ABSTRACT
Cancer stem cells (CSCs) contribute to malignant features. Long non-coding RNA (LncRNA) FENDRR has been shown to regulate tumor proliferation, migration, and invasion. However, the effects of FENDRR on the CSC-like traits of colorectal cancer cells remain to be elucidated. Here, we identified that lncRNA FENDRR level was remarkably lower in spheres formed by colorectal cancer cells compared to that in parental cancer cells. Further functional experiments showed that FENDRR overexpression attenuated the CSC-like traits of colorectal cancer spheres, while FENDRR knockdown conferred the CSC-like traits for colorectal cancer cells, as characterized by the alteration of ALDH activity, sphere-formation ability, and expression of stemness markers (Oct4, Sox2, and KLF4). RNA–RNA interaction in vitro analysis combined with mRNA stability assay revealed that lncRNA FENDRR directly interacted with Sox2 mRNA 3’UTR, reduced its mRNA stability and thus inhibited Sox2 expression. In addition, lncRNA FENDRR-mediated effects on the CSC-like traits of colorectal cancer cells depended on Sox2 expression. This work suggests that lncRNA FENDRR can block the CSC-like traits in colorectal cancer cells through directly interacting with Sox2 mRNA 3’UTR.
Introduction
Long non-coding RNA (LncRNA) is a kind of RNAs longer than 200 nucleotides [Citation1]. LncRNA can not or can only encode limited proteins, but it can modulate gene expression at the post-transcriptional or transcriptional level, thus affecting the biological process, for example, lncRNA plays a critical role in transcriptional silencing or activation, nuclear transport, chromosome modification, and so on [Citation2]. In recent years, many abnormal lncRNAs have been found in colorectal cancer and have been regarded as specific biomarkers for prognosis, diagnosis, and even treatment prediction [Citation3]. Additionally, lncRNAs are closely related to the growth and proliferation, invasion and metastasis, apoptosis, and drug resistance of colorectal cancer cells [Citation4,Citation5]. However, the mechanisms contributing to lncRNA-mediated effects on colorectal cancer progression are still confusing.
The theory of cancer stem cells (CSCs) was put forward in 2001 when CSC was regarded to be a critical effector for tumor occurrence, recurrence, and drug resistance [Citation6]. Recently, lncRNAs are also found to be involved in CSC progression or the CSC-like traits of cancer cells, for example, lncRNA LUCAT1 has been found to be related to the metastasis and TNM staging and promote the self-renewal of breast CSCs [Citation7]; LncRNA Sox2OT is shown to promote the CSC-like traits of bladder cancer cells by sponging miR-200 c and thus positively regulating Sox2 expression [Citation8]; And lncRNA NEAT1 confers cancer stemness and sensitizes cells to chemotherapy in triple-negative breast cancer (TNBC) [Citation9]. LncRNA FENDRR has been confirmed to be related to tumorigenesis in various tumors, such as migration, invasion, apoptosis, and chemoresistance [Citation10,Citation11]. Notably, it is found that lncRNA FENDRR attenuates adriamycin resistance by inhibiting MDR1 expression [Citation12] and in agreement with this rationale, FENDRR reduces the CSC-like traits of non-small cell lung cancer (NSCLC) cells via suppressing MDR1 expression [Citation13]. However, its effects in colorectal cancer cell stemness remain to be elucidated.
Here, we aimed to explore the effects of lncRNA FENDRR in colorectal cancer stemness. We constructed a colorectal CSC model through collecting spheres by 3D non-adherent culture and found that FENDRR was lowly expressed in colorectal cancer sphere. Then, gain- and loss-of functional experiments revealed that FENDRR negatively regulated the CSC-like traits of colorectal cancer cells. Mechanistic studies showed that lncRNA FENDRR directly interacted with Sox2 mRNA 3’UTR, but not Oct4 and KLF4 mRNA and thus increased Sox2 mRNA stability and expression. At last, we demonstrated that lncRNA FENDRR conferred the CSC-like traits and chemoresistance of colorectal cancer cells dependent on Sox2 expression.
Material and methods
Cell culture
Colorectal cancer cell lines HCT-116 and HT-29 were purchased from Fenghui Biotechnology Co., Ltd (Changsha, China) and cultured in 1640 medium (MeilunBio, Dalian, China) plus 10% FBS (Fetal bovine serum, Oricell, Suzhou, China) as well as 1% penicillin (Sangon, Shanghai, China) and streptomycin (Sangon). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2. 5-Fu-resistant HT-29 cells were established by culturing HT-29 cells with 5-Fu (2 μM) for at least three months and then the resistant clones were collected and expanded for a long culture with 100 nM 5-Fu.
Real-time quantitative PCR (RT-qPCR)
1 mL Trizol reagent (Cat # R401-01, Vazyme, Nanjing, China) was used to extract total RNA of 106 cells. The absorbance value of A260 and A280 of RNA should be 1.8≤ A260/A280 ≤ 2.0, and the RNA concentration should be calculated. M-MLV reverse transcriptase (Cat # R021-01, Vazyme) was used to reversely transcribe RNA at 37°C for 1 h in 10 μL system and inactivate the reverse transcriptase at 95°C for 3 min. The synthesized cDNA was saved at −20°C. 20 μL reaction system containing 2 × Quantitative PCR buffer (Cat # Q711-02, Vazyme) 10 μL, 2 μL upstream and downstream primers was used to detect the relative expression levels of transcripts. The reaction conditions were as follows: pre-denaturation at 93°C for 3 min, 93°C for 30 s, 56°C for 40 s. GAPDH was served as an endogenous control.
Western blot
Cells were digested with 0.05% trypsin and 0.02% EDTA for 30 s – 2 min. Then, a complete medium was added to terminate the digestion. The supernatant was centrifuged at 4°C 800 r/min, and added 200 μL 1% SDS protease inhibitor to lyse cells, and the lysate was repeatedly pumped (on ice bath). Protein concentration was quantified following the instructions of Pierce protein assay kit. Before use, freeze-thaw samples on ice, take a certain volume (including 50 μg protein) into a clean Eppendorf tube and add 4 μL 5 × SDS loading buffer, 1% SDS to 20 μL. Denatured at 95°C for 5 min, the samples were placed on ice and loaded as soon as possible. 10% SDS-PAGEs were used to separate the proteins, which were then transferred to the ECL membranes (100 V, 1 h), sealed with 10% no-fat milk for 2 h at 5% PBST, and then incubated with primary antibodies overnight at 4°C. PBST solution was used to wash membranes three times, which were then reacted with horseradish peroxidase labeled Goat anti-mouse Ig at room temperature for 1.5 h. Finally, membranes were exposed using an ECL kit (Cat # E411-03, Vazyme, Nanjing, China) to detect protein expression. GAPDH served as an endogenous control.
Lentivirus and plasmid construction, infection, and transfection
The FENDRR overexpression (FENDRR-oe) and knockdown (FENDRR-kd) lentivirus and control lentivirus vector were purchased from HANBIO (Shanghai, China). The siRNA against Sox2 (Sox2-kd) and Sox2 overexpression (Sox2-oe) plasmid were purchased from GenePharma (Shanghai, China). The transfection procedure was performed using jetPRIME (Polyplus, New York, USA) following the manufacturer’s protocols.
Sphere-formation assay
Sphere formation analysis was performed to evaluate the CSC-like traits of colorectal cancer cells. Briefly, cells were cultured in 37°C, 5% CO2 incubator, low-adherent culture plates with sphere-culturing medium containing DMEM/F12 (Cat # 31,331,093, Thermo Fisher Scientific, Waltham, MA, USA) with 1% methylcellulose (Cat # M0512, Sigma) and 10 ng/ml FGF-β (Cat # 11,343,623, ImmunoTools), 10 ng/ml EGF (Cat # 11,343,406, ImmunoTools) and 1 × B27 (Cat # 17,504,044, Thermo Fisher Scientific). Ten days later, sphere size and number were observed under microscope. For experiments of spheres, spheres were collected, re-digested, and subjected to further experiments.
ALDH activity detection
The Aldehyde Dehydrogenase Activity Colorimetric Assay Kit (Sigma–Aldrich) was used to detect ALDH activity according to the manufacturer’s recommendation.
Luciferase reporter analysis
The 5’UTR (Untranslated Region), CDS (Coding sequence), and 3’UTR sequences of Sox2 were cloned into PMIR-Reporter plasmid, referred as PMIR-Sox2-5’UTR, PMIR-Sox2-CDS, and PMIR-Sox2-3’UTR, which were co-transfected into 293 T cells with β-gal plasmid. Then, the luciferase activity and related analysis were referred to the previous study [Citation14].
RNA–RNA interaction in vitro analysis
The detailed protocols were referred to the previous study [Citation15]. BrU-labeled RNAs (FENDRR and FENDRR-Anti-sense) were synthesized from Genepharma (Shanghai, China).
Analysis on mRNA stability
Colorectal cancer cells were infected with FENDRR-oe and vector, respectively. 48 h later, cells were treated with actinomycin D (Act D) for another 2 h, 4 h, and 6 h, respectively, RNA was extracted and Sox2 relative expression level was determined by RT-qPCR analysis.
Cell viability assay
Colorectal cancer cells were seeded into 96-well plates at 4 × 103 cells/well. Cell Counting Kit-8 (CCK-8) kit (GLPBIO, Shanghai, China) was used to measure cell viability. Add 10 μL CCK8 solution to each well on day 1, 2, and 3, respectively. Incubate the plates at 37°C for 2 h. Mix gently on the track vibrator for 1 min to ensure uniform color distribution. Then, the absorbance at 450 nm was measured using a microplate reader to evaluate the cell viability.
Statistical analysis
Graphpad Prism version 8.0 statistical software was used to analyze the significance between groups. Data were expressed as χ ± s. T test was used to compare the two groups. P < 0.05 was statistically significant.
Results
Overexpression of lncRNA FEDNRR attenuates the CSC-like traits of colorectal cancer spheres
To investigate the effects of lncRNA FENDRR on the CSC-like traits of colorectal cancer cells, the spheres formed by colorectal cancer cells were collected through 3D non-adherent culture which has been confirmed to retain the CSC-like traits in various tumors [Citation16] (). The CSC-like traits were confirmed through examining the sphere-formation capacity (), ALDH activity () and expression of stemness markers () comparing to the parental colorectal cancer cells. And we identified that lncRNA FENDRR level was remarkably lower in colorectal cancer spheres compared to that in the parental cancer cells (). We further evaluated FENDRR expression through TCGA data using the Tumor, Normal and Metastatic tissues tool (https://tnmplot.com/analysis/) including normal samples from non-cancerous patients and further pediatric tissues, or paired tumor and adjacent normal tissues, and found that FENDRR was indeed lowly expressed in colorectal cancer tissues (). Then, FENDRR was overexpressed in colorectal cancer spheres through lentivirus infection and the overexpression efficiency was validated by RT-qPCR (). It was found that FENDRR overexpression attenuated the sphere-formation ability (), ALDH activity () and expression of stemness markers () in colorectal cancer spheres.
Figure 1. LncRNA FEDNRR level is significantly downregulated in colorectal cancer spheres. (a) The representative images of spheres formed by colorectal cancer cells. (b) Sphere size was examined in colorectal cancer spheres and cells. (c) Sphere number was measured in colorectal cancer spheres and cells. (d) ALDH activity was evaluated in colorectal cancer spheres and cells. (e and f) The mRNA level of stemness markers (Sox2, Oct4, KLF4) was determined in colorectal cancer spheres and cells. (g) The protein level of stemness markers was detected in colorectal cancer spheres and cells. (h) FENDRR level was examined in colorectal cancer spheres and cells. (i – l) FENDRR expression was detected in data from TCGA using the Tumor, Normal and Metastatic tissues tool (https://tnmplot.com/analysis/). n ≥ 3, **P < 0.01 vs. control
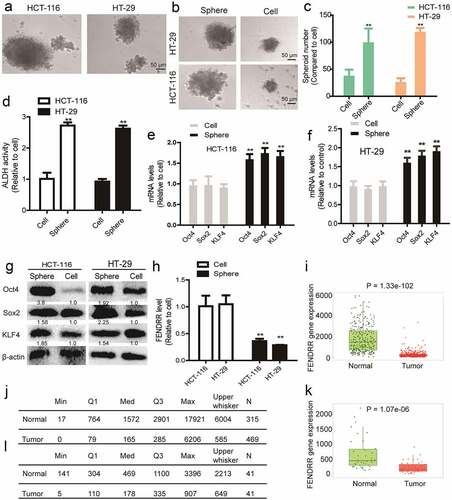
Figure 2. Overexpression of lncRNA FEDNRR attenuates the CSC-like traits of colorectal cancer spheres. (a) The overexpression efficiency of FENDRR-oe was validated by RT-qPCR. (b and c) Sphere number and size were measured in colorectal cancer spheres with FENDRR overexpression or not. (d) Colorectal cancer spheres with or without FENDRR overexpression were subjected to ALDH activity detection. (e and f) The mRNA levels of stemness markers (Sox2, Oct4, KLF4) were detected in colorectal cancer spheres with or without FENDRR overexpression. (g) The protein levels of stemness markers (Sox2, Oct4, KLF4) were detected in colorectal cancer spheres with or without FENDRR overexpression. n ≥ 3, **P < 0.01 vs. control
Knockdown of lncRNA FENDRR confers the CSC-like traits of colorectal cancer cells
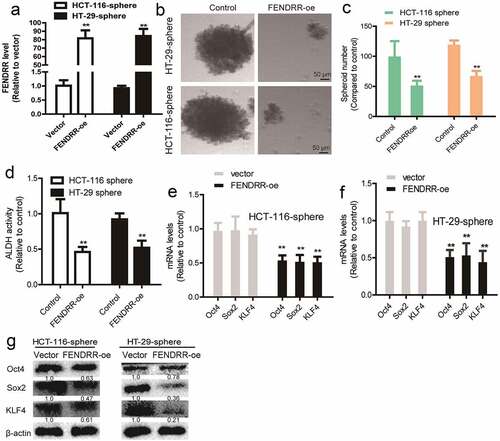
In contrast, FENDRR was knocked down in colorectal cancer cells and knockdown efficiency was validated (). We found that FENDRR knockdown enhanced the sphere-formation capacity, as characterized by the increase of sphere number and size (). In addition, ALDH activity was increased by FENDRR knockdown in colorectal cancer cells (). Furthermore, the expression of stemness markers (Oct4, Sox2, and KLF4) was increased by FENDRR knockdown (). Thus, these results demonstrate that FENDRR can suppress the CSC-like traits of colorectal cancer cells.
Figure 3. Knockdown of lncRNA FENDRR confers the CSC-like traits of colorectal cancer cells. (a) The knockdown efficiency of FENDRR-kd was confirmed by RT-qPCR. (b and c) Sphere number and size were determined in colorectal cancer cells with FENDRR knockdown or not. (d) ALDH activity was determined in colorectal cancer cells with FENDRR knockdown or not. (e and f) The mRNA levels of stemness markers were detected in colorectal cancer cells with FENDRR knockdown or not. (g) The protein levels of stemness markers were examined in colorectal cancer cells with or without FENDRR knockdown. n ≥ 3, **P < 0.01 vs. control
LncRNA FENDRR directly interacts with Sox2 mRNA 3’UTR, decreases its stability and expression
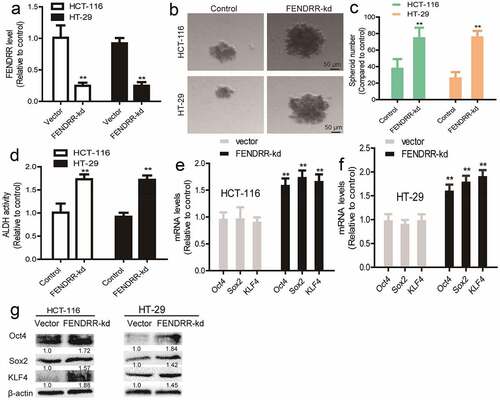
As lncRNAs have been confirmed to act as RNA partners, we wondered whether lncRNA FENDRR can act as a partner for the stemness markers (Oct4, Sox2, and KLF4). As shown in , lncRNA FENDRR directly interacted with Sox2 mRNA, but not Oct4 and KLF4 mRNA through RNA–RNA interaction in vitro analysis. Consistently, we found that FENDRR overexpression decreased the mRNA stability of Sox2, but not Oct4 and KLF4 mRNA (). Additionally, to gain insight about the concrete regions of Sox2 mRNA bound by FEDNRR, RNA–RNA interaction in vitro analysis was further performed and we found that FENDRR interacted with Sox2 3’UTR, but not its 5’UTR and CDS (). Furthermore, luciferase reporter analysis revealed that FENDRR overexpression decreased the luciferase activity of Luc-Sox2-3’UTR, but failed to change the activity of Luc-sox2-CDS and Luc-Sox2-5’UTR (). Therefore, our results indicate that lncRNA FENDRR directly interacts with Sox2 3’UTR and thus decreases its mRNA stability and expression.
Figure 4. LncRNA FENDRR directly interacts with Sox2 mRNA 3’UTR, enhances its stability and expression. (a – c) The interaction between FENDRR and Sox2, Oct4, or KLF4 was examined through the RNA–RNA interaction in vitro assay. (d – f) The mRNA stability of Sox2, Oct4 and KLF4 was determined in colorectal cancer cells with FENDRR overexpression or not. (g and h) The interaction between FENDRR, and Sox2 CDS, Sox2 5’UTR, or Sox2 3’UTR was evaluated in colorectal cancer cells. (i and j) The activity of Luc-Sox2-3’UTR, Luc-Sox2-CDS, and Luc-Sox2-5’UTR was measured in colorectal cancer cells with FENDRR overexpression or not. n ≥ 3, **P < 0.01 vs. control
LncRNA FENDRR inhibits the CSC-like traits of colorectal cancer cells dependent on Sox2
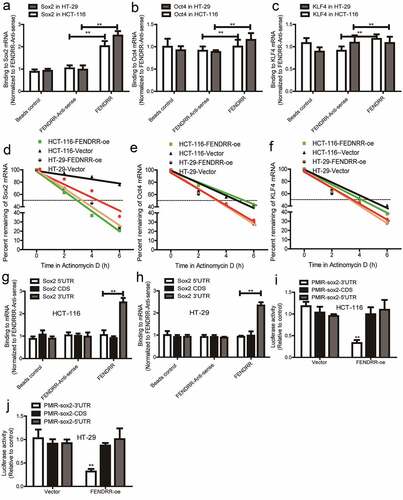
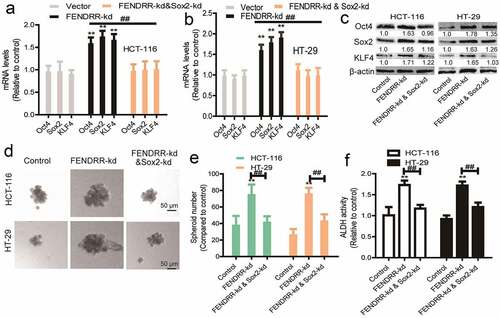
We then determined whether FENDRR inhibited the CSC-like traits of colorectal cancer cells through Sox2. Sox2 was knocked down in colorectal cancer cells with FEDNRR knockdown. Firstly, it was found that Sox2 knockdown attenuated the promoting effects of FENDRR knockdown on the expression of stemness markers (KLF4, Oct4) (). Secondly, FENDRR knockdown-induced enhancement of sphere-formation capacity was partially abrogated by knocking down Sox2 (). Thirdly, the increase of ALDH activity mediated by FENDRR knockdown was reduced by Sox2 knockdown (). Thus, these results confirm that lncRNA FENDRR suppresses the CSC-like traits of colorectal cancer cells dependent on Sox2 mRNA.
Figure 5. LncRNA FENDRR promotes the CSC-like traits of colorectal cancer cells dependent on Sox2. (a and b) The stemness markers’ mRNA levels were examined in cells with FENDRR overexpression as well as Sox2 knockdown or not. (c) The stemness markers’ protein levels were detected in cells with FENDRR overexpression as well as Sox2 knockdown or not. (d and e) Sphere number and size were determined in cells with FENDRR overexpression plus Sox2 knockdown or not. (f) ALDH activity was evaluated in cells with FENDRR overexpression as well as Sox2 knockdown or not. n ≥ 3, **P < 0.01 vs. control, ##P < 0.01 vs. FENDRR-oe
LncRNA FENDRR inhibits chemoresistance of colorectal cancer cells dependent on Sox2
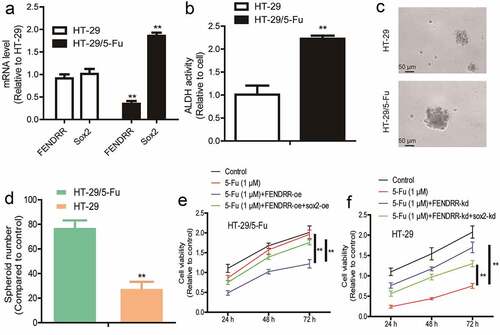
Finally, as CSCs can result in drug resistance of tumors, we explored the roles of the FENDRR/Sox2 axis in chemotherapeutic sensitivity. We initially detected the levels of FENDRR and Sox2 in 5-Fu resistant (HT-29/5-Fu) and sensitive (HT-29) colorectal cancer cells and found that HT-29/5-Fu exhibited a lower level of FENDRR and higher level of Sox2 compared to HT-29 cells, respectively (). Notably, HT-29/5-Fu displayed a stronger CSC-like trait than HT-29 cells, as evident by the increase of ALDH activity, sphere-formation ability (). Then, FENDRR was overexpressed in HT-29/5-Fu cells as well as Sox2 overexpression or not, it was shown that FENDRR overexpression attenuated 5-Fu resistance in HT-29/5-Fu cells, which was rescued by Sox2 overexpression (). Furthermore, FENDRR was knocked down in HT-29 cells as well as Sox2 knockdown or not, it was identified that FENDRR knockdown decreased 5-Fu sensitivity in HT-29 cells, which was partially abrogated by Sox2 knockdown (). Taken together, our results suggest that the FEDNRR/Sox2 axis suppresses the CSC-like traits and thus drug resistance in colorectal cancer cells.
Figure 6. LncRNA FENDRR confers chemoresistance of colorectal cancer cells dependent on Sox2. (a) The mRNA levels of FENDRR and Sox2 were detected in HT-29/5-Fu and HT-29 cells. (b) ALDH activity was measured in HT-29/5-Fu and HT-29 cells. (c and d) Sphere number and size were determined in HT-29/5-Fu and HT-29 cells. (e) HT-29/5-Fu cells with FENDRR knockdown as well as Sox2 overexpression or not were subjected to cell viability detection. (f) Cell viability was examined in HT-29 cells with FENDRR overexpression plus Sox2 knockdown or not. n ≥ 3, **P < 0.01 vs. control
Discussion
Here, to explore the roles of lncRNA FENDRR in the CSC-like traits of colorectal cancer cells, the spheres formed by colorectal cancer cells through 3D non-adherent culture, which has been shown to enrich CSCs in cancer cells, were collected as a colorectal CSC model [Citation17]. We found that FENDRR was lowly expressed in spheres, this promotes us to assume that FENDRR can suppress the stemness of colorectal cancer cells. Then, we performed the gain-of functions in spheres and loss-of functions in colorectal cancer cells, respectively. Through analyzing sphere-formation ability, detecting ALDH activity and stemness marker expression, it was found that FENDRR negatively regulated the CSC-like traits of colorectal cancer cells. So far, this work, for the first time, revealed the roles of FENDRR in the CSC-like traits of colorectal cancer cells.
LncRNAs have been revealed to function through different ways, such as acting as miRNA sponges, RNA or protein partners [Citation1,Citation18]. Additionally, a recent study indicates that lncRNA can encode a small peptide, through which lncRNA suppresses colon cancer progression [Citation19]; A novel primate-specific long non-coding RNA (lncRNA), named FLANC, was identified to promote CRC cell metastasis [Citation20]. And a transcription coactivator, Yes-associated protein 1 (YAP1) – mediated regulation on lncRNA LINC00152 could promote the proliferation and metastasis of CRC cells [Citation21]. Furthermore, a recent work indicated that lncRNA FEZF1-AS1 could facilitate CRC cell proliferation and metastasis by activating STAT3 signaling [Citation22]. In this work, we revealed that lncRNA FENDRR directly interacted with Sox2 mRNA, a critical stemness regulator, and thus decreased Sox2 mRNA stability and expression, this effect of FENDRR on Sox2 mRNA is similar with that of miRNAs on transcripts. Notably, FENDRR has been shown to exert similar inhibitory effects on MDR1 3’UTR mRNA through competitively binding to MDR1 mRNA with RNA binding protein HuR [Citation12,Citation13], these results remind us to investigate which areas of Sox2 mRNA were bound by FENDRR. We then performed luciferase reporter assay combined with RNA–RNA interaction in vitro analysis, and revealed that lncRNA FENDRR directly binds to Sox2 mRNA 3’UTR. However, it is still unclear whether RNA binding proteins are involved in FENDRR-mediated effects on Sox2 mNRA stability, such as RNA binding protein HuR, this should be explored in the future. In addition, we found that lncRNA FENDRR directly interacted with Sox2 mRNA, not Oct4 and KLF4, the other stemness regulators, and FENDRR regulated the expression of Oct4 and KLF4 in a Sox2-dependent manner, this is consistent with the previous studies showing that Sox2 is necessary for CDK1- and HIF1α/HIF2α-induced effects on other stemness marker expression [Citation23,Citation24]. Moreover, the previous studies have demonstrated that FENDRR is necessary for mammalian embryogenesis [Citation25,Citation26], the procedure of which is similar with that of CSCs, this effect further strengthens the inhibitory effects of FENDRR on the CSC-like traits of colorectal cancer cells. Notably, lncRNA FENDRR has been shown to repress the protein expression of Sox4, which is another stemness marker and belongs to the same family as Sox2; thus, we wonder whether FENDRR can suppress the CSC-like traits of colorectal cancer cells through repressing Sox4 protein, this could be investigated in the future. Furthermore, the effects of FENDRR on the protein stability of Sox2 are still unclear.
Conclusion
All in all, although in vivo experiments are needed, this study reveals a novel FENDRR/Sox2 axis necessary for the CSC-like traits and chemoresistance of colorectal cancer cells, which might be a novel biomarker for colorectal cancer and chemotherapeutic efficiency.
Contributors’ statement
Feng Ye and Chunyue Wang designed the research. Xin Zhao, Jincheng Wu, and Yongwen Li performed the research. Xin Zhao and Jincheng Wu wrote the manuscript.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Yang L, Chen Y, Liu N, et al. Low expression of TRAF3IP2-AS1 promotes progression of NONO-TFE3 translocation renal cell carcinoma by stimulating N(6)-methyladenosine of PARP1 mRNA and downregulating PTEN. J Hematol Oncol. 2021;14:46.
- Xue J, Zhong S, Sun BM, et al. Lnc-THOR silencing inhibits human glioma cell survival by activating MAGEA6-AMPK signaling. Cell Death Dis. 2019;10:866.
- Zheng YJ, Zhao JY, Liang TS, et al. Long noncoding RNA SMAD5-AS1 acts as a microRNA-106a-5p sponge to promote epithelial mesenchymal transition in nasopharyngeal carcinoma. FASEB J. 2019;33:12915–12928.
- Liao Z, Nie H, Wang Y, et al. The emerging landscape of long non-coding RNAs in colorectal cancer metastasis. Front Oncol. 2021;11:641343.
- Li S, Lin L. Long noncoding RNA MCF2L-AS1 promotes the cancer stem cell-like traits in non-small cell lung cancer cells through regulating miR-873-5p level. Environ Toxicol. 2021;36:1457–1465.
- Reya T, Morrison SJ. Clarke MF and Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111.
- Zheng A, Song X, Zhang L, et al. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2019;38:305.
- Zhan Y, Chen Z, He S, et al. Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol Cancer. 2020;19:25.
- Shin VY, Chen J, Cheuk IW, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10:270.
- Zheng Q, Zhang Q, Yu X, et al. FENDRR: a pivotal, cancer-related, long non-coding RNA. Biomed Pharmacother. 2021;137:111390.
- Cheng C, Li H, Zheng J, et al. FENDRR sponges miR-424-5p to inhibit cell proliferation, migration and invasion in colorectal cancer. Technol Cancer Res Treat. 2020;19:1533033820980102.
- Zhang F, Ni H, Li X, et al. FENDRR attenuates adriamycin resistance via suppressing MDR1 expression through sponging HuR and miR-184 in chronic myelogenous leukaemia cells. FEBS Lett. 2019;593:1993–2007.
- Gong F, Dong D, Zhang T, et al. Long non-coding RNA FENDRR attenuates the stemness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR. Eur J Pharmacol. 2019;853:345–352.
- Zeng R, Wang C, Wang W, et al. Long non-coding RNA DUXAP9 promotes hepatocellular carcinoma cell stemness via directly interacting with sox9. Environ Toxicol. 2021;36:1793–1801.
- Song H, Xu Y, Shi L, et al. THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed Pharmacother. 2018;108:338–346.
- Gupta PB, Onder TT, Jiang G, et al. Weinberg RA and Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659.
- Guo Q, Wang T, Yang Y, et al. Transcriptional factor Yin Yang 1 promotes the stemness of breast cancer cells by suppressing miR-873-5p transcriptional activity. Mol Ther Nucleic Acids. 2020;21:527–541.
- Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12:81.
- Huang JZ, Chen M, Chen D, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68:171–184.e176.
- Pichler M, Rodriguez-Aguayo C, SY N, et al. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut. 2020;69:1818–1831.
- Ou C, Sun Z, He X, et al. Targeting YAP1/LINC00152/FSCN1 signaling axis prevents the progression of colorectal cancer. Adv Sci (Weinh). 2020;7:1901380.
- Bian Z, Zhang J, Li M, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24:4808–4819.
- Wang P, Zhao L, Gong S, et al. HIF1α/HIF2α-Sox2/Klf4 promotes the malignant progression of glioblastoma via the EGFR-PI3K/AKT signalling pathway with positive feedback under hypoxia. Cell Death Dis. 2021;12:312.
- Huang Z, Shen G, Gao J. CDK1 promotes the stemness of lung cancer cells through interacting with Sox2. Clin Transl Oncol. 2021;23:1743–1751.
- Szafranski P and Stankiewicz P. Long non-coding RNA FENDRR: gene structure, expression, and biological relevance. Genes (Basel). 2021;12:177.
- Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579–1585.
