.
- Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197–214.
- Gmerek L, Martyniak K, Horbacka K, et al. MicroRNA regulation in colorectal cancer tissue and serum. PLoS One. 2019;14:e0222013.
- Naeli P, Pourhanifeh MH, Karimzadeh MR, et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. 2020;145:102854.
- Xu H, Wang C, Song H, et al. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18:8.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18.
- Gu Q, Hou W, Shi L, et al. Circular RNA ZNF609 functions as a competing endogenous RNA in regulating E2F transcription factor 6 through competitively binding to microRNA-197-3p to promote the progression of cervical cancer progression. Bioengineered. 2021;12:927–936.
- Chen H, Gu B, Zhao X, et al. Circular RNA hsa_circ_0007364 increases cervical cancer progression through activating methionine adenosyltransferase II alpha (MAT2A) expression by restraining microRNA-101-5p. Bioengineered. 2020;11:1269–1279.
- Shuwen H, Qing Z, Yan Z, et al. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. 2018;645:157–162.
- Xie L, Pan Z. Circular RNA circ_0000467 regulates colorectal cancer development via miR-382-5p/EN2 axis. Bioengineered. 2021;12:886–897.
- Liu K, Mou Y, Shi X, et al. Circular RNA 100146 promotes colorectal cancer progression by the MicroRNA 149/HMGA2 axis. Mol Cell Biol. 2021;41:e00445–20.
- Fang G, Ye BL, Hu BR, et al. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;504:184–189.
- Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246:166–179.
- Chen P, Yao Y, Yang N, et al. Circular RNA circCTNNA1 promotes colorectal cancer progression by sponging miR-149-5p and regulating FOXM1 expression. Cell Death Dis. 2020;11:557.
- Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol. 2017;10:79.
- Huang L, Zhang Y, Li Z, et al. MiR-4319 suppresses colorectal cancer progression by targeting ABTB1. United European Gastroenterol J. 2019;7:517–528.
- Iseki Y, Shibutani M, Maeda K, et al. MicroRNA-96 promotes tumor invasion in colorectal cancer via RECK. Anticancer Res. 2018;38:2031–2035.
- Iida M, Hazama S, Tsunedomi R, et al. Overexpression of miR‑221 and miR‑222 in the cancer stroma is associated with malignant potential in colorectal cancer. Oncol Rep. 2018;40:1621–1631.
- Wang M, Wang J, Kong X, et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep. 2014;4:6145.
- Li LX, Lam IH, Liang FF, et al. MiR-198 affects the proliferation and apoptosis of colorectal cancer through regulation of ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:1487–1493.
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
- Mirecka A, Morawiec Z, Wozniak K. Genetic polymorphism of SUMO-specific cysteine proteases - SENP1 and SENP2 in breast cancer. Pathol Oncol Res. 2016;22:817–823.
- Ma C, Wu B, Huang X, et al. SUMO-specific protease 1 regulates pancreatic cancer cell proliferation and invasion by targeting MMP-9. Tumour Biol. 2014;35:12729–12735.
- Zhang X, Wang H, Wang H, et al. SUMO-specific cysteine protease 1 promotes epithelial mesenchymal transition of prostate cancer cells via regulating SMAD4 deSUMOylation. Int J Mol Sci. 2017;18:808.
- Xu Y, Li J, Zuo Y, et al. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett. 2011;309:78–84.
- Zhou M, Bian Z, Liu B, et al. Long noncoding RNA MCM3AP-AS1 enhances cell proliferation and metastasis in colorectal cancer by regulating miR-193a-5p/SENP1. Cancer Med. 2021;10:2470–2481.
- Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848.
- Jaca A, Govender P, Locketz M, et al. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J Clin Pathol. 2017;70:331–356.
- Buranasin P, Mizutani K, Iwasaki K, et al. High glucose-induced oxidative stress impairs proliferation and migration of human gingival fibroblasts. PLoS One. 2018;13:e0201855.
- Bahmad HF, Cheaito K, Chalhoub RM, et al. Sphere-formation assay: three-dimensional in vitro culturing of prostate cancer stem/progenitor sphere-forming cells. Front Oncol. 2018;8:347.
- Gentile MT, Pastorino O, Bifulco M, et al. HUVEC tube-formation assay to evaluate the impact of natural products on angiogenesis. J Vis Exp. 2019;24:148.
- Liu X, Zhang Z, Ruan J, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158.
- Zhang Q, Lu S, Li T, et al. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J Exp Clin Cancer Res. 2019;38:173.
- Iwakawa HO, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665.
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121.
- Liu J, Qiao X, Liu J, et al. Identification of circ_0089153/miR-608/EGFR p53 axis in ameloblastoma via MAPK signaling pathway. Oral Dis. 2021.
- Hassani B, Mollanoori H, Pouresmaeili F, et al. Constructing MRNA, miRNA, circRNA and lncRNA regulatory network by analysis of microarray data in breast cancer. 2021.
- Li X, Tian Y, Hu Y, et al. CircNUP214 sponges miR-145 to promote the expression of ZEB2 in thyroid cancer cells. Biochem Biophys Res Commun. 2018;507:168–172.
- De Francesco EM, Sotgia F, Lisanti MP. Cancer stem cells (CSCs): metabolic strategies for their identification and eradication. Biochem J. 2018;475:1611–1634.
- Munro MJ, Wickremesekera SK, Peng L, et al. Cancer stem cells in colorectal cancer: a review. J Clin Pathol. 2018;71:110–116.
- Leone P, Buonavoglia A, Fasano R, et al. Insights into the regulation of tumor angiogenesis by Micro-RNAs. J Clin Med. 2019;8:2030.
- Li T, Kang G, Wang T, et al. Tumor angiogenesis and anti-angiogenic gene therapy for cancer. Oncol Lett. 2018;16:687–702.
- Liu W, Zhao J, Jin M, et al. circRAPGEF5 contributes to papillary thyroid proliferation and metastatis by regulation miR-198/FGFR1. Mol Ther Nucleic Acids. 2019;14:609–616.
- Ray J, Hoey C, Huang X, et al. MicroRNA198 suppresses prostate tumorigenesis by targeting MIB1. Oncol Rep. 2019;42:1047–1056.
- Wu S, Zhang G, Li P, et al. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol. 2016;37:5193–5202.
- Wang G, Li Y, Zhu H, et al. Circ-PRKDC facilitates the progression of colorectal cancer through miR-198/DDR1 regulatory axis. Cancer Manag Res. 2020;12:12853–12865.
- Zhou GQ, Han F, Shi ZL, et al. miR-133a-3p targets SUMO-specific protease 1 to inhibit cell proliferation and cell cycle progress in colorectal cancer. Oncol Res. 2018;26:795–800.
