ABSTRACT
Bladder cancer is one of the most severe genitourinary cancers, causing high morbidity worldwide. However, the underlying molecular mechanism is not clear, and it is urgent to find target genes for treatment. G-protein-coupled receptors are currently a target of high interest for drug design. Thus, we aimed to identify a target gene-related to G-protein-coupled receptors for therapy. We used The Cancer Genome Atlas (TCGA) and DepMap databases to obtain the expression and clinical data of RGS19. The results showed that RGS19 was overexpressed in a wide range of tumor, especially bladder cancer. We also explored its effect on various types of cancer. High expression of RGS19 was also shown to be significantly associated with poor prognosis. Cell models were constructed for cell cycle detection. shRGS19 can halt the cell cycle at a polyploid point. RGS19 is a G-protein-coupled receptor signaling pathway-related gene with a significant effect on survival. We chose RGS19 as a therapeutic target gene in bladder cancer. The drug GSK1070916 was found to inhibit the effect of RGS19 via cell rescue experiments in vitro.
Introduction
Bladder cancer (BLCA) is one of the most severe genitourinary cancers. It is estimated that in 2018, approximately 550,000 people were diagnosed with BLCA, and it caused approximately 200,000 deaths [Citation1,Citation2]. Depending on the cellular infiltration of cancer cells in the muscular layer, the 8th edition of the AJCC manual classified BLCA into non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC). MIBC accounts for 25% of newly diagnosed cases, with a high propensity to metastasize and a 5-year survival rate of only 15% [Citation3,Citation4]. The treatment for BLCA usually depends on the type and stage. Radical cystectomy with pelvic lymph node dissections is considered the first-line treatment, and platinum-based combined chemotherapy is the essential protocol during the perioperative period to prolong survival [Citation4]. Before surgery, cisplatin-based neoadjuvant chemotherapy may shrink the tumor and destroy cancer cells that are spreading to the surroundings [Citation5,Citation6]. Currently, the primary challenge is resistance to chemotherapeutic drugs, as studies demonstrate that more than 50% of MIBC therapy is not curative [Citation7]. Adverse effects, including but not limited to mucositis, neutropenia, neutropenic fever, and neutropenic sepsis, are another problem for chemotherapy and cannot be overlooked [Citation8]. Once metastasis occurs, there is no scheme to cure patients permanently. In this situation, a combined method of chemotherapy, immunotherapy and targeted therapy can prolong the life span [Citation9]. In terms of targeted therapy, the FDA approved erdafitinib for patients who do not respond to chemotherapy. However, the short duration and toxicity to the eyes should be considered [Citation10]. Two other targeted drugs, enfortumab vedotin-ejfv and sacituzumab govitecan, may also cause serious side effects in patients, such as anemia, and diarrhea [Citation11]. To enhance therapeutic efficacy and reduce adverse effects, there is an urgent need for novel therapeutic targets.
As a type of seven-transmembrane proteins located on the cell surface, G-protein-coupled receptors (GPCRs) are the largest family of receptors in many organisms, including humans. Their main functions are detecting compounds on the cell surface and initiating several signaling cascades [Citation12]. GPCRs are related to the pathogenesis of many diseases and are therefore widely used as targets for drug design. The vital role of GPCRs in multiple physiological functions and pathological mechanisms establishes them as targets for 34% of drugs approved by the Food and Drug Administration (FDA) [Citation13]. GPCR-targeting drugs can act on multiple cancer-related processes, such as cell division, growth, differentiation, apoptosis, angiogenesis and microenvironment construction, which suggests great untapped potential in identifying new targets related to GPCRs and relevant pathways for new drugs to treat BLCA in the future [Citation14]. However, there is no research focusing on GPCR-related pathways in BLCA therapy.
In this study, we aimed to identify a target gene involved in the GPCR pathway in BLCA and offer a novel treatment option for BLCA patients. We performed whole-genome screening in silico to investigate the genetic alterations associated with the G protein pathway in BLCA. We evaluated the abnormally high expression level and effects on the prognosis of RGS19 in many types of cancer through integrated data analysis. Bioinformatic analysis and in vitro tests demonstrated the effect of RGS19 on the cell cycle and the inhibitory effect of GSK1070916 on BLCA with abnormally high expression of RGS19. We hypothesized that RGS19 is a potential therapeutic target for BLCA and that GSK1070916 is an alternative drug for its clinical treatment.
Materials and methods
Data acquisition and analysis
We downloaded the RNA-seq data of BLCA from The Cancer Genome Atlas (TCGA) database [Citation15]. Based on the TCGA database, the gene expression of candidate genes and clinicopathological characteristics data were obtained from 16 types of cancer whose tumor and nontumor sample sizes were both more than 10. In the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), we selected two profiles and downloaded the original (.CEL file) and platform files.
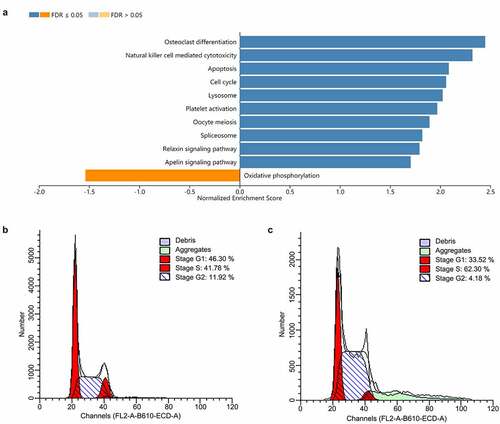
The DEGs between BLCA and normal samples were identified by the limma package [Citation16]. The cutoff criteria were |log2 fold-change|>2 and false discovery rate (FDR) < 0.05. To identify potential biological pathways distinguishing low-risk and high-risk patients in the progression of BLCA, we analyzed the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using Gene Set Enrichment Analysis (GSEA) [Citation17]. FDR < 0.05 and p-value < 0.05 were deemed statistically significant. The GPCR signaling pathway data were downloaded from the Gene Ontology Annotation (GOA) database at the cBioPortal for Cancer Genomics (www.cbioportal.org).
Cell culture
The BLCA cell line T24, SW780, 5637 and cell line SV-HUC-1, were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Cell Counting Kit-8 (MedChemExpress, Shanghai, China) supplemented with 10% fetal bovine serum (Gibco, Gaithersburgh, MD, USA) and 100 U/mL penicillin–streptomycin (Beyotime Biotech, Beijing, China) in an incubator with 5% CO2 at 95% humidity and 37°C.
RNA purification and quantitative RT-PCR
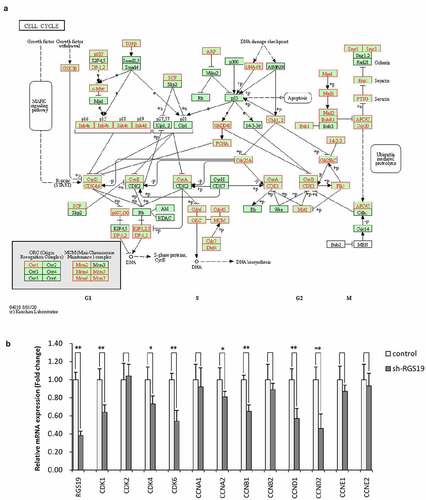
RNA was extracted using TRIzol reagent (Invitrogen). The complementary cDNA was transcribed using TaqMan Reverse Transcription Reagents (Applied Biosystems, Branchburg, New Jersey, USA). Real-time quantitative PCR (qRT-PCR) was performed using a two-step SYBR Green II fluorescent chimaeric real-time PCR system [Citation18]. The sequence of the RGS19 forward primer was 5ʹ-CCGTCTGACTT-GAGTCCCTG-3ʹ, and the sequence of the reverse primer was 5ʹ-CGTGGTACCAGCTCTCAGAC-3ʹ. GAPDH was used as an internal reference. The sequence of the GAPDH forward primer was 5ʹ-CCG-TCTGACTTGAGTCCCTG-3ʹ, and the sequence of the GAPDH reverse primer was 5ʹ-CGTGGTACC-AGCTCTCAGAC-3ʹ. The primers for cell cycle-related genes were indicated in Supplementary Table 1.
Construction of shRNA and transfection
Oligonucleotides 5ʹ-CCGGGAGGCTCATCTAC-GAGGACTACTCGAGTAGTCCTCGTAGATGAGCCTCTTTTTG-3ʹ and 5ʹ-CCGGCCCTTCAA-TGTCCAGTCATGACTCGAGTCATGACTGGACATTGAAGGGTTTTTG-3ʹ were synthesized and annealed to produce a double-stranded shRNA template, which was amplified by PCR. Purified shRNA was digested with BglII and HindIII and inserted into pSUPER-retro-GFP/Neo. Constructs were verified by sequencing.
Western blotting
Cells were lysed in RIPA buffer (Cell Signaling Technologies, Beverly, MA, USA). Proteins were separated on a 10% gel using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Protein was transferred to polyvinylidene difluoride membranes (Millipore, MA, USA). The membranes were immunoblotted with primary antibodies overnight at 4°C followed by the respective secondary antibodies. Primary antibodies against RGS19 were obtained from Amylet Scientific (Wuhan, Hubei, China). The secondary antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). GAPDH was used as a loading control. The relative expression was calculated as the ratio of drug-treated samples vs. control (DMSO) and corrected using the quantified level of RGS19 expression.
Cell cycle detection and flow cytometry analysis
BLCA cells were harvested, centrifuged and then washed twice with cold PBS. A DNA Content Quantitation Assay (Cell Cycle) (Solarbio, Beijing, China) was used to analyze the cell cycle process. The cell cycle distribution of each group of cells was analyzed by flow cytometry. Data were acquired with an Arial III flow cytometer (BD Bioscience) and analyzed with Tree Star FlowJo software.
Colony formation assay
T24 cells (1,000/well) were cultured for 24 h in the presence or absence of germacrene. The medium was changed, and the culture continued until clear cell colonies were formed. Cells were then stained using the Wright–Giemsa Stain Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China). Transfected cells were harvested, washed twice with cold PBS, and fixed in cold 70% ethanol.
Statistical analysis
The clinical data and expression data are expressed as the means ± SD. Differences between groups were estimated using the χ2-test or Student’s t-test. Overall survival and disease-free survival analyses were calculated according to the Kaplan–Meier method with the log-rank test to examine the differences in the incidence of death between the high-RGS19 group and the low-RGS19 group. A Cox regression analysis (proportional hazards model) was performed for the multivariate analyses of prognostic factors. All analyses were conducted using SPSS 16.0 software (IBM, Armonk, NY, USA), and significance was defined as a two-tailed value of P < 0.05.
Results
In this study, we sought to identify a gene related to the GPCR signaling pathway in BLCA as a potential treatment target. Based on the data from the TCGA and DepMap databases, comprehensive analysis proved significantly high expression of RGS19 in BLCA, which was related to tumor progression and poor prognosis. The influence of RGS19 expression on cancer pathogenesis was explored, and pathway analysis and cell models verified its function in the cell cycle. GSK1070916 may serve as a potential drug to treat abnormally high expression of RGS19 in BLCA.
Identification of RGS19 from GPCR-related genes
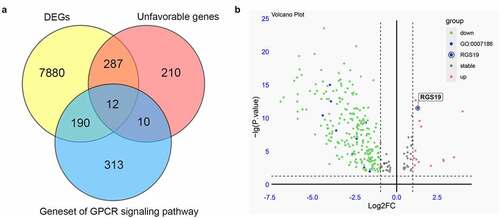
We collected RNA-seq data from the TCGA database to screen for essential GPCR-related genes in BLCA. Whole-genome data and ten-year survival data were downloaded, and differential expression analysis and survival analysis were performed. Finally, 8,369 DEGs were identified from 411 BLCA samples and 19 nontumor samples. In addition, a survival analysis of BLCA-related genes was performed, and 519 unfavorable genes were identified whose overexpression was related to poor prognosis. As mentioned above, due to the importance of the GPCR pathway in drug design, we finally collected a gene set of the GPCR signaling pathway containing 525 genes. We overlapped the above three gene groups and obtained 12 genes, including ADCYAP1R1, AGTR1, AVPR1A, PTGER3, S1PR1, ARHGEF17, ADRA2A, ADRA1D, MC4R, ADCY5, RGS19, and DRD2 (). Only RGS19 was significantly overexpressed among these 12 essential GPCR-related genes (P = 4.2x10−13, ). Thus, we concluded that RGS19 plays a very important role in BLCA and set it as our target.
Figure 1. Screening essential GPCR-related genes for BLCA. (a) Through the integration of differential expression analysis, pathway analysis and survival analysis, we identified 12 essential GPCR-related genes that play an important role in BLCA. (b) The volcano plot was constructed based on differential expression analysis. The expression of RGS19 in BLCA was significantly upregulated (P = 4.2x10−13)
The effects of RGS19 in many types of cancer and survival analysis
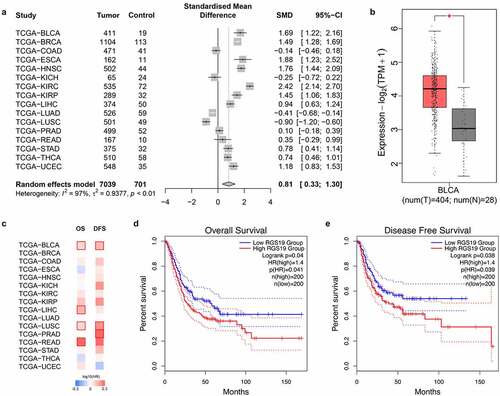
We aimed to explore the effects and prognosis of RGS19 in various types of tumor. Therefore, we collected expression data on 7,740 samples from 16 types of tumor and 701 corresponding nontumor samples. The expression of RGS19 was high in most types of tumor (P < 0.01, ), including BLCA (P < 0.01, ). To further confirm the abnormally high expression of RGS19 in BLCA, we evaluated the expression levels of RGS19 in BLCA cell lines T24, SW780 and 5637 b, compared with those in normal human bladder cells, SV-HUC-1 cell line. The qRT-PCR results presented a consistent trend with bioinformatic analysis (Supplementary Figure 1).
We also performed survival analysis of RGS19 in these 16 types of tumor, and the results for BLCA showed significant association with both OS and DFS (P < 0.05, ). According to the median RGS19 expression level, the patients were divided into high- and low-RGS19 groups to draw a Kaplan–Meier curve. High RGS19 expression was negatively correlated with OS (P = 0.041, ) and DFS (P = 0.039, ). The data of TCGA_BLCA, GSE32894 and GSE31684 were used for meta-analysis and survival analysis to validate the prognostic value of RGS19. The forest plot showed that RGS19 was a steadily unfavorable factor (I2 = 7%, t2 = 0.0010, p = 0.34, Supplementary Figure 2). The nomogram plot was built based on four independent prognostic factors in BLCA. The results showed that RGS19 could predict survival more accurately combined with the TNM stage (Supplementary Figure 3).
Figure 2. Overview of the expression levels of RGS19 between tumor and nontumor samples and summary of clinical analysis based on the TCGA. (a) The forest plot of the expression levels among 16 types of tumor, including breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe carcinoma (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC). (P < 0.01). (b) We analyzed the expression levels of RGS19 in 404 BLCA tumor samples and 28 nontumor samples. The box plot showed that RGS19 was highly expressed in BLCA tissue (P < 0.01). (c) The heatmap of the expression levels. (d) Overall survival analysis showed that the group of high-RGS19 patients had a worse prognosis (P = 0.041). (e) Disease-free survival analysis revealed that the survival time of high-RGS19 patients was shorter than that of low-RGS19 patients (P = 0.039)
The effect of RGS19 on cell proliferation and clone formation
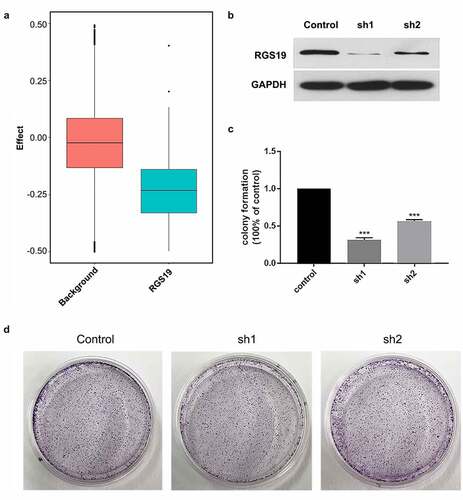
Cell models were established to study the effects of RGS19 in BLCA. Based on the DepMap database, we evaluated the knockout effect of RGS19 in 1,000 cell lines. The background group consisted of 1,000 random knockout genes in 1,000 cell lines. The results showed that the survival ratio of RGS19 knockout was significantly lower (). This result indicated that RGS19 was an essential gene for cells.
Cell models were established to study the effects of RGS19 in BLCA. We conducted in vitro experiments on T24 cell lines to explore the silencing effect of shRGS19 on BLCA. We constructed two shRNA plasmids and detected their silencing effects using Western blotting (), which revealed that shRGS19-1 had a better effect (). The two shRNA plasmids were also introduced separately into the T24 cell line, and then clone formation of the cells was observed. The results showed that shRGS19-1 had a significant inhibitory effect on the number of clones ().
Figure 3. Colony formation experiments with shRGS19. (a) Compared to the background group, the knockout effect of RGS19 in 1,000 cell lines was significantly stronger. (b) The Western blot results of two constructed shRNA plasmids. shRGS19-1 showed a more significant effect on RGS19 silencing. (c and d). After the knockdown, the number of colonies was decreased significantly. The effect of shRGS19-1 was more potent
The molecular mechanism of RGS19 related to the cell cycle
To further explore the biological significance of RGS19 in BLCA, we performed pathway analysis based on the RNA-seq data of BLCA. We found that it was related to the activation of the cell cycle based on GSEA (). We also constructed cell models to explore the effect of RGS19 on the cell cycle. The results of cell cycle analysis showed that the cell cycle of T24 cells was disordered, with more polyploid cells appearing (). Under normal conditions, polyploidy will trigger apoptosis [Citation19,Citation20]. However, apoptosis was inhibited because of kinase inhibition at this checkpoint. Therefore, after RGS19 knockdown, a large number of BLCA cells will be stalled as polyploids. In addition, the results of qPCR showed that RGS19 promoted the expression of 8 important genes that participated in the cell cycle pathway according to the data from the KEGG database ().
Drug screening for BLCA treatment
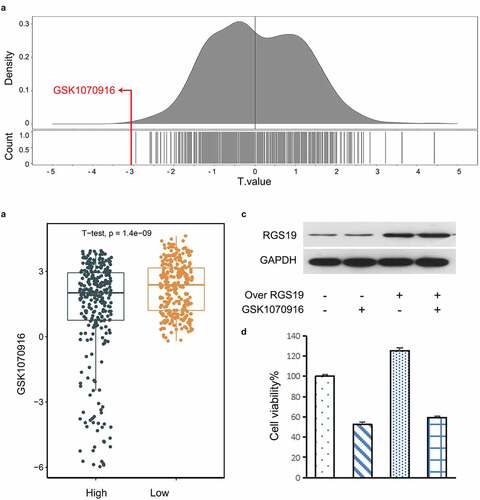
Based on the Genomics of Drug Sensitivity in Cancer (GDSC) database, we explored possible drugs that might affect RGS19. We used the 50% inhibitory concentration (IC50) as the standard for screening. The results showed that BLCA cells with high expression of RGS19 were most sensitive to GSK1070916, with the smallest t value based on the IC50 (). We used the median RGS19 expression level as the standard and divided the cell lines into ‘High’ and ‘Low’ groups. The t value in the ‘High’ group was significantly lower than that in the ‘Low’ group (P = 1.4 x 10−9, ).
To further verify the inhibitory effect of GSK1070916 on RGS19, we conducted a cellular experiment. We induced RGS19 overexpression in T24 cells. The downregulation of RGS19 caused by GSK1070916 was abrogated at both the protein and mRNA levels (). We analyzed the cell proliferation rate and found that it was increased in RGS19°ver+GSK1070916− T24 cells, but this effect was abrogated in RGS19°ver+GSK1070916+ T24 cells (), suggesting that GSK1070916 can inhibit T24 cell proliferation by silencing the overexpression of RGS19.
Figure 6. Drug screening for targeting RGS19. (a) In the GDSC database, we found that GSK1070916 was the most significant with the smallest t value. The negative t value indicated that overexpression of this gene was highly sensitive to this drug. (b) Targeted drug screening for RGS19 based on IC50. T24 cells were most sensitive to GSK1070916, with the smallest t value (P = 1.4 x 10−9). (c and d) The western blot and cell proliferation results of the inhibitory effect of GSK1070916 on RGS19 overexpression
Discussion
The value and novelty of our study was the discovery that RGS19 is a critical gene in BLCA through genome-wide screening, cell experiments and drug screening in cell lines with high RGS19 expression. There were three key points and main aims in our research. We first performed screening and identified a GPCR-related gene, RGS19, as a candidate for therapeutic targeting of BLCA. Then, we systematically showed the importance of RGS19 for the oncogenesis of BLCA at different levels by comprehensive multi-omics analysis. Moreover, we demonstrated the association of RGS19 with the cell cycle and the inhibitory effect of GSK1070916 on cells with high RGS19 expression.
RGS19 (regulator of G protein signaling 19) belongs to the regulators of the G-protein signaling (RGS) family. RGS proteins are negative regulators of GPCR signaling through their ability to act as GTPase-activating proteins (GAPs) for activated Gα subunits [Citation21]. The RGS domain modulates signaling pathways initiated by GPCRs in a way that facilitates the deactivation of heterotrimeric G-proteins [Citation22]. Previous studies have shown that RGS proteins participate in diverse cellular activities, including cell proliferation, differentiation, and apoptosis [Citation23]. In addition to a conserved RGS domain responsible for GAP activities, different RGS proteins have different signaling motifs [Citation24]. RGS19 has a C-terminal PDZ-binding motif used to interact with GIPC, which in turn associates RGS19 with downstream signals [Citation25]. A GIPN, E3 ubiquitin ligase, binds to the N-terminus to induce the degradation of Gia through a ubiquitin proteasome-dependent pathway [Citation26]. In addition, RGS19 has been associated with the carcinogenesis of several other cancers, including ovarian cancer, primary kidney tumor, gastric cancer, prostate cancer and colorectal tumor [Citation22,Citation27,Citation28]. In our study, we first confirmed the overexpression and adverse effects of RGS19 in BLCA. Next, our bioinformatic analysis showed that upregulation occurred in the pancancer dataset and was associated with poor prognosis as an unfavorable factor. Among multiple cancers, high expression of RGS19 showed a particularly strong impact on BLCA, because both OS and DFS were significantly lower in BLCA patients with high RGS19 expression than in those with low RGS19 expression. Pancancer analysis reinforced the importance of RGS19 in BLCA. All these facts suggest that RGS19 is a potential therapeutic target in BLCA. It is reported that the expression of CD44 is lower in BLCA than in none tumor tissue and it is associated with TNM staging [Citation29]. Another study found that TERT promoter mutations may predict BLCA recurrence and become a novel target for BLCA treatment [Citation30]. Previous studies had explored the associations between RGS family members and bladder cancer risk, including RGS1, RGS2, RGS4, RGS5, RGS6, and RGS20. Our study is the first research that identified the risk of RGS19 on BLCA by means of bioinformatic analysis in combination with experiment. In addition, we found a potential therapeutic drug, GSK1070916, for BLCA patients with high expression of RGS19 [Citation31–34].
In this report, pathway analysis showed the involvement of RGS19 in the cell cycle. Furthermore, subsequent cellular experiments supported this idea. After RGS19 knockdown, cell cycle arrest led to polyploid cells. Previous research in non-small cell lung carcinoma (NSCLC) also supported the role of RGS19 in the cell cycle, as the suppression of tumorigenesis after knocking down RGS19 cell line H1299 suggested that RGS19 can facilitate the process of oncogenesis [Citation35]. However, the mechanism by which RGS19 dysregulates the cell cycle in BLCA is not fully understood. Past work has pointed out that RGS19 promotes the cell cycle by upregulating cyclin D1/3 and cyclin-dependent kinase 6, the molecules responsible for the G1/S transition, through the phosphatidylinositol 3ʹ-kinase (PI3K)-Akt pathway, and this mechanism is independent of its role as a GAP [Citation25,Citation36].
In our study, we found that GSK1070916, an inhibitor of Aurora kinase B/C, significantly suppressed BLCA when the expression level of RGS19 was upregulated. An inhibitor of Aurora kinase B was reported to disturb normal chromosome segregation and resulted in the appearance of polyploids [Citation37]. Aurora B is highly expressed in various cancers, including NSCLC [Citation38]. As mentioned above, RGS19 knockdown interfered with the cell cycle in NSCLC. These two facts together suggested a possible mechanism involving the RGS19-AURKB pathway. In the cell cycle assay, after RGS19 knockdown, BLCA cells transformed into polyploids. AURKB can upregulate the expression of CCND1, which encodes cyclin D1 [Citation39]. All of these clues suggest the novel conclusion that RGS19 regulates AURKB through the Akt-PI3K pathway.
GSK1070916 is a reversible and ATP-competitive inhibitor of Aurora B/C, which is encoded by an essential gene controlling multiple events in the cell cycle and mitosis [Citation40]. Its inhibition of proliferation has a broad-spectrum effect on more than 100 cell lines from multiple tumor types [Citation41]. Some previous studies confirmed the anti-tumor effect of GSK1070916 in human tumor xenograft models, including colon, breast, lung and leukemia [Citation41,Citation42]. The Aurora kinase family, which was initially identified in Drosophila, consists of Aurora kinase A, Aurora kinase B and Aurora kinase C [Citation43]. The main functions of Aurora kinase are linked to the regulation of cellular mitosis [Citation44]. Aurora kinase B is a chromosomal passenger protein that forms a chromosomal passenger protein complex (CPC) with three other chromosomal passenger proteins, inner-centromere protein (INCENP), borealin, and survin. In the early stage of mitosis, Aurora B is distributed along the chromosome arm and then aggregates on the centromere of the chromosome and remains until the middle of the division. The complex helps Aurora B instigate spatial displacement [Citation45–47]. The function of CPC is mostly regulated by Aurora kinase B, whereas Aurora C requires further exploration. Therefore, we focused on the regulation of Aurora kinase B [Citation44]. We infer that RGS19 may influence the expression of Aurora kinase B or its partner proteins, including INCENP, borealin, and survin. A number of studies have demonstrated that Aurora kinase is highly expressed in many tumor tissues. However, it is activated only during mitosis, and its expression in nonproliferating cells is low. In the human body, most normal cells do not proliferate at a rapid rate. Therefore, inhibitors targeting Aurora kinase have an advantage than nonspecific drugs cannot match.
Based on the existing literature and experimental results, we propose the following assumptions. The RGS19-AURKB pathway is a pivotal process in the regulation of the BLCA phenotype, and RGS19 is a promising new target for targeted BLCA therapy. We hypothesized that RGS19 regulates AURKB through the PI3K-Akt pathway. However, this hypothesis requires further exploration and verification. RGS19 is a potential biomarker for predicting the effect of AURKB/C inhibitors in BLCA therapy. Finally, several limitations to this study should be considered. We did not investigate the concrete molecular mechanism of how the RGS19-AURKB/C pathway influences the phenotype of BLCA cells in clinical trials or in vitro models.
Conclusion
The upregulated expression of RGS19 showed a significant association with poor prognosis in BLCA. RGS19 was found to participate in cell cycle regulation. GSK1070916, an inhibitor of Aurora kinase B/C, inhibited the proliferation of BLCA with high RGS19 expression. Therefore, RGS19 might serve as a promising therapeutic target for BLCA, and GSK1070916 has potential as a BCLA treatment option.
Highlights
(1) RGS19 is a potential therapeutic target gene involved in the GPCR pathway in BLCA.
(2) RGS19 is associated with the regulation of the cell cycle.
(3) GSK1070916 was proven to be a promising antitumor drug in BLCA treatment.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Sridhar SS. Evolving treatment of advanced urothelial cancer. J Oncol Pract. 2017;13(5):309–315.
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41.
- Becker REN, Meyer AR, Brant A, et al. Clinical Restaging and Tumor Sequencing are Inaccurate Indicators of Response to Neoadjuvant Chemotherapy for Muscle-invasive Bladder Cancer. Eur Urol. 2021;79(3):364–371.
- Chang SS, Bochner BH, Chou R, et al. Treatment of Non-Metastatic Muscle-Invasive Bladder Cancer: AUA/ASCO/ASTRO/SUO Guideline. J Urol. 2017;198(3):552–559.
- Witjes JA, Bruins HM, Cathomas R, et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: summary of the 2020 Guidelines. Eur Urol. 2021;79(1):82–104.
- Westergren DO, Gårdmark T, Lindhagen L, et al. A Nationwide, Population Based Analysis of Patients with Organ Confined, Muscle Invasive Bladder Cancer Not Receiving Curative Intent Therapy in Sweden from 1997 to 2014. J Urol. 2019;202(5):905–912.
- von der Maase H, Hansen SW, Roberts JT, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol. 2000;18(17):3068–3077.
- Lenis AT, Lec PM, Chamie K, et al. Bladder Cancer: a Review. JAMA. 2020;324(19):1980–1991.
- Montazeri K, Bellmunt J. Erdafitinib for the treatment of metastatic bladder cancer. Expert Rev Clin Pharmacol. 2020;13(1):1–6.
- Tagawa ST, Balar AV, Petrylak DP, et al. TROPHY-U-01: a Phase II Open-Label Study of Sacituzumab Govitecan in Patients With Metastatic Urothelial Carcinoma Progressing After Platinum-Based Chemotherapy and Checkpoint Inhibitors. J Clin Oncol. 2021;39(22):2474–2485.
- Ross EM, Gilman AG. Biochemical properties of hormone-sensitive adenylate cyclase. Annu Rev Biochem. 1980;49(1):533–564.
- Hauser AS, Attwood MM, Rask-andersen M, et al. Trends in GPCR drug discovery: new agents, targets and indications Alexander. Nat Rev Drug Discov. 2019;16(12):829–842.
- Sriram K, Insel PA, Protein-Coupled G. Receptors as Targets for Approved Drugs: how Many Targets and How Many Drugs? Mol Pharmacol. 2018;93(4):251–258.
- Hutter C, Zenklusen JC. The Cancer Genome Atlas: creating Lasting Value beyond Its Data. Cell. 2018;173(2):283–285.
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361.
- Gu Z, You Z, Yang Y, et al. Inhibition of MicroRNA miR-101-3p on prostate cancer progression by regulating Cullin 4B (CUL4B) and PI3K/AKT/mTOR signaling pathways. Bioengineered. 2021;12(1):4719–4735.
- Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25(3):486–541.
- Vitale I, Galluzzi L, Castedo M, et al. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12(6):385–392.
- De Vries L, Mousli M, Wurmser A, et al. GAIP, a protein that specifically interacts with the trimeric G protein Gαi3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci U S A. 1995;92(25):11916–11920.
- Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009;78(10):1289–1297.
- De Vries L. RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol. 1999;9(4):138–144.
- Soundararajan M, Willard FS, Kimple AJ, et al. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits. Proc Natl Acad Sci U S A. 2008;105(17):6457–6462.
- Tso PH, Wang Y, Wong SY, et al. RGS19 enhances cell proliferation through its C-terminal PDZ motif. Cell Signal. 2010;22(11):1700–1707.
- Fischer T, De Vries L, Meerloo T, et al. Promotion of Gαi3 subunit down-regulation by GIPN, a putative E3 ubiquitin ligase that interacts with RGS-GAIP. Proc Natl Acad Sci U S A. 2003;100(14):8270–8275.
- Hurst JH, Mendpara N, Hooks SB. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell Mol Biol Lett. 2009;14(1):153–174.
- Kirikoshi H, Katoh M. Expression of human GIPC1 in normal tissues, cancer cell lines, and primary tumors. Int J Mol Med. 2002;9(5):509–513.
- Hu Y, Zhang Y, Gao J, et al. The clinicopathological and prognostic value of CD44 expression in bladder cancer: a study based on meta-analysis and TCGA data. Bioengineered. 2020;11(1):572–581.
- Wan S, Liu X, Hua W, et al. The role of telomerase reverse transcriptase (TERT) promoter mutations in prognosis in bladder cancer. Bioengineered. 2021;12(1):1495–1504.
- Lee EK, Ye Y, Kamat AM, et al. Genetic variations in regulator of G-protein signaling (RGS) confer risk of bladder cancer. Cancer. 2013;119(9):1643–1651.
- Deng M, Wei W, Duan J, et al. ZHX3 promotes the progression of urothelial carcinoma of the bladder via repressing of RGS2 and is a novel substrate of TRIM21. Cancer Sci. 2021;112(5):1758–1771.
- Berman DM, Wang Y, Liu Z, et al. A functional polymorphism in RGS6 modulates the risk of bladder cancer. Cancer Res. 2004;64(18):6820–6826.
- Li G, Wang M, Ren L, et al. Regulator of G protein signaling 20 promotes proliferation and migration in bladder cancer via NF-κB signaling. Biomed Pharmacother. 2019;117:109112.
- Wang Y, Tong Y, Tso PH, et al. Regulator of G protein signaling 19 suppresses Ras-induced neoplastic transformation and tumorigenesis. Cancer Lett. 2013;339(1):33–41.
- Tso PH, Yung LY, Wang Y, et al. RGS19 stimulates cell proliferation by deregulating cell cycle control and enhancing Akt signaling. Cancer Lett. 2011;309(2):199–208.
- Sadaie M, Dillon C, Narita M, et al. Cell-based screen for altered nuclear phenotypes reveals senescence progression in polyploid cells after Aurora kinase B inhibition. Mol Biol Cell. 2015;26(17):2971–2985.
- Goldenson B, Crispino JD. The aurora kinases in cell cycle and leukemia. Oncogene. 2015;34(5):537–545.
- Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl). 2016;94(12):1313–1326.
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2(1):21–32.
- Hardwicke MA, Oleykowski CA, Plant R, et al. GSK1070916, a potent Aurora B/C kinase inhibitor with broad antitumor activity in tissue culture cells and human tumor xenograft models. Mol Cancer Ther. 2009;8(7):1808–1817.
- Adams ND, Adams JL, Burgess JL, et al. Discovery of GSK1070916, a potent and selective inhibitor of Aurora B/C kinase. J Med Chem. 2010;53(10):3973–4001.
- Carmena M, Earnshaw WC, Glover DM. The Dawn of Aurora Kinase Research: from Fly Genetics to the Clinic. Front Cell Dev Biol. 2015;3:73.
- Willems E, Dedobbeleer M, Digregorio M, et al. The functional diversity of Aurora kinases: a comprehensive review. Cell Div. 2018;13(1):7.
- Vader G, Kauw JJ, Medema RH, et al. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006;7(1):85–92.
- Vader G, Medema RH, Lens SM. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173(6):833–837.
- Kelly AE, Sampath SC, Maniar TA, et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12(1):31–43.