ABSTRACT
Compound mutations in the additional sex combs-like 3 (ASXL3) gene greatly impact the expression of long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs) in mouse myocardial tissues. Little is known about ASXL3 mutation effects on lncRNAs and mRNAs expression in the cerebrum and cerebellum. This study aims to clarify this point using quantitative real-time polymerase chain reaction and Western blotting. Transcriptome analysis based on RNA-seq followed by bioinformatics analysis were used to compare lncRNA and mRNA expression profiles. Cell proliferation, cell cycle progression, and apoptosis were evaluated after silencing of ASXL3 expression using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4- sulfophenyl)-2 H-tetrazolium method and flow cytometry. Results showed that ASXL3 gene expression was decreased in the cerebrum and cerebellum of mice with ASXL3 P723R*P1817A mutations. We identified 319 lncRNAs and 252 mRNAs differentially expressed in the cerebrum of ASXL3 P723R*P1817A mutant mice. In the cerebellum of ASXL3 P723R*P1817A mutant mice, 5330 lncRNAs and 2204 mRNAs were differentially expressed. Differentially expressed lncRNAs and mRNAs were widely distributed across the mouse genome and were associated with various biological processes and pathways. ASXL3 silencing by siRNA transfection affected the proliferation, cell cycle progression, and apoptosis of neural cells. Therefore, the ASXL3 P723R*P1817A mutations greatly modify the lncRNA and mRNA expression profiles in the mouse cerebrum and cerebellum.
1. Introduction
The cerebrum, which contains the cerebral cortex and subcortical structures, and the cerebellum are two major components of the mammalian central nervous system, responsible for the control of all the body voluntary actions and most cognitive functions, including language, attention, and fear or pleasure responses [Citation1,Citation2]. The development and the physiological functions of neurons and glial cells in the cerebrum and cerebellum are precisely controlled by complex networks of signal transduction and gene expression [Citation3,Citation4]. The deregulation of the functional expression of genes, especially those associated with major signaling cascades, in neurons and glial cells has also been involved in the pathogenic processes of various severe disorders of the human brain such as glioma [Citation5,Citation6]. However, the mechanisms controlling the gene expression in neuronal and glial cells and associated with development, functioning, and pathogenesis of the human cerebrum and cerebellum are remain poorly understood.
Long non-coding RNAs (lncRNAs) are transcripts containing more than 200 nucleotides, which are associated with various biological processes and disease pathogenesis [Citation7]. The synthesis of lncRNAs is carried out by RNA Polymerase II-mediated genome transcription and can be followed by 5′ capping, alternative splicing, and polyadenylation, similarly to the mechanisms involved in the transcription and processing of protein coding messenger RNAs (mRNAs) [Citation8]. Additionally, lncRNAs can be encoded by genomic sequences overlapping with protein-coding genes or intergenic sequences. The different genomic locations and orientations of lncRNAs might influence their mechanism of action [Citation9]. Currently, some lncRNAs are known to modulate the transcription of other genes by forming ribonucleoprotein complexes together with chromatin-regulating proteins [Citation10]. For instance, Xist, a well-studied lncRNA, was reported to silence the gene transcription in one X chromosome by interacting with different proteins, such as SPEN, heterogeneous nuclear ribonucleoprotein U family proteins, and CDKN1A interacting zinc finger protein 1 [Citation11]. The potent capability of lncRNAs to influence gene expression suggests that they have a role in various biological and pathogenic processes.
Recent studies revealed that many lncRNAs are prevalently expressed in both the developing and adult central nervous systems, especially in the brain [Citation12]. More importantly, accumulating evidence shows that an abnormal lncRNAs expression in neural tissues are closely associated with the initiation and progression of various severe neurological diseases such as glioma, autism, Alzheimer’s disease, Parkinson’s disease, and schizophrenia [Citation13–17]. Transcriptomic analysis showed that a group of lncRNAs are acutely and dynamically expressed during neuronal activation processes [Citation17]. Gomafu, a lncRNA associated with retinal development, directly binds several splicing factors and influences alternative splicing, being thus involved in the pathogenesis of neurological disorders including schizophrenia [Citation17]. Recent single cell analyses based on strand-specific RNA-seq technique demonstrated that lncRNAs are expressed in the developing human brain in a cell type-specific and tissue-specific manner [Citation18]. For instance, the lncRNA LOC646329 is highly enriched in individual radial glia cells and affects glial cell proliferation [Citation18]. Despite the recent progress in understanding lncRNA expression and functions in the nervous system, the overall role, expression regulation, and specific mechanisms of action of lncRNAs during the cerebrum and cerebellum development and pathogenic events deserve further investigations.
The additional sex combs-like 3 (ASXL3) gene, is a protein associated with the polycomb repressive complex 2 and is upregulated in small cell lung cancer cells [Citation19]. The patients suffering from ASLX3-related disorder rather show signs or features of autism, sleep disturbance, and feeding problem [Citation20]. The ASXL3 gene is differentially expressed in human neural stem cells treated with dihydrotestosterone, indicating ASXL3 association with neurodevelopmental disorders including autism spectrum disorder [Citation21]. However, the molecular events leading by ASXL3 pathogenic expression and mutation are still unclear.
Our previous investigations characterized the compound heterozygous mutations (c.2168 C > G*c.5449 C > G) of the ASXL3 gene from a Chinese family of congenital heart disease [Citation22]. Our transcriptome study revealed significant alterations in the lncRNA and mRNA expression profiles in mouse myocardial tissues caused by the compound mutations of ASXL3 gene. Therefore, we speculated that ASXL3 compound mutations induce changes in the lncRNA and mRNA expression profiles in the nervous system. The present work aims to investigate the expression profiles of lncRNAs and mRNAs in the cerebrum and cerebellum of mice with ASXL3 mutations. We also analyzed the influence of the ASXL3 gene on neural cell functions. This study provides novel insights into the roles of the ASXL3 gene and lncRNA–mRNA interactions in brain functions and neurological disorders.
1. Material and Methods
1.1 Animals and brain tissue collection
The mutant mouse line with a heterozygous double mutation (P723R*P1817A) in the ASXL3 gene was established by the Cyagen Biosciences (Suzhou, China) as previously described [Citation22]. Cerebrum and cerebellum tissues from double mutant and wild-type C57BL/6 mice were collected at the age of 4–6 weeks, after the mice were euthanized by anesthesia with isoflurane inhalation and instant cervical dislocation. The weights of the cerebrum and cerebellum tissues were measured separately. All experimental procedures in this study were carried out following the guideline of the European Union Directive 2010/63/EU on the protection of animals used for scientific purposes and the NIH guidelines and were approved by the Animal Care and Use Committee of the Guangzhou Medical University.
2.2. qRT-PCR
The relative mRNA and lncRNA expression levels in mouse tissues were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). The experimental protocol was identical to that from our previous study [Citation22]. The sequences of the primers used in this study are listed in .
Table 1. Primers sequences used for quantitative real-time polymerase chain reaction
2.3 RNA sequencing and quantification
The alterations of the mRNA and lncRNA expression profiles in the cerebrum and cerebellum of wild-type and mutant mice were analyzed by RNA sequencing as described previously [Citation22]. The lncRNAs or mRNAs were considered differentially expressed in mutant mice compared to the levels in wild-type animals for a fold change ≥ 2 and FDR < 0.001.The genomic distribution and length of differentially expressed lncRNAs and mRNAs were also analyzed separately.
2.4 Bioinformatics
Gene ontology (GO) enrichment analysis was carried out using the Gene Ontology Consortium website (http://www.geneontology.org), according to the predicted molecular functions, biological processes, and cellular components. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was done using the routine settings (http://www.genome.jp/kegg/). The predicted interactions between differentially expressed lncRNAs and mRNAs were further analyzed based on their specific expression levels by construction of the Coding–Non-Coding (CNC) Network using the Cytoscape software.
2.5 Cell culture and transfection
SK-N-SH cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 4 mM glutamine (Thermo Fisher Scientific) at 37°C in a humidified atmosphere supplied with 5% CO2. The suppression of ASXL3 gene expression was realized by transfecting cells with ASXL3-siRNAs or the corresponding siRNA negative control using the Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The ASXL3 siRNAs, including ASXL3-Homo-2743, ASXL3-Homo-3617, and ASXL3-Homo-5299, were purchased from the GenePharma (Shanghai, China) and are listed in . The ASXL3 gene expression was evaluated by qRT-PCR 48 h after the transfection.
Table 2. ASXL3 siRNA sequences used for cell transfection
2.6 Cellular function analysis
We performed a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2 H-tetrazolium (MTS) assay and a flow cytometry apoptosis assay as described in our previous study [Citation22]. Cell cycle progression of cultured cells were analyzed by flow cytometry using Tali™ Cell Cycle Kit (Thermo Fisher Scientific). After incubation with Cell Cycle solution for 25 min in the dark, the percentage of cells at the different cell cycle stages were determined using a flow cytometer (BD Biosciences, San Jose, CA, USA).
2.7 Immunofluorescence detection
The expression of Class III beta-tubulin isotype (betaIII-tubulin) in cultured cells was assessed using the FITC Immunofluorescence Detection Kit (Sangon Biology, Shanghai, China) following the manufacturer’s instructions. Briefly, cell slides were incubated with the blocking solution for 30 min at room temperature and with the primary antibody (Anti-beta III Tubulin antibody, #ab25770, Abcam, Cambridge, MA, USA) for 1 h. After four washes with phosphate-buffered saline containing Tween 20, the slides were incubated with the fluorescent-labeled secondary antibody and the nuclei were stained with DAPI. The slides were observed under a fluorescence microscope XSP-63B (Shanghai optical instrument factury, Shanghai, China).
2.8 Western blotting
Western blotting was performed as described in our previous study [Citation22].
2.9 Statistical analyses
Data from at least three biological replicates are presented as mean ± standard deviation. The significance of the differences was evaluated by Student T-test (for two groups) or analysis of variance (for more than three groups) using the SPSS 18.0 software. The differences were considered significant for a P-value < 0.05.
3. Results
Significant alterations of the lncRNA and mRNA expression profiles in mouse myocardial tissues are caused by compound mutations of the ASXL3 gene. Therefore, we speculated that compound mutations of the ASXL3 gene induced changes in the lncRNA and mRNA expression profiles in the nervous system. In this study, we showed that the expression levels of many lncRNAs and mRNAs were changed in the cerebrum and cerebellum of mice with ASXL3 mutations. ASXL3 silencing inhibited neural cell proliferation and promoted cell apoptosis. These finding provide novel insights into the roles of the ASXL3 gene and lncRNA-mRNA interactions in brain functions and neurological disorders.
3.1 ASLX3 gene expression in the cerebrum and cerebellum of double mutant mice
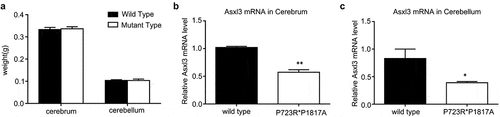
To analyze the effect of ASXL3 compound mutations on lncRNA and mRNA expression profiles in the cerebrum and cerebellum, ASXL3 double mutant mice (P723R*P1817A) were established as mentioned above. The weights of the mouse cerebrum and cerebellum were measured immediately after tissue collection. There was no significant change in the weights of the cerebrum and cerebellum between double mutant and wild-type C57BL/6 mice (). Subsequently, the expression of the ASXL3 gene in cerebrum and cerebellum tissues was determined by qRT-PCR. The ASXL3 mRNA levels in the cerebrum and cerebellum of ASXL3 P723R*P1817A mutant mice were significantly decreased compared with those in the wild-type mice (). These mouse cerebrum and cerebellum tissues were used for the following lncRNA and mRNA profile analysis.
Figure 1. Weight and ASL3 expression analyses in the mouse cerebrum and cerebellum. (a) Weight of the cerebrum and cerebellum of wild-type C57BL/6 control and ASXL3 P723R*P1817A mutant mice. (b) ASXL3 mRNA levels in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. (c) ASXL3 mRNA expression in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice. ASXL3 expression was evaluated by qRT-PCR. ASXL3: additional sex combs-like 3; qRT-PCR: quantitative real-time polymerase chain reaction; mRNA: messenger RNA; * and ** indicate P < 0.05 and P < 0.01, respectively
3.2 Alteration of the lncRNA and mRNA expression profiles in the mouse cerebrum
To explore the effects of ASXL3 P723R*P1817A mutations on the expression of genes and lncRNAs, we used RNA-seq to compare lncRNA and mRNA expression profiles in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. We identified 319 lncRNAs differentially expressed between the wild-type and mutant mice. Specifically, 212 lncRNAs were upregulated and 107 were downregulated in mutant mice compared to the levels in wild-type animals ( and Supplemental ). Additionally, 252 mRNAs, 91 upregulated and 161 downregulated, were differentially expressed in the cerebrum of ASXL3 P723R*P1817A mutant mice compared with their expression in wild-type mice ( and Supplemental ). The great number of lncRNAs and mRNAs affected by ASXL3 compound point mutations in the mouse cerebrum suggests a multifaceted role of the ASXL3 gene during cerebrum development and functioning.
Figure 2. Differential expression of lncRNAs and mRNAs in the ASXL3 mutant mouse cerebrum. (a, b) Differential expression of lncRNAs (a) and mRNAs (b) in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. The differential expression of lncRNAs and mRNAs are presented as heatmap (left) and volcano plot (right). (c, d) Genomic distributions of lncRNAs (c) and mRNAs (d) differentially expressed in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. Upregulated and downregulated transcripts are shown as red and green bars, respectively. (e, f) Lengths of lncRNAs (e) and mRNAs (f) differentially expressed in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. ASXL3: additional sex combs-like 3; WT: wild-type; MT: mutant; mRNAs: messenger RNAs; lncRNAs: long non-coding RNAs
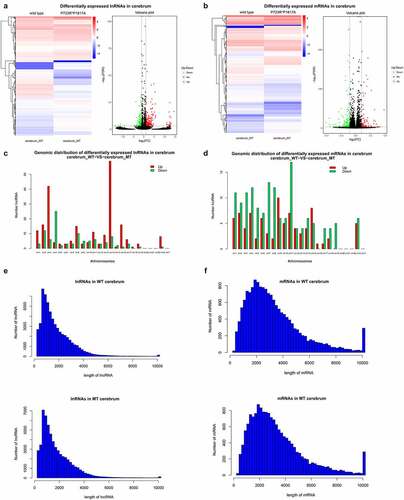
Furthermore, the differentially expressed lncRNAs in the cerebrum of mutant mice were encoded by genomic sequences distributed in most mouse chromosomes (). Upregulated lncRNAs were highly enriched in Chromosomes 3 and 14, while downregulated lncRNAs were significantly present in Chromosome 4 (). Differentially expressed mRNAs in mouse cerebrum were also widely distributed on mouse chromosomes, most of them being encoded by Chromosome 11 (). The lncRNA lengths were variable, with most lncRNAs being shorter than 2000 bp (). Similarly, the lengths of mRNAs in the cerebrum were also variable, with most mRNAs were shorter than 6000 bp ().
3.3 Functional categorization of differentially expressed mRNAs in the mouse cerebrum
To obtain more functional information regarding the effects of ASLX3 mutations, the mRNAs differentially expressed in the cerebrum of wild-type and mutant mice were further analyzed by bioinformatics. These mRNAs were associated with multiple cellular components including protein-containing complexes, supermolecular complexes, cell junctions, membranes, extracellular regions, membrane-enclosed lumen, and synapses (). The differentially expressed mRNAs possessed various molecular activities including catalytic, signaling transduction, binding, structural, transcriptional regulating, transporter, cargo receptor, and molecular carrier activities (). In addition, the mRNAs differentially expressed in the mouse cerebrum were involved in several biological processes such as growth, development, immune system, responses to stimulus, metabolic processes, biological adhesion, cell aggregation, proliferation, reproduction, locomotion, rhythmic process, and pigmentation (). The large number of functional categories containing the differentially expressed mRNAs suggested the prevalent involvement of the ASXL3 gene in physiological and pathogenic processes in cerebrum.
Figure 3. Functional annotation of mRNAs differentially expressed in the ASXL3 mutant mouse cerebrum. (a) GO categorization of mRNAs differentially expressed in the cerebrum of ASXL3 P723R*P1817A mutant mice. The mRNAs were categorized according to their annotated subcellular components, molecular functions, and biological processes. (b) Signaling pathways associated with the mRNAs differentially expressed in the cerebrum of ASXL3 P723R*P1817A mutant mice. These signaling pathways were analyzed by the KEGG method. (c) Coding-Non-Coding network based on the co-expression of lncRNAs and mRNAs differentially expressed in the cerebrum of mice with ASXL3 P723R*P1817A mutations. The network was constructed using upregulated (red) and downregulated (green) lncRNAs in the mutant mouse cerebrum. Plod3: 2-oxoglutarate 5-dioxygenase 3; EZH2: enhancer of zeste human homolog 2; SUV39H1: suppressor of variegation 3–9 homolog 1; KEGG: Kyoto encyclopedia of genes and genomes; GO: gene ontology; mRNAs: messenger RNAs
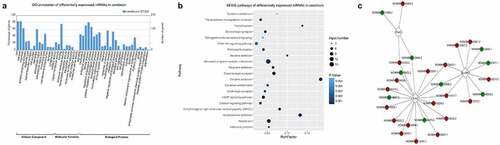
KEGG annotation revealed that mRNAs differentially expressed in the mutant mouse cerebrum were enriched in a number of signaling pathways including tyrosine metabolism, transcriptional mis-regulation in cancer, thyroid cancer, serotonergic synapse, retrograde endocannabinoid signaling, PI3K-Akt signaling, Parkinson’s disease, nicotine addiction, neuro-active ligand-receptor interaction, morphine addiction, dopaminergic synapse, cocaine addiction, circadian entrainment, cholinergic synapse, cAMP signaling, calcium signaling, amphetamine addition, and alcoholism (). By constructing CNC Network based on the co-expression of lncRNAs and mRNAs, we found that the expression of 2-oxoglutarate 5-dioxygenase 3 (Plod3), enhancer of zeste human homolog 2 (EZH2), and suppressor of variegation 3–9 homolog 1 (SUV39H1), as well as many of their interacting lncRNAs, were significantly affected by the ASXL3 P723R*P1817A mutations ().
3.4 Differential expression of lncRNAs and mRNAs in the mouse cerebellum
To study the influence of ASXL3 P723R*P1817A mutations on cerebellum genes and lncRNAs expression, lncRNA and mRNA expression profiles in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice were analyzed by RNA-seq. We detected 5330 lncRNAs differentially expressed in the cerebellum of wild-type and mutant mice, with 3891 upregulated and 1439 downregulated lncRNAs in ASXL3 mutant mice compared with the levels in wild-type mice ( and Supplemental Table 3). Moreover, compared with those in wild-type mice, 2204 mRNAs were differentially expressed in the cerebellum of the ASXL3 P723R*P1817A mutant mice. Among those, 1180 mRNAs were significantly upregulated and 1024 mRNAs were downregulated ( and Supplemental Table 4). These remarkable changes in lncRNA and mRNA expression profiles suggested an essential function of the ASXL3 gene in cerebellum biology and pathogenesis.
Figure 4. Differential expression of lncRNAs and mRNAs in the cerebellum of ASXL3 mutant mice. (a, b) Differential expression of lncRNAs (a) and mRNAs (b) in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice. The expression of lncRNAs and mRNAs in mutant mice was compared with that of wild-type mice and are represented as heatmap (left) and volcano plot (right). (c, d) Chromosomal distribution of differentially expressed lncRNAs (c) and mRNAs (d) in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice. Upregulated and downregulated transcripts are indicated by red and green bars, respectively. (e, f) Statistical analysis of the length of differentially expressed lncRNAs (e) and mRNAs (f) in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice. ASXL3: additional sex combs-like 3; WT: wild-type; MT: mutant; mRNAs: messenger RNAs; lncRNAs: long non-coding RNAs
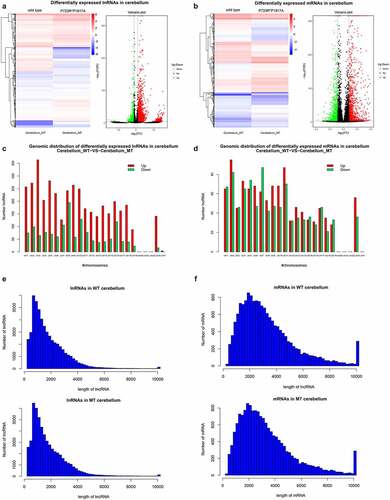
In addition, our genomic distribution analysis revealed that the lncRNAs differentially expressed in the mouse cerebellum were encoded on various mouse chromosomes (). Upregulated lncRNAs were mainly distributed in Chromosome 3 and downregulated lncRNAs were greatly enriched in Chromosome 8 (). On each chromosome there were significantly more lncRNAs upregulated than downregulated ones (). Similarly, mRNAs differentially expressed in the mutant mouse cerebellum were distributed across most mouse chromosomes, especially the Chromosome 2, which harbored the largest number of upregulated mRNAs and the second larger numbers of downregulated mRNAs (). The analysis of lncRNAs and mRNAs lengths in the mouse cerebellum provided similar results to those obtained in ASXL3 P723R*P1817A mutant mouse cerebrum ().
3.5 Functional categorization of mRNAs differentially expressed in the mouse cerebellum
To provide insights into the effects of ASLX3 mutations in the cerebellum, mRNAs differentially expressed between the wild-type and mutant mouse cerebellum tissues were subsequently categorized by bioinformatics. The GO analysis revealed that mRNAs differentially expressed in the cerebellum belonged to many cellular components including membrane-enclosed lumen, cell junction, synapse, supramolecular complex, extracellular region, and nucleoid (). Regarding the molecular functions, mRNAs differentially expressed in the mouse cerebellum were greatly enriched in catalytic, molecular function regulator, binding, structural molecule, signaling transducer, transcription regulator, transporter, translation regulator, antioxidant, molecular carrier, and cargo receptor activities (). Additionally, the mRNA differentially expressed in the cerebellum were linked with various biological processes including development, cellular component organization, biogenesis, localization, signaling, responses to stimulus, metabolism, immune system, reproduction, locomotion, behavior, cell proliferation and aggregation, rhythmic process, growth, chemical synaptic transmission, detoxification, pigmentation, cell killing, and protein complex biogenesis ().
Figure 5. Functional annotation of mRNAs differentially expressed in ASXL3 mutant mouse cerebellum. (a) GO annotation of mRNAs differentially expressed in the cerebellum of mice with ASXL3 P723R*P1817A mutations. Differential mRNAs were categorized according to their subcellular components, molecular functions, and biological processes. (b) KEGG pathways of mRNAs differentially expressed in the cerebellum of ASXL3 P723R*P1817A mutant mice. KEGG signaling pathways with a significant enrichment of the differentially expressed mRNAs are shown. (c) Coding-Non-Coding network of lncRNAs and mRNAs differentially expressed in the cerebellum of ASXL3 P723R*P1817A mutant mice. LncRNAs upregulated (red) and downregulated (green) in the mutant mouse cerebellum were used for network construction. Plod3: 2-oxoglutarate 5-dioxygenase 3; EZH2: enhancer of zeste human homolog 2; SUV39H1: suppressor of variegation 3–9 homolog 1; KEGG: Kyoto encyclopedia of genes and genomes; GO: gene ontology; mRNAs: messenger RNAs
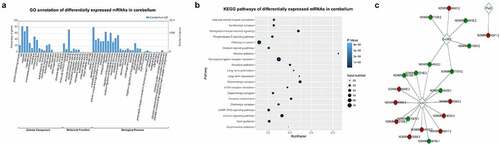
We also showed by KEGG analysis that mRNAs differentially expressed in the mutant mouse cerebellum were significantly associated with multiple biological pathways, including vascular smooth muscle contraction, serotonergic synapse, retrograde endocannabinoid signaling, phospholipase D signaling, pathways in cancer, oxytocin signaling, nicotine addiction, neuro-active ligand–receptor interaction, morphine addiction, long-term potentiation, long-term depression, glutamatergic synapse, ECM–receptor interaction, dopaminergic synapse, circadian entrainment, cholinergic synapse, cGMP-PKG signaling, calcium signaling and axon guidance, and amphetamine addiction (). The construction of a CNC Network also showed that the differential expression of Plod3, EZH2, and SUV39H1 induced by ASXL3 mutations correlated with their interacting lncRNAs ().
3.6 Validation of lncRNA and mRNA expression profiles in the mouse cerebrum and cerebellum
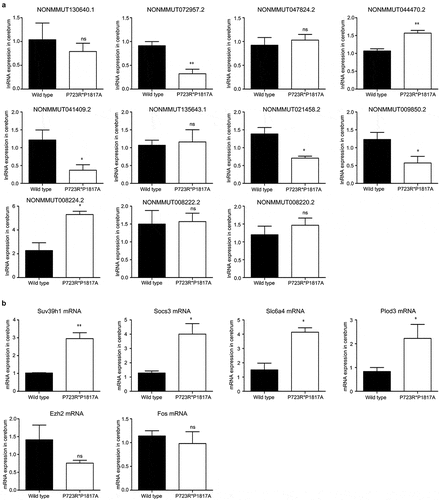
To validate the lncRNA and mRNA expression profiles detected by RNA-seq, eleven lncRNAs differentially expressed in the mouse cerebrum were first confirmed by qRT-PCR. The levels of four lncRNAs were significantly decreased in the cerebrum of ASXL3 mutant mice, whereas the expression of two other lncRNAs was remarkably increased in the mutant mouse cerebrum (). In addition, the mRNA levels of four functional genes, namely, SUV39H1, suppressor of cytokine signaling 3 (SOCS3), serotonin transporter gene Slc6a4, and Plod3, were also confirmed to be greatly elevated in the cerebrum of mice with P723R*P1817A mutations in the ASXL3 gene ().
Figure 6. Validation of lncRNA and mRNA expression levels in the ASXL3 mutant mouse cerebrum. (a) Relative expression of eleven lncRNAs in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. We analyzed the LncRNAs levels by qRT-PCR. (b) Relative expression of six functional genes in the cerebrum of wild-type and ASXL3 P723R*P1817A mutant mice. A qRT-PCR analysis was performed to compare the mRNA levels. Plod3: 2-oxoglutarate 5-dioxygenase 3; EZH2: enhancer of zeste human homolog 2; SUV39H1: suppressor of variegation 3–9 homolog 1; SOCS3: suppressor of cytokine signaling 3; Slc6a4: serotonin transporter gene; qRT-PCR: quantitative real-time polymerase chain reaction; ns: no significant; mRNAs: messenger RNAs; lncRNAs: long non-coding RNAs; * and ** indicate P < 0.05 and P < 0.01, respectively
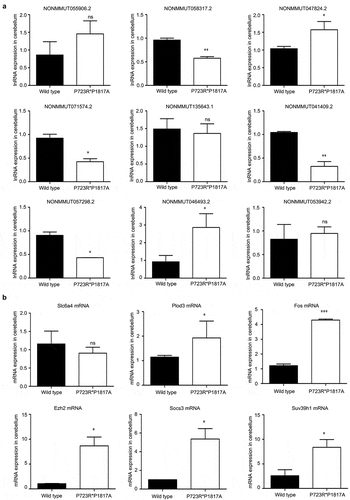
The expression levels of nine lncRNAs differentially expressed in the cerebellum of mutant mice were also confirmed by qRT-PCR. The expression of four of these lncRNAs was significantly reduced in the cerebellum of ASXL2 mutant mice compared with that in wild-type mice (). The expression of two other lncRNAs was remarkably enhanced in the cerebellum of mutant mice compared with wild-type levels (). The mRNA levels of five functional genes, Plod3, Fos, Ezh2, SOCS3, and SUV39H1, in the cerebellum of ASXL3 P723R*P1817A mutant mice were significantly greater than those in the wild-type animals ().
Figure 7. Validation of lncRNA and mRNA expression levels in the ASXL3 mutant mouse cerebellum. (a) Relative expression levels of nine lncRNAs in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice. We analyzed the lncRNAs levels by qRT-PCR. (b) Relative mRNA expression levels of six genes in the cerebellum of wild-type and ASXL3 P723R*P1817A mutant mice. The mRNA levels were determined by qRT-PCR. Slc6a4: serotonin transporter gene; Plod3: 2-oxoglutarate 5-dioxygenase 3; EZH2: enhancer of zeste human homolog 2; SOCS3: suppressor of cytokine signaling 3; SUV39H1: suppressor of variegation 3–9 homolog 1; qRT-PCR: quantitative real-time polymerase chain reaction; ns: no significant; mRNAs: messenger RNAs; lncRNAs: long non-coding RNAs; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively
3.7 Modulation of neural cell functions by ASXL3 knockdown
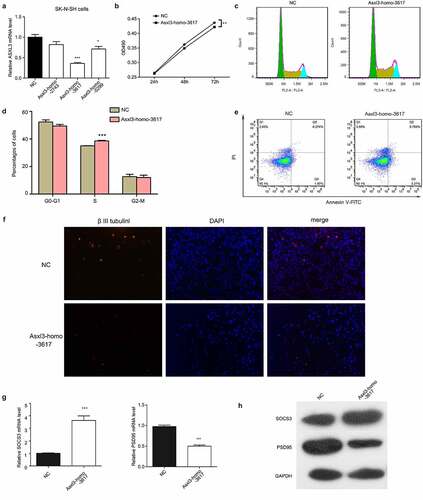
To obtain more insights into the biological roles of ASXL3, SK-N-SH neuroblastoma cells were transfected with siRNAs targeting the ASXL3 gene. The siRNAs, especially Asxl3-homo-3617, greatly downregulated ASXL3 expression (). The proliferation of SK-N-SH cells transfected with ASXL3 siRNAs was decreased compared with that of cells transfected with the negative control (). In addition, ASXL3 knockdown induced an increased number of SK-N-SH cells in S stage (). The apoptosis levels of SK-N-SH cells was also remarkably increased by the ASXL3 knockdown (). Furthermore, the suppression of ASXL3 gene expression in SK-N-SH cells caused a significant repression of betaIII-tubulin expression (), promoted Socs3 expression, and inhibited postsynaptic density-95 (PSD95) expression (). These results revealed a significant effect of ASXL3 expression on neural cell functions, which is consistent with the significant changes in lncRNAs and mRNAs expression profiles induced by ASXL3 point mutations and the resultant decreased ASXL3 expression levels.
Figure 8. Neural cell functions affected by siRNA-mediated ASXL3 depletion. (a) Relative expression levels of ASXL3 in SK-N-SH neuroblastoma cells transfected with ASXL3 siRNAs was assessed by qRT-PCR. (b) Proliferation rates of SK-N-SH neuroblastoma cells transfected with ASXL3 siRNAs was measured using the MTS method. (c, d) Cell cycle progression of SK-N-SH neuroblastoma cells transfected with ASXL3 siRNAs determined by flow cytometry. (e) Apoptosis of SK-N-SH neuroblastoma cells transfected with ASXL3 siRNAs evaluated by flow cytometry. (f) Immunofluorescence analysis of the expression of betaIII-tubulin in SK-N-SH neuroblastoma cells. (g) Relative mRNA levels of Socs3 and PSD96 in SK-N-SH neuroblastoma cells was determined by qRT-PCR. (h) Western blot analysis of Socs3 and PSD96 protein abundance in SK-N-SH neuroblastoma cells. NC: negative control; ASXL3: additional sex combs-like 3; betaIII-tubulin: Class III beta-tubulin isotype; PSD95: postsynaptic density-95; qRT-PCR: quantitative real-time polymerase chain reaction; MTS: 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4- sulfophenyl)-2 H-tetrazolium; mRNAs: messenger RNAs; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively
4. Discussion
It has been established that lncRNAs constitute a critical regulatory module in various processes associated with physiological functions and pathogenesis [Citation23]. Large-scale transcriptome analysis based on RNA-seq is an efficient strategy for investigating lncRNA profiles because it allows instant identification and quantification of the expression of large amounts of lncRNAs [Citation24]. Regarding the great influence of lncRNAs on gene expression [Citation11], the simultaneous characterization of lncRNA and mRNA expression profiles by transcriptome sequencing has provided valuable insights into the mechanistic properties of key lncRNAs during several pathogenic processes [Citation25,Citation26]. Our previous study identified compound heterozygous mutations in the ASXL3 gene in a Chinese family presenting congenital heart disease. Further investigations using a mouse model showed that these ASXL3 mutations caused great alterations of the lncRNA and mRNA expression profiles in heart tissues [Citation22], suggesting that ASXL3 mutations might be a critical event regulating lncRNAs and mRNAs expression. The ASXL3 gene has been previously involved in neural cell function and neurological processes [Citation20,Citation21]. However, little is known about lncRNAs and mRNAs expression regulation in brain tissues.
To provide a general view of the influence of ASXL3 compound mutations on lncRNAs and mRNAs expression in the brain, we generated an ASXL3 P723R*P1817A mutant mice, in which ASXL3 gene expression is greatly decreased. A transcriptome analysis based on the RNA-seq technique revealed that the expression of many lncRNAs and mRNAs were affected by the introduction of ASXL3 P723R*P1817A mutations. Moreover, these differentially expressed lncRNAs and mRNAs were widely distributed across almost all mouse chromosomes. We identified much more upregulated than downregulated lncRNAs in the ASXL3 mutant mouse cerebellum. This observation suggested that the upregulation of lncRNAs expression in the mouse cerebellum are an important molecular mechanism underlying cerebellum physiology and pathogenic processes.
The bioinformatics analysis revealed an enrichment of the differentially expressed lncRNAs and mRNAs in many cellular components and molecular functions. The great variety of lncRNAs and mRNAs affected by the ASXL3 gene mutations suggest that transcriptome alterations regulated by ASXL3 are essential for neural cell functions. For instance, synapses are critical intercellular structures passing electrical or chemical signals from one neuron to another or to an effector cell during neurotransmission, which is often disturbed in neurological disorders such as Alzheimer’s disease and spinal cord injury [Citation27]. The lncRNA GM12371 was recently reported to regulate synapse morphology, synaptic communication, and synapse density by modulating the transcription of a group of genes associated with neuronal development and differentiation [Citation28]. In the present study, we identified many lncRNAs distributed in synapses, including dopaminergic, cholinergic, serotonergic, and glutamatergic synapses, and involved in synapse-related biological processes. These observations indicate that ASXL3-regulated lncRNAs and mRNAs might be involved in the functioning of various synapse types. This deserves further investigations.
The KEGG annotation showed that numerous functional genes associated with many signaling pathways in the cerebrum and cerebellum were regulated by ASXL3 mutations. For instance, PI3K/AKT signaling is involved in the regulation of functions such as neuronal migration and apoptosis [Citation29,Citation30]. Moreover, the PI3K/AKT pathway was previously shown to be regulated by lncRNAs [Citation31]. Our analysis further suggests that the interaction of lncRNA with the PI3K/AKT signaling pathway plays physiological and pathogenic roles in neuronal cells. The CNC Networks predicted the interaction of numerous differentially expressed lncRNAs with three functional genes, including EZH2, Plod3, and SUV39H1. The inhibition of EZH2 activity was shown to suppress bupivacaine-induced neuronal apoptosis in spinal cord dorsal root ganglia [Citation32]. Overexpression of the PLOD3 gene, which encodes a lysyl hydroxylase, significantly promotes the progression of gliomas and is associated with a poor prognosis in glioma patients [Citation33]. Suv39h1 is a methyltransferase that was characterized as a chromatin modifier regulating neuron survival, neuron death, and trauma progression [Citation34]. However, the molecular mechanisms regulating the role of these three genes in neuron physiology and neurological disorders remains poorly understood. The identification of many lncRNAs influenced by the ASXL3 mutation in the mouse cerebrum and cerebellum suggests that lncRNA-mediated transcription regulation controls the expression of these genes in neural cells during neurological disorder progression.
Finally, our cellular assays based on ASXL3 gene silencing confirmed ASXL3 involvement in neural cell proliferation, cell cycle progression, and apoptosis. ASXL3 gene depletion greatly altered the expression of betaIII-tubulin, Socs3, and PSD96 genes, which are all closely linked to synapse formation and neuronal functions [Citation35,Citation36]. This further supports a potential role for ASXL3-regulated lncRNAs and mRNAs in neural cells and requires further investigations. Whether the alterations of lncRNA and mRNA expression profiles caused by the compound mutations in the ASXL3 gene are involved in other biological and pathogenic processes is also worth studying. For instance, previous reports showed that ASXL3 acts as a novel pluripotency factor in respiratory epithelial cells associated with small cell lung cancer [Citation19]. Moreover, lncRNAs were recently linked with invasion and metastasis of small cell lung cancer [Citation37,Citation38]. Based on the significant changes in lncRNAs and mRNAs expression induced by ASXL3 mutations reported here, lncRNAs and their interacting mRNAs networks might constitute biomarkers, treatment targets, or critical molecular events mediating the pathogenesis of diseases. Their therapeutic potential might be exploited in a similar fashion than miRNA and fungi are used in the fight against cancer and COVID-19 [Citation39–41].
This study is a preliminary exploration and additional work is needed to confirm our findings by overexpression or inhibition of lncRNAs or mRNAs in animal models.
5. Conclusion
In summary, many lncRNAs and mRNAs were differentially expressed in the cerebrum and cerebellum of an ASXL3 mutant mouse. Our work provides novel insights into the roles of the ASXL3 gene and lncRNA-mRNA interactions in brain functions and neurological disorders.
Ethics approval
All experimental procedures in this study were carried out following the guideline of the European Union Directive 2010/63/EU on the protection of animals used for scientific purposes and the NIH guidelines and were approved by the Animal Care and Use Committee of the Guangzhou Medical University.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Highlights
The expression of lncRNAs and mRNAs is altered in ASXL3 mutant mouse cerebrum.
The expression of lncRNAs and mRNAs is also altered in ASXL3 mutant mouse cerebellum.
ASXL3 is involved in the proliferation, cell cycle, and apoptosis of neural cells
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Coombes SA, Misra G. Pain and motor processing in the human cerebellum. Pain. 2016;157(1):117–127.
- Wolf U, Rapoport MJ, Schweizer TA. Evaluating the affective component of the cerebellar cognitive affective syndrome. J Neuropsychiatry Clin Neurosci. 2009;21(3):245–253.
- Abdi K, Kuo CT. Laminating the mammalian cortex during development: cell polarity protein function and Hippo signaling. Genes Dev. 2018;32(11–12):740–741.
- Su CH DD, Tarn WY. Alternative splicing in neurogenesis and brain development. Front Mol Biosci. 2018;5:12.
- Gritsenko PG, Atlasy N, Dieteren CEJ, et al. p120-catenin-dependent collective brain infiltration by glioma cell networks. Nat Cell Biol. 2020;22(1):97–107.
- Menet R, Lecordier S, ElAli A. Wnt pathway: an emerging player in vascular and traumatic mediated brain injuries. Front Physiol. 2020;11:565667.
- Latowska J, Grabowska A, Zarębska Ż, et al. Non-coding RNAs in brain tumors, the contribution of lncRNAs, circRNAs, and snoRNAs to cancer development-their diagnostic and therapeutic potential. Int J Mol Sci. 2020;21(19):7001.
- Taniue K, Akimitsu N. The functions and unique features of LncRNAs in cancer development and tumorigenesis. Int J Mol Sci. 2021;22(2):632.
- Ramakrishnaiah Y, Kuhlmann L, Tyagi S. Towards a comprehensive pipeline to identify and functionally annotate long noncoding RNA (lncRNA). Comput Biol Med. 2020;127:104028.
- Liu SJ, Dang HX, Lim DA, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–460.
- Lu Z, Guo JK, Wei Y, et al. Structural modularity of the XIST ribonucleoprotein complex. Nat Commun. 2020;11(1):6163.
- Andersen RE, Lim DA. Forging our understanding of lncRNAs in the brain. Cell Tissue Res. 2018;371(1):55–71.
- Tao C, Luo H, Chen L, et al. Identification of an epithelial-mesenchymal transition related long non-coding RNA (LncRNA) signature in Glioma. Bioengineered. 2021;12(1):4016–4031.
- Zhang Y, Zhang Y, Wang S, et al. SP1-induced lncRNA ZFPM2 antisense RNA 1 (ZFPM2-AS1) aggravates glioma progression via the miR-515-5p/Superoxide dismutase 2 (SOD2) axis. Bioengineered. 2021;12(1):2299–2310.
- Tong Z, Zhou Y, Wang J. Identification and functional analysis of long non-coding RNAs in autism spectrum disorders. Front Genet. 2020;11:849.
- Moreno-García L, López-Royo T, Calvo AC, et al. Competing endogenous RNA networks as biomarkers in neurodegenerative diseases. Int J Mol Sci. 2020;21(24):9582.
- Barry G, Briggs JA, Vanichkina DP, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19(4):486–494.
- Liu SJ, Nowakowski TJ, Pollen AA, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17(1):67.
- Shukla V, Rao M, Zhang H, et al. ASXL3 is a novel pluripotency factor in human respiratory epithelial cells and a potential therapeutic target in small cell lung cancer. Cancer Res. 2017;77(22):6267–6281.
- Balasubramanian M, Schirwani S. ASXL3-Related Disorder. In: geneReviews(®). Seattle (WA): University of Washington, Seattle; 1993-2020.
- Quartier A, Chatrousse L, Redin C, et al. Genes and pathways regulated by androgens in human neural cells, potential candidates for the male excess in autism spectrum disorder. Biol Psychiatry. 2018;84(4):239–252.
- Fu F, Li R, Lei TY, et al. Compound heterozygous mutation of the ASXL3 gene causes autosomal recessive congenital heart disease. Hum Genet. 2021;140(2):333–348.
- Ang CE, Trevino AE, Chang HY. Diverse lncRNA mechanisms in brain development and disease. Curr Opin Genet Dev. 2020;65:42–46.
- Jiang Q, Ma R, Wang J, et al. LncRNA2 function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics. 2015;16 Suppl 3(Suppl3):S2.
- Ye Z, Fang J, Wang Z, et al. Bioinformatics-based analysis of the lncRNA-miRNA-mRNA and TF regulatory networks reveals functional genes in esophageal squamous cell carcinoma. Biosci Rep. 2020;40(8):BSR20201727.
- Lv Z, Sun L, Xu Q, et al. Joint analysis of lncRNA m(6)A methylome and lncRNA/mRNA expression profiles in gastric cancer. Cancer Cell Int. 2020;20(1):464.
- Suzuki K, Elegheert J, Song I, et al. A synthetic synaptic organizer protein restores glutamatergic neuronal circuits. Science. 2020;369(6507):eabb4853.
- Raveendra BL, Swarnkar S, Avchalumov Y, et al. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc Natl Acad Sci U S A. 2018;115(43):E10197–e10205.
- Jover-Mengual T, Miyawaki T, Latuszek A, et al. Acute estradiol protects CA1 neurons from ischemia-induced apoptotic cell death via the PI3K/Akt pathway. Brain Res. 2010;1321:1–12.
- Luo T, Ou JN, Cao LF, et al. The autism-related lncRNA MSNP1AS regulates moesin protein to influence the RhoA, Rac1, and PI3K/Akt pathways and regulate the structure and survival of neurons. Autism Res. 2020;13(12):2073–2082.
- Ding X, Xu X, XF H, et al. Muscleblind-like 1 antisense RNA 1 inhibits cell proliferation, invasion, and migration of prostate cancer by sponging miR-181a-5p and regulating PTEN/PI3K/AKT/mTOR signaling. Bioengineered. 2021;12(1):803–814.
- Zheng J, Chen J, Wu G, et al. Inhibiting EZH2 rescued bupivacaine-induced neuronal apoptosis in spinal cord dorsal root ganglia in mice. J Anesth. 2018;32(4):524–530.
- Tsai CK, Huang LC, Tsai WC, et al. Overexpression of PLOD3 promotes tumor progression and poor prognosis in gliomas. Oncotarget. 2018;9(21):15705–15720.
- Liu DX, Nath N, Chellappan SP, et al. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev. 2005;19(6):719–732.
- Li S, He Q, Wang H, et al. Injured adult retinal axons with Pten and Socs3 co-deletion reform active synapses with suprachiasmatic neurons. Neurobiol Dis. 2015;73:366–376.
- Hu Z, Liu Z, Li X, et al. Stimulation of synapse formation between stem cell-derived neurons and native brainstem auditory neurons. Sci Rep. 2017;7(1):13843.
- Chen S, Wu H, Lv N, et al. LncRNA CCAT2 predicts poor prognosis and regulates growth and metastasis in small cell lung cancer. Biomed Pharmacother. 2016;82:583–588.
- Huang C, Liu S, Wang H, et al. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res. 2016;8(11):5025–5034.
- Shin Low S, Pan Y, Ji D, et al. Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept. Sens Actuators B Chem. 2020;308:127718.
- How CW, Ong YS, Low SS, et al. How far have we explored fungi to fight cancer? Semin Cancer Biol. 2021; S1044-579X(21)00059-6.
- Chia WY, Kok H, Chew KW, et al. Can algae contribute to the war with Covid-19? Bioengineered. 2021;12(1):1226–1237.
