ABSTRACT
Long non-coding RNAs (lncRNAs) emerge as vital modulators and tissue-specific biomarkers of multiple cancers, including gastric cancer (GC). Instead, the expression characteristics, biological function and molecular mechanism of lncRNA PCED1B antisense RNA 1 (PCED1B-AS1) in GC await more elaboration. In this study, 48 cases of GC tissues and matched non-cancerous tissues were collected, and PCED1B-AS1, microRNA-215-3p (miR-215-3p) and C-X-C motif chemokine receptor 1 (CXCR1) expression levels were detected by qRT-PCR. Besides, CCK-8, EdU, Transwell and Western blot assays were conducted to assess the impact of PCED1B-AS1 or miR-215-3p on cell growth, migration, invasion and epithelial-mesenchymal transition (EMT). The interaction between genes was verified by bioinformatics analysis, rna immunoprecitipation (RIP) and dual-luciferase reporter gene assays. We demonstrated that, PCED1B-AS1 expression level was raised in GC tissues and cell lines, and increased expression of PCED1B-AS1 was in association with tumor size, TNM stage and lymph node metastasis in GC patients. Additionally, PCED1B-AS1 overexpression promoted GC cells proliferation, migration, invasion and EMT, and miR-215-3p overexpression counteracted the biological effects of PCED1B-AS1. Mechanistically, PCED1B-AS1 specifically inhibited miR-215-3p expressions, thus up-regulating CXCR1 expressions. In conclusion, PCED1B-AS1 accelerates GC progression via adsorbing miR-215-3p and up-regulating CXCR1, indicating that PCED1B-AS1 is a novel therapeutic target for treating GC.
KEYWORDS:
Introduction
Gastric cancer (GC) is a common malignancy, posing a threat to global public health [Citation1]. GC is also one of the main causes of cancer-related deaths in China [Citation2]. Although there is great progression in diagnosis and treatment in recent years, patients who suffered from advanced or metastatic GC still have poor prognosis [Citation3,Citation4]. Thus, searching for novel biomarkers and understanding the potential mechanism of GC are urgent for the improving the diagnosis and prognosis of GC.
Non-coding RNAs (ncRNAs) are widely expressed in human cells, including microRNAs (miRNAs) with approximately 22nt and long non-coding RNAs (lncRNAs) with exceeding 200nt, and they all feature prominently in post-transcriptional regulation [Citation5]. LncRNA is at play in various biological activities of tumor cells, including proliferation, apoptosis, metastasis, and drug resistance [Citation6–8]. To date, diverse abnormally expressed lncRNAs have been unveiled in GC [Citation9]. LncRNA, as a competitive endogenous RNA (ceRNA), can modulate downstream target gene expressions via decoying miRNAs. For example, lncRNA LINC00511 represses miR-625-5p and up-regulates transducers and activators of transcription 3 (STAT3) to facilitate the progression of GC [Citation10]. Another study reports that, lncRNA AL139002.1 promotes GC progression by sponging miR-490-3p to regulate hepatitis A virus cellular receptor 1 [Citation11]. LncRNA PCED1B antisense RNA 1 (PCED1B-AS1) mediates macrophage apoptosis and autophagy via targeting the miR-155 axis in active tuberculosis [Citation12]. Additionally, PCED1B-AS1 is a tumor-promoter, and for example, PCED1B-AS1 expression level is markedly raised in glioblastoma tissues and cell lines, which is in association with larger tumor size and higher grade [Citation13]. Instead, the role of PCED1B-AS1 in GC is still unclear.
Chemokines, as a family of small proteins (8–11kDa), which are divided into four classes (C, CC, CXC, and CX3C), can promote directional chemotaxis of leukocytes and play important roles in inflammation and cancers [Citation14]. CXC chemokines bind to the G-protein-coupled receptors (GPCR) e.g. C-X-C motif chemokine receptor 1 (CXCR1) and C-X-C motif chemokine receptor 2 (CXCR2), to exert their biological effects [Citation15,Citation16]. CXCR1 is reported to be up-regulated in GC tissues, and its high expression indicates poor prognosis of GC patients; besides, CXCR1 expedites the growth, migration, and invasion of GC cells [Citation17,Citation18]. Instead, its regulatory mechanism in GC is not yet clear.
In this work, we supposed that PCED1B-AS1 could probably participate in GC progression. Our study was performed to investigate the expression characteristics, biological function and underlying mechanism of PCED1B-AS1 in GC. We demonstrated that, PCED1B-AS1 expression level was markedly up-regulated in GC tissues, which was closely associate with larger tumor size, higher TNM stage and lymph node metastasis. Also, PCED1B-AS1 strengthened cell viability, migration and invasion, and epithelial-mesenchymal transition (EMT) through the microRNA-215-3p (miR-215-3p)/CXCR1 axis.
Materials and methods
Clinical samples collection
48 pairs of GC tissues and normal tissues adjacent to cancer were available from the First Affiliated Hospital of Kunming Medical University. Besides, this work was endorsed by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University with informed consent available from patients. All volunteers did not receive any treatment before operation, and the clinicopathological characteristics of the sufferers are detailed in . Immediately after the removal, the tissue samples were positioned in liquid nitrogen and subsequently stored at −80°C.
Table 1. The correlations of PCED1B-AS1 with clinicopathological features of patients with gastric cancer
Cell culture and transfection
The Cell Bank of Chinese Academy of Sciences (Shanghai, China) was the supplier of four human GC cell lines (HGC-27, KATO III, NCI-N87, and AGS), normal gastric epithelial cell line (GES-1) and the human embryo kidney epithelial cell line HEK293T. The cells were cultured in Roswell Park Memorial Institute-1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) in 5% CO2 at 37°C.
Small interfering RNA (siRNA) targeting PCED1B-AS1 (si-PCED1B-AS1-1, si-PCED1B-AS1-2 and si-PCED1B-AS1-3) and negative control (si-NC), miR-215-3p-mimics/NC-mimics, miR-215-3p inhibitor/NC inhibitor, PCED1B-AS1 overexpression vector (pc-PCED1B-AS1), CXCR1 overexpression vector (pc-CXCR1)/pcDNA empty vector (Vector) were available from GenePharma (Shanghai, China). The GC cells were subsequently transfected with the plasmids or the oligonucleotides by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) as protocols, and 48 h later, quantitative real-time polymerase chain reaction (qRT-PCR) was executed to estimate the transfection efficiency.
qRT-PCR
The total RNA from GC tissues and cell lines was extracted by a RNAiso Plus kit (TaKaRa, Dalian, China) and reverse-transcribed into complementary DNA (cDNA) with a PrimeScript First Strand cDNA Synthesis Kit (Takara, Dalian, China). Next, qRT-PCR amplifications were accomplished by a SYBR® Premix ExTaq kit (TaKaRa, Dalian, China) on ABI PRISM 7300 system (Applied Biosystems, Foster City, CA, USA). Besdes, Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was adopted as the internal reference for PCED1B-AS1 and CXCR1, and U6 snRNA as that for miR-215-3p. The relative expressions of the genes involved in this study were calculated by 2−ΔΔCt. Specifically, the primer sequences are listed in . To identify the sub-cellular localization of PCED1B-AS1 in GC cells, nuclear/cytoplasmic separation of GC cells was performed with a PARIS™ Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instruction. After the cytoplasmic RNA and nuclear RNA were isolated, PCED1B-AS1 level was measured by qRT-PCR, with GAPDH as the cytoplasmic endogenous control and the U6 as the nuclear endogenous control.
Table 2. The primer sequence used in this study
Cell counting kit-8 (CCK-8) assay
GC cells were positioned in 96-well plates at 2 × 103 cells per well, and cultured. At the 24th, 48th, and 72nd h, 10 μL of CCK-8 reagent (Dojindo Molecular Technologies, Japan) was added into each well respectively, and the cells were cultured at 37°C for 2 h. Then the OD450 nm value was estimated by a microplate reader (Bio-Rad, Hercules, CA, USA).
EdU proliferation assay
Cell proliferation was examined with an EdU assay kit (RiboBio, Guangzhou, China). Cells were cultured with 50 μM EdU for 2 h, then were fixed in 4% paraformaldehyde and subsequently stained with Apollo staining solution away from light for 2 h at room temperature. Subsequently, the cells were stained with DAPI staining solution (Beyotime, Shanghai, China) away from light for 30 min at room temperature. Next, the cells were washed with phosphate buffer saline (PBS), the EdU positive cells were photographed under a fluorescence microscopy (Olympus, Tokyo, Japan) in five randomly selected fields, and counted.
Cell migration and invasion assay
48 h after transfection, cells in serum-free medium were transferred into the upper compartment of Transwell chambers (pore size: 8 μm, Millipore, Bedford, MA, USA). 600 μL of medium containing 10% FBS was dripped into the lower compartment. After the cells were cultured for 24 h, the cells which passed through membrane were fixed with 4% formaldehyde and stained with 0.5% crystal violet. Ultimately, cells were observed and the numbers were subsequently counted under the microscope (Olympus, Tokyo, Japan). Notably, Matrigel (Sigma-Aldrich, Louis, MO, USA) was used to cover the membrane to mimic the extracellular matrix in invasion analysis, but it was not used in migration assay.
Dual-luciferase reporter assay
The sequences of PCED1B-AS1 or CXCR1 3ʹUTR containing the wild type (WT) or mutant type (MUT) miR-215-3p binding sites were cloned into the pmirGLO luciferase reporter vector (Promega, Madison, WI, USA) to generate the reporter plasmids (PCED1B-AS1-WT, CXCR1-WT, PCED1B-AS1-MUT and CXCR1-MUT). HEK293T cells were inoculated in 96-well plate and cultured for 24 h, then co-transfected with the reporter plasmids and miR-215-3p mimics or miR-NC by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). 48 h later, the luciferase activity was measured by the Dual-Luciferase Reporter System (Promega, Madison, WI, USA).
Western blot assay
Total proteins in tissues and cells were respectively extracted by RIPA lysis buffer (Beyotime, Shanghai, China), and protein concentration was detected by a bicinchoninic acid (BCA) kit (Beyotime, Shanghai, China). The same amount of protein in each group was subsequently separated by SDS-PAGE and then transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Subsequently, the membranes was blocked by 5% skimmed milk for 2 h at room temperature, and then incubated at 4°C overnight with primary antibodies anti-E-cadherin (1:1000; ab40772; Abcam), anti-N-cadherin (1:1000; ab18203; Abcam), anti-Vimentin (1:1000; ab92547; Abcam), CXCR1 (1:1000; ab124344; Abcam) and anti-GAPDH (1:1000; ab9485; Abcam), and incubated with Goat Anti-Rabbit IgG H&L (HRP) (1:2000; ab205718; Abcam) secondary antibody for 1 h. Besides, the protein bands were developed by an electrochemiluminescence kit (Pierce Biotechnology, Rockford, IL, USA), with GAPDH as the internal control.
RNA immunoprecipitation (RIP) assay
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was utilized for RIP assay. Cell was lysed with RIP lysis buffer, and cell lysate was incubated with human anti-Argonaute 2 (Ago 2) antibody (Millipore, Billerica, MA, USA) or negative control mouse IgG (Millipore, Billerica, MA, USA) coupled with magnetic bead overnight at 4°C. Then the proteins in the immunoprecipitate were degraded with proteinase K and the immunoprecipitated RNA was then isolated and purified, followed by qRT-PCR detection.
Statistical analysis
SPSS 22.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA) were employed for statistical analysis. Each experiment was repeated for three times or more, with the data expressed as the mean ± standard deviation (SD). Additionally, student’s t-test and one-way analysis of variance (ANOVA) were performed for comparisons. Chi-squared test was executed to analyze the association between PCED1B-AS1 expression and the clinicopathological characteristics. Besides, Spearman’s correlation analysis was used to analyze the correlation between gene expressions in GC tissues. Statistically, P < 0.05 is significant.
Results
In the present study, with in vitro experiments, we investigate the expression pattern and biological functions of PCED1B in GC. We also investigated the regulatory effects of PCED1B-AS1 on the expression of miR-215-3p and CXCR1.
1. PCED1B-AS1 is highly expressed in GC tissues and cell lines
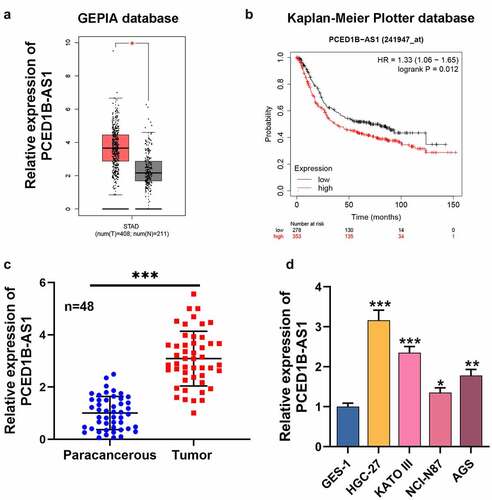
First of all, we searched GEPIA database (http://gepia.cancer-pku.cn/) and observed that PCED1B-AS1 expression in GC patients was demonstrably higher than that in normal tissues (). Besides, Kaplan-Meier Plotter database (http://kmplot.com/analysis/index.php?p=service) highlighted that the overall survival of patients with low PCED1B-AS1 expression was longer than that of patients with high PCED1B-AS1 expression (). qRT-PCR revealed that PCED1B-AS1 expression in GC tissues was significantly higher compared with that in normal tissues adjacent to cancer (). In addition, as against normal gastric epithelium cell line GES-1, PCED1B-AS1 expression in GC cell lines (HGC-27, KATO III, NCI-N87, and AGS) was dramatically increased (). Additionally, it was also revealed that higher PCED1B-AS1 expression in GC tissues was associated with larger tumor size, higher TNM stage and lymph node metastasis ().
Figure 1. PCED1B-AS1 is highly expressed in GC tissues and cell lines
2. Effects of PCED1B-AS1 on GC cell viability, migration, invasion and EMT
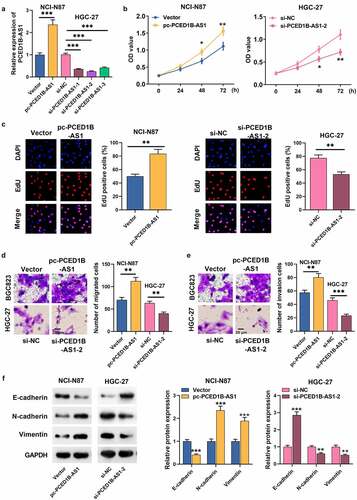
As mentioned above, among the GC cells, PCED1B-AS1 expression was the highest in HGC-27 and the lowest in NCI-N87, so we transfected siRNA (si-PCED1B-AS1-1, si-PCED1B-AS1-2, and si-PCED1B-AS1-3) and PCED1B-AS1 overexpression vector (pc-PCED1B-AS1) targeting PCED1B-AS1 into HGC-27 and NCI-N87, respectively (). CCK-8 and EdU assays revealed that the proliferation rate of GC cells with PCED1B-AS1 overexpression was higher, while PCED1B-AS1 knockdown worked oppositely (). Transwell assay revealed that the metastatic potential of GC cells with PCED1B-AS1 overexpression was evidently stronger than that of the control group, and PCED1B-AS1 knockdown suppressed the migration and invasion of GC cells (). Additionally, Western blot assay depicted that PCED1B-AS1 overexpression promoted N-cadherin and Vimentin expressions and inhibited the protein expression of E-cadherin, while PCED1B-AS1 knockdown had the opposite effect ().
Figure 2. The effect of PCED1B-AS1 on GC cells’ proliferation, migration, invasion and EMT
3. PCED1B-AS1 works as a ceRNA via adsorbing miR-215-3p in GC
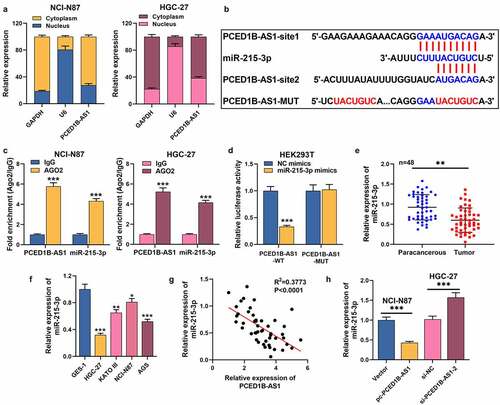
To expound the hidden regulatory mechanism of PCED1B-AS1 in GC, we first performed nucleocytoplasmic separation experiments and observed that PCED1B-AS1 was mainly distributed in the cytoplasm of GC cells (). DIANA database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted) highlighted that miR-215-3p may be a promising downstream target of PCED1B-AS1 (). In the 48 patients, low miR-215-3p expression was associated with larger tumor size, higher TNM stage and poor differentiation status of the tumor tissues (). RIP assay revealed that PCED1B-AS1 and miR-215-3p were specifically enriched in anti-Ago2 group, but not in anti-IgG group (). Dual-luciferase reporter gene assay depicted that the luciferase activity of HEK293T cells co-transfected with PCED1B-AS1-WT and miR-215-3p mimics was greatly lower than that of NC mimics group, but there was no significant change in PCED1B-AS1-MUT group (). Besides, miR-215-3p was markedly reduced in GC tissues and cell lines (). Furthermore, miR-215-3p expression was negatively correlated with PCED1B-AS1 expression in GC tissues (). Additionally, miR-215-3p was inhibited in GC cells with PCED1B-AS1 overexpression, but overexpressed in cell lines with PCED1B-AS1 knockdown (). These highlights that PCED1B-AS1, as a miRNA sponge, has a negative regulatory effect on miR-215-3p expression.
Table 3. The correlations of miR-215-3p with clinicopathological features of patients with gastric cancer
Figure 3. PCED1B-AS1 serves as a ceRNA by sponging miR-215-3p in GC
4. CXCR1 emerges as a target of miR-215-3p
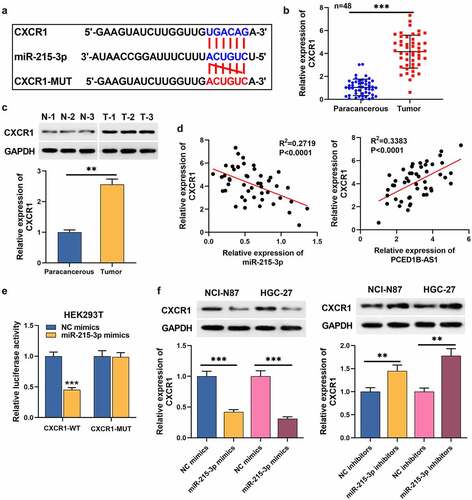
TargetScan database (http://www.targetscan.org/vert_71/) predicted that miR-215-3p had a putative binding site in CXCR1 3ʹUTR (). qRT-PCR and Western blot assays uncovered that CXCR1 expression in GC tissues was higher than that in adjacent normal tissues (). In the 48 patients, higher CXCR1 expression was associated with the distant metastasis after surgery (). Notably, CXCR1 expression was negatively interrelated with miR-215-3p expression in GC tissues, but positively interrelated with PCED1B-AS1 expression (). Dual-luciferase reporter gene assay showed that miR-215-3p mimics remarkably repressed the luciferase activity of CXCR1-WT (). Western blot assay uncovered that miR-215-3p could inhibit CXCR1 expression in GC cells (). These data suggests that CXCR1 is the target of miR-215-3p.
Table 4. The correlations of CXCR1 with clinicopathological features of patients with gastric cancer
Figure 4. CXCR1 is a target mRNA of miR-215-3p
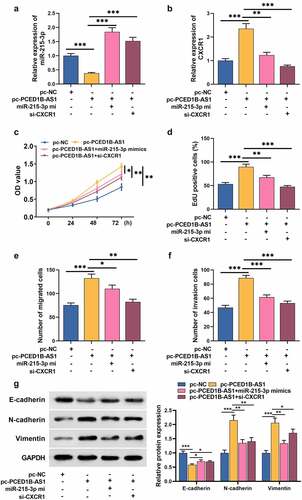
5. PCED1B-AS1 facilitates the growth, migration, invasion, and EMT process of GC cells via regulating the miR-491-5p/CXCR1 axis
To confirm whether PCED1B-AS1 participates in the GC progression by adsorbing miR-215-3p and up-regulating CXCR1 expression, rescue assays were performed. MiR-215-3p mimic reversed the inhibitory effect of pc-PCED1B-AS1 on miR-215-3p, and PCED1B-AS1 overexpression could promote the expression of CXCR1, but the si-CXCR1 markedly inhibited CXCR1 expression (). Compared with GC cells transfected with pc-PCED1B-AS1, the growth of GC cells transfected with pc-PCED1B-AS1+ miR-215-3p mimics or pc-PCED1B-AS1+ si-CXCR1 was greatly inhibited (). Transwell and Western bolt assays revealed that pc-PCED1B-AS1 enhanced cell migration, invasion, and EMT process, while miR-215-3p mimics or si-CXCR1 transfection counterracted the effect of pc-PCED1B-AS1 on GC cells ().
Figure 5. PCED1B-AS1 promotes the proliferation, migration, invasion and EMT of GC cells by regulating the miR-491-5p/CXCR1 axis
Discussion
Reportedly, lncRNA features prominently in regulating epigenetics, transcriptional and post-transcriptional processes to modulate gene expression. LncRNA, abnormally expressed in cancer cells, participates in regulating the progression of multiple cancers, including GC. For example, lncRNA cancer susceptibility 11 can promote GC development by facilitating cell cycle progression [Citation19]. LncRNA SOX2 overlapping transcript is in high expression in GC, and it contributes to the growth and metastasis of GC cells [Citation20]. There are few studies on the role of PCED1B-AS1 in tumors, specifically, PCED1B-AS1, as a tumor-promoter, partakes in modulating glioma cell proliferation and apoptosis [Citation13]. Additionally, it also facilitates the progression of clear cell renal cell carcinoma and pancreatic cancer [Citation21,Citation22]. Here, we proved that PCED1B-AS1 expression was greatly raised in GC tissues, which was closely associated with larger tumor size, higher TNM stage and lymph node metastasis. Besides, functional experiments further confirmed that PCED1B-AS1 overexpression strengthened malignant biological behaviors of GC cells, while PCED1B-AS1 inhibition showed the opposite effects, implying that PCED1B-AS1 was a tumor-promoter in GC.
As reported, lncRNAs, as a ceRNA, can exert its function by sponging endogenous miRNA to inhibit mRNA translation, and this mechanism is involved in tumorigenesis [Citation23,Citation24]. For example, small nucleolar RNA host gene 5/microRNA-32/Kruppel like factor 4 (KLF4) axis regulates GC cell migration, which contributes to improving the diagnosis and treatment of GC [Citation25]. LncRNA TMPO antisense RNA 1 can sponge miR-140-5p and indirectly regulate SRY-box transcription factor 4 expression to accelerate the progression of GC [Citation26]. Reportedly, PCED1B-AS1 is elevated in gliomas and head and neck squamous cell carcinoma [Citation13,Citation27], and PCED1B-AS1 modulates the multiplication and apoptosis of gliomas via serving as ceRNA and regulating the miR-194-5p/PC-esterase domain containing 1B (PCED1B) axis [Citation28]. In this work, we confirmed that miR-215-3p was a direct target of PCED1B-AS1. According to previous reports, miR-215-3p is pivotal in cancer biology as a tumor suppressor [Citation29,Citation30]. This study revealed that miR-215-3p level was dramatically declined in GC tissues and cell lines, and miR-215-3p and PCED1B-AS1 expression were negatively correlated in GC tissues; the transfection of miR-215-3p mimics partially impeded the promoting effect of PCED1B-AS1 overexpression on the malignant biological behaviors of GC cells. These data suggest that PCED1B-AS1 can probably function as a ceRNA to regulate miR-215-3p and its downstream genes.
In the present work, CXCR1 was identified as a target gene of miR-215-3p in GC cells. CXCR1 mediates the progression of multiple cancers, including GC [Citation15,Citation18]. For example, IL-8 promotes the migration of liver cancer cells through CXCR1 and CXCR2, and targeting CXCR1/2 may be a strategy for treating liver cancer [Citation31]. Another study reports that miR-761 can impede osteosarcoma cell growth and invasion partly via targeting CXCR1 [Citation32]. In colorectal cancer cells, miR-215-3p improves the 5-Fu sensibility of cancer cells via regulating CXCR1 expression [Citation16]. In GC, CXCR1 regulates the activation of AKT and ERK1/2 signal pathways to modulate the malignant biological behaviors of cancer cells [Citation18]. Besides, E-cadherin is a cell adhesion molecule involved in cell-cell and cell-matrix interactions and EMT process of cancer cells, and it reduces the aggressiveness of cancer cells [Citation33]; reportedly, CXCR1 knockdown increases E-cadherin expression in GC cells [Citation18]. Herein, our rescue experiments revealed that inhibition of CXCR1 demonstrably restrained N-cadherin and Vimentin expressions, increased the expression of E-cadherin, and reversed the promoting effect of PCED1B-AS1 overexpression on the malignant biological behaviors of GC cells. In short, these data suggest that PCED1B-AS1 can facilitate the growth, migration, invasion and EMT of GC cells via modulating the miR-215-3p/CXCR1 axis.
Conclusion
This study confirms the tumor-promoting effect of PCED1B-AS1 in GC. PCED1B-AS1 is highly expressed in GC cancer tissues and cell lines, and PCED1B-AS1 expression is associated with the clinicopathological characteristics of GC patients. Furthermore, PCED1B-AS1, as ceRNA, up-regulates CXCR1 through competitively binding to miR-215-3p to promote the malignancy of GC cells, suggesting that PCED1B-AS1/miR-215-3p/CXCR1 axis is a novel mechanism involved in GC progression.
Highlights
LncRNA PCED1B-AS1 is up-regulated in gastric cancer tissues, and its high expression indicates poor prognosis.
PCED1B promotes the malignancy of GC cell lines.
PCED1B can increase the expression of CXCR1 via repressing miR-215-3p in GC cells.
Author’s contribution
Conceived and designed the experiments: LWL, RJY;Performed the experiments: RJY, XN, and ZRZ;Statistical analysis: ZRZ, HFC, and ZHB;Wrote the paper: RJY, LWL, XN.All authors read and approved the final manuscript.
Ethics statement
Our study was approved by the Ethics Review Board of the First Affiliated Hospital of Kunming Medical University.
Disclosure statement
The authors declare that they have no competing interests.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin.2019;69 (1):7-34.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Quadri HS, Hong YK, Al-Refaie WB. Approach to the surgical management of resectable gastric cancer. Clin Adv Hematol Oncol. 2016;14(4):249–257.
- Sun W, Yan L. Gastric cancer: current and evolving treatment landscape, Chinese. J Cancer. 2016;35(1):83.
- Wei JW, Huang K, Yang C, et al. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9.
- Tao F, Tian X, Lu M, et al. A novel lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and migration by sponging miR-34a and miR-34c. J Genet Genome. 2018;45(3):137–145.
- Peng X, Wei F, Hu X. Long noncoding RNA DLGAP1-AS1 promotes cell proliferation in hepatocellular carcinoma via sequestering miR-486-5p. J Cell Biochem. 2020;121(2):1953–1962.
- Zhang Z, Li M, Zhang Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther. 2020;13:1343–1354.
- Sun W, Yang Y, Xu C, et al. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142(11):2231–2237.
- Cui N, Sun N,Q, Liu Q,H, et al. Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered. 2021;12(1):2915–2927.
- Chen Y, Zhang R. Long non-coding RNA AL139002.1 promotes gastric cancer development by sponging microRNA-490-3p to regulate Hepatitis A Virus Cellular Receptor 1 expression. Bioengineered. 2021;12(1):1927–1938.
- Li M, Cui J, Niu W, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun. 2019;509(3):803–809.
- Yao Z, Zhang Q, Guo F, et al. Long noncoding RNA PCED1B-AS1 promotes the Warburg effect and tumorigenesis by upregulating HIF-1α in glioblastoma. Cell Transplant. 2020;29:963689720906777.
- Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics. 2017;7(6):1543–1588.
- Liu Q, Li A, Tian Y, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61–71.
- Li XW, Qiu SJ, Zhang X. Overexpression of miR-215-3p sensitizes colorectal cancer to 5-fluorouracil induced apoptosis through regulating CXCR1. Eur Rev Med Pharmacol Sci. 2018;22(21):7240–7250.
- Wang JP, Hu WM, Wang KS, et al. Expression of C-X-C chemokine receptor types 1/2 in patients with gastric carcinoma: clinicopathological correlations and significance. Oncol Lett. 2013;5(2):574–582.
- Wang J, Hu W, Wu X, et al. CXCR1 promotes malignant behavior of gastric cancer cells in vitro and in vivo in AKT and ERK1/2 phosphorylation. Int J Oncol. 2016;48(5):2184–2196.
- Zhang L, Kang W, Lu X, et al. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17(15):1886–1900.
- Qu F, Cao P. Long noncoding RNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2. Exp Cell Res. 2018;369(2):187–196.
- Qin J, Zhu T, Wu W, et al. Long non-coding RNA PCED1B-AS1 promotes the progression of clear cell renal cell carcinoma through miR-484/ZEB1 axis. Onco Targets Ther. 2021;14:393–402.
- Zhang Y, Ma H, Chen C. Long non-coding RNA PCED1B-AS1 promotes pancreatic ductal adenocarcinoma progression by regulating the miR-411-3p/HIF-1alpha axis. Oncol Rep. 2021;46(1):134.
- Guo LL, Song CH, Wang P, et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21(41):11680–11687.
- Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
- Zhao L, Han T, Li Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31(3):893–903.
- Sun Y, Han C. Long non-coding RNA TMPO-AS1 promotes cell migration and invasion by sponging miR-140-5p and inducing SOX4-mediated EMT in gastric cancer. Cancer Manag Res. 2020;12:1261–1268.
- Zhong Z, Hong M, Chen X, et al. Transcriptome analysis reveals the link between lncRNA-mRNA co-expression network and tumor immune microenvironment and overall survival in head and neck squamous cell carcinoma. BMC Med Genomics. 2020;13(1):57.
- Yang J, Yu D, Liu X, et al. LncRNA PCED1B-AS1 activates the proliferation and restricts the apoptosis of glioma through cooperating with miR-194-5p/PCED1B axis. J Cell Biochem. 2020;121(2):1823–1833.
- Liu CQ, Chen Y, Xie BF, et al. MicroRNA-215-3p suppresses the growth and metastasis of cervical cancer cell via targeting SOX9. Eur Rev Med Pharmacol Sci. 2019;23(13):5628–5639.
- Zhang F, Wang XS, Tang B, et al. Long non-coding RNA FTX promotes gastric cancer progression by targeting miR-215. Eur Rev Med Pharmacol Sci. 2020;24(6):3037–3048.
- Bi H, Zhang Y, Wang S, et al. Interleukin-8 promotes cell migration via CXCR1 and CXCR2 in liver cancer. Oncol Lett. 2019;18(4):4176–4184.
- Wang S, Zhang J, Chen G, et al. miR-761 inhibits human osteosarcoma progression by targeting CXCR1. Int J Clin Exp Pathol. 2018;11(11):5327–5334.
- Liu X, Chu KM. E-cadherin and gastric cancer: cause, consequence, and applications. Biomed Res Int. 2014;2014:637308.
