ABSTRACT
Schisandrin A (SchA) has been reported to have good anti-cancer effects. However, its anti-cancer mechanism in breast cancer remains unknown. This study aimed to explore the mechanism of SchA in breast cancer treatment using bio-informatics analysis and in vitro experiments. The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx), Gene Cards, and PharmMapper databases were used to screen the candidate targets of SchA against MDA-MB-231 cells selected as the tested cell line through MTT analysis. The functions and pathways of the targets were identified using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and further analyzed using DAVID 6.8.1 database. Network pharmacology analysis revealed 77 candidate targets, 31 signal pathways, and 208 GO entries (P < 0.05). The targets regulated serine-type endopeptidase and protein tyrosine kinase activities, thereby promoting the migration and inhibiting the apoptosis of MDA-MB-231 cells. Comprehensive analysis of the ‘Protein–Protein Interaction’ (PPI) and ‘Component-Targets-Pathways’ (C-T-P) networks constructed using Cytoscape 3.7.1 software revealed four core targets: EGFR, PIK3R1, MMP9 and Caspase 3. Their docking scores with SchA were subsequently investigated through molecular docking. The wound healing, Hoechst 33342/PI, and western blot assays confirmed that SchA significantly down-regulated EGFR, PIK3R1, and MMP9, but up-regulated cleaved-caspase 3, thus inhibiting the migration and promoting the apoptosis of MDA-MB-231 cells. Reckoning the findings of the study, SchA is a potential adjuvant treatment for breast cancer.
Introduction
Breast cancer is the leading cancer affecting women and is characterized by high morbidity and mortality [Citation1]. There is an annual increase of about 2 million breast cancer patients and 685 thousand deaths attributed to the cancer. Modern treatment measures have not significantly improved the overall survival and prognosis of breast cancer patients. Therefore, there is an urgent need for high-efficiency and low toxicity drugs for breast cancer.
The remarkable curative effects of traditional Chinese medicines (TCMs) against breast cancer have attracted researchers’ attention in recent years [Citation2]. For instance, SchA, an important bio-active lignan extracted from Schisandra chinensis (Turcz.) Baill possesses inhibitory effects on the growth of ovarian cancer by regulating endoplasmic reticulum stress [Citation3,Citation4]. Moreover, it can reverse the multidrug resistance in human osteosarcoma and colon cancers [Citation5]. But how SchA affects the progress of breast cancer remains unclear.
Significant progress has been made in studying the pathogenesis of cancers with the development of bioinformatics. Messenger RNAs (mRNAs) play an important role in the occurrence and development of numerous cancers, including breast cancer [Citation6]. Identifying the differentially expressed genes is an important route to analyze the mechanisms of drug resistance in breast cancer. Researchers have identified differentially expressed genes in emphysema, atherosclerosis, and lung squamous cell carcinoma from clinical samples in the GEO and TCGA databases, thus providing invaluable insights for their clinical treatment [Citation7–9]. The network pharmacology concept is consistent with the characteristics of TCMs, which involve treating diseases through multi-components, multi-targets, and multi-pathways. Combining network pharmacology with molecular docking, R software, and other bio-informatics tools is thus suitable for studying the active components and mechanism of TCMs in disease treatment [Citation10].
This study aimed to identify the mechanism of SchA in breast cancer treatment. The cell metabolic activity of the tested cell lines was determined using the MTT assay, and the candidate targets of SchA against them were subsequently analyzed using bio-informatics tools. The core targets of SchA were obtained after analyzing the targets’ biological functions and their relationships with SchA. The results of these analyses were subsequently confirmed using AutoDock4 and PyMOL tools. The core genes were deemed to play essential roles in the mechanisms of SchA against the MDA-MB-231 cells. In vitro assays further revealed that SchA significantly regulated the core targets, inhibited migration, and promoted the apoptosis of MDA-MB-231 cells. The graphical abstract accentuates the work of this study.
Material and methods
Cell culture
Breast cancer cell lines, MCF-7, and MDA-MB-231 were obtained from Wuhan University (Wuhan, China). The cells were cultured in Dulbecco’s modified Eagle medium basic (DMEM, 1×, Gibreast cancero, MD, USA) supplemented with 10% fetal bovine serum (FBS) (Gibreast cancero, MD, USA), and 100 U/mL penicillin G, and 100 µg/mL of streptomycin. The culture was maintained at 37°C in an incubator (DHP-9270, Shandong OLABO Instrument Equipment Co., Ltd, China) under 5% CO2.
Cell proliferation assay
The MCF-7 and MDA-MB-231 cells were incubated in 96-well plates for 24 h at a density of 8.0 × 103 cells per well. Once the cells had adhered to the well walls, they were treated with different concentrations of SchA (0, 10, 20, 40, 80, and 160 µM) for 24 h. After that, 20 µL MTT (Sigma, MO, USA) was added to each well. After 4 h, the culture medium in each well was sucked away. The formazan product was dissolved in 150 µL dimethyl sulfoxide (DMSO) (Sigma) and stirred for 10 min on a QB-9002 microporous quick shaker (Beyotime Biotechnology Co., Ltd, Shanghai, China). The absorbance was read at 490 nm using a Thermo scientific varioskan LUX microplate reader (Thermo, Shanghai, China).
Schisandrin A information collection
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (http://lsp.nwu.edu.cn/tcmsp.php) collects the pharmacokinetics parameters of numerous components and provides valuable reference for the research and development of new drugs [Citation11]. Pubchem database (https://pubchem.ncbi.nlm.nih.gov/), Pubreast cancerhem CID, and Canonical SMILES contains common names of components, and data on various components, which helps to ensure correct identification of components [Citation12]. SwissADME (http://www.swissadme.ch/index.php) and pkCSM (http://biosig.unimelb.edu.au/pkcsm/prediction) databases contain many ADMET component parameters, which have been confirmed through various experiments [Citation13,Citation14]. It lays a foundation for ADMET parameters analysis of components by the algorithm themselves [Citation15]. This study used these four databases to collect information on SchA, hence analyzing its ADMET parameters.
Schisandrin A-related targets
The SwissTargetPrediction database (https://labworm.com/tool/swisstargetprediction) contains more than 3,000 proteins of 370,000 known active drugs [Citation16]. PharmMapper (http://www.lilab-ecust.cn/pharmmapper/submitfile.html) database identifies potential targets of candidate components by pharmacophore mapping [Citation17]. Consequently, the 3D structure of SchA was imported into both of databases to match the related targets after setting the species as human. The Retrieve/ID mapping of Uniprot (https://www.uniprot.org/) was used to obtain the Gene official symbol formats of the targets [Citation18].
Candidate targets of Schisandrin A against breast cancer
TCGA (https://portal.gdc.cancer.gov/) database collects various clinical data on 33 types of cancer [Citation19]. GTEx (https://gtexportal.org/) is a specialized database containing information on normal human clinical samples [Citation20]. The RNA sequencing data of 1,480 breast cancer samples and 572 normal samples were downloaded from the TCGA and GTEx databases. All the samples were classified and compared by R software where ggplot2. P < 0.05 was considered statistically significant. Limma package was used to study the differentially expressed mRNAs with the thresholds that adjusted P < 0.05 and |Log (Fold Change)| >1. Gene Cards database (https://www.genecards.org/) includes comprehensive information on human genes sourced from 150 web sources [Citation21]. Drug Bank (https://www.drugbank.ca/) is a professional tool for analyzing the interactions between drugs and proteins [Citation22]. The breast cancer-related genes were obtained from these two databases by searching for ‘breast cancer’. Venn 2.1.0 database (https://bioinfogp.cnb.csic.es/tools/venny/) was used to obtain the overlapping targets for them [Citation23].
GO Entries and KEGG pathways enrichment
The candidate targets of SchA against breast cancer were imported into the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/), which is widely used for gene function annotations and enrichment [Citation24]. The Identifier as ‘GENE OFFICIAL SYMBOL’ and species as ‘Homo sapiens’ were selected for GO entries and KEGG pathways enrichment. In order to intuitively visualize enrichment results, the Omicshare database (http://www.omicshare.com) was used to plot the top ten KEGG pathways and GO entries (P < 0.05) as bubble plot and circus, respectively [Citation25].
Construction of ‘C-T-P’ network
Cytoscape 3.7.1 software (https://cytoscape.org/download.html) is an analytical tool frequently used to visualize the network constructed by bio-informatics [Citation26]. A ‘C-T-P’ network was constructed by Cytoscape 3.7.1 to analyze the association among SchA, candidate targets, and the top ten KEGG pathways. After the analysis of each node, the critical candidate targets were selected according to their degrees.
Construction of ‘PPI’ network and acquisition of core targets
STRING database (https://string-db.org/) contains over than 52 million proteins, and has open access to analyze their interactions [Citation27]. The PPI network of 77 targets was created using STRING and Cytoscape tools, which helped decipher their relationship [Citation28–30]. Cytoscape 3.7.1 software was used to download the TSV format of the ‘PPI’ network for visualization analysis. According to the median principle, the node with a higher degree value than the median is considered an essential target. In order to ensure the accuracy of this study, the CytoHubba plug-in was used to screen the top 20 targets according to their degrees. Based on the results from the ‘PPI’ and ‘C-T-P’ networks, the potential core targets of SchA against breast cancer were obtained.
Molecular docking analysis
The potential core targets were imported into the RSCBPDB (https://www1.rcsb.org/) database to download the PDB formats of their structures [Citation31]. Then, the 3D structure of SchA was downloaded from the Pubchem database, and its energy was minimized by Chem Draw software [Citation32].
AutoDock4 and PyMOL are used for visualization analysis of molecular docking using the principle of semi-flexibility [Citation33]. Therefore, processed SchA structures and targets were imported into AutoDock4 and PyMOL software to be hydrogenated and charged. The docking conformations were set to 20, and the molecular docking results were classified according to the clusters. Then, the conformation with the lowest energy was selected for analysis. Via PyMOL software, the 3D docking results of SchA and its core targets were created. The amino acid residues, functional groups and hydrogen bonds were displayed in different colors and shapes. In order to make the docking results more reliable, the length of hydrogen bond was set below 3 nm, and the docking scores threshold was −4 Kcal/mol. Comprehensive analysis from the above results created the hub targets of SchA against breast cancer.
Wound healing assay
Logarithmic growth phase MDA-MB-231 cells were inoculated into 6-well plates at 6.0 × 104 cells/well. After the cells had adhered to the walls, a 1 mL pipette tip was used to mark the bottom of each well gently. Then, the floating cells were moderately washed with PBS, and each well photographed under a fluorescence microscope. The MDA-MB-231 cells were thereafter preincubated with SchA (0, 40 and 80 µM) for 24 h, photos were taken under fluorescence microscope. Changes in the scratch area of the cells were analyzed and compared.
Hoechst 33342/PI staining assay
Logarithmic growth phase MDA-MB-231 cells in logarithmic growth phase were inoculated into 6-well plates at 6.0 × 104 cells/well. After the cells had adhered to the walls, they were incubated in different concentrations of SchA (0, 40 and 80 µM) for 24 h. After this, the cells were fixed with 4% polyoxymethylene, washed 3 times with PBS, incubated in 10 µg/mL Hoechst 33342 (Beyotime) staining solution for 15 min at 4 °C. Subsequently, 1 mL PI (30 µg/mL) was added to each well for 15 minutes at room temperature. Cells were washed twice with PBS and observed using a fluorescence microscope. Image J software was used to analyze the results.
Western blot assay
The MDA-MB-231 cells were seeded in 6-well plates at a concentration of 6.0 × 104 cells/well and treated with SchA (0, 40 and 80 µM) for 24 h. RIPA lysis kit (Beyotime) and protein analysis kit (Takara, Kyoto, Japan) were used for cell lysis and total protein quantification analysis, respectively. The total protein was then separated on 10% SDS-PAGE gel electrophoresis and afterward transferred onto a nitrocellulose membrane by electroblotting (Millipore, Billerica, MA, USA). After blocking the membrane using 5% BSA for 2 h at room temperature, it was incubated overnight at 4°C with primary antibodies (p-EGFR, PIK3R1, Cleaved-caspase 3, AKT1, p-AKT1 and MMP9), in a dilution of 1:1000, (Cell Signaling Technology, Danvers, MA, USA). They were then incubated with secondary antibodies at room temperature for 1 h. Later, the membrane was washed in three tris buffered saline + Tween (TBST) cycles for 10 minutes. The proteins were then visualized by adding ECL (Thermo) and subsequently scanned and imaged using a FluorChem FC 3 system (ProteinSimple, USA). Finally, image J software was used to analyze the results.
Statistical analysis
Statistical significance was assessed using the students’ t-test. All statistical analyses were performed using SPSS 22.0 software (IBM, USA). Data were expressed as the mean ± SD. All experiments were repeated in triplicate and P < 0.05 was set as a significant threshold.
Results
This study aimed to explore the mechanism of SchA against breast cancer via bio-informatics analysis and in vitro experimentations. Based on the data from TCGA, GTEx, Gene cards and Drug bank databases, differentially expressed genes associated with breast cancer were obtained through the R software. By analyzing the ‘C-T-P’ and ‘PPI’ networks constructed by Cytoscape 3.7.1, we hypothesized that EGFR, PIK3R1, MMP9 and Caspase 3 were the core genes of SchA against breast cancer. Molecular docking results showed that SchA had increased affinity binding to the core genes. In vitro experiments also verified that SchA significantly regulated the expression of core targets, besides inhibiting the migration and promoting the apoptosis of MDA-MB-231 cells.
Schisandrin A decreased proliferation of MCF-7 and MDA-MB-231 cells
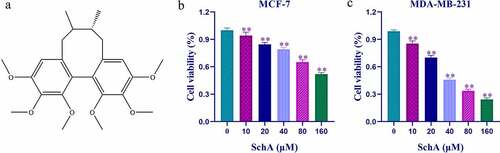
MTT analysis showed a significant decrease in cell viability of the treatment groups compared to the control group (P < 0.01) as the concentrations of SchA increased. Therefore, SchA inhibited the proliferation of MCF-7 and MDA-MB-231 cells in a concentration-dependent manner (). It was noteworthy that SchA had a better inhibitory effect on MDA-MB-231 than MCF-7 cells, and the half-maximal inhibitory concentration (IC 50) values for them were 26.6092 µM and 112.6672 µM respectively.
Schisandrin A information data
The TCMSP, Pubreast cancerhem, SwissADME, and pkCSM databases were used to establish an information table of SchA. The two databases with the essential information, Pubchem CID and Canonical SMILES were used to identify the ADMET parameters and potential targets of SchA. The parameters of ADMET revealed that SchA provided good pharmacokinetics properties in vivo. Absorption and distribution parameters confirmed that SchA is easily absorbed orally and utilization, and evenly distributed in the body. CNS and total clearance analysis confirmed its metabolism in vivo. Toxicological study results showed that SchA was nontoxic to skin and liver, and the max accelerated human dose was 0.058 (log mg/kg/day). Based on these characteristics, SchA can be developed into a drug. All the SchA data is shown in , and its structural formula is shown in ).
Table 1. Information about SchA
Acquisition of candidate targets
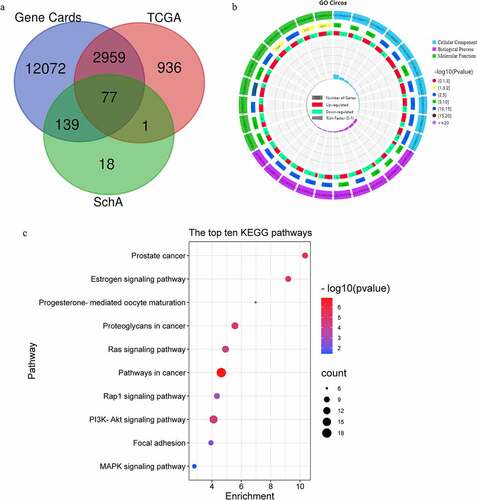
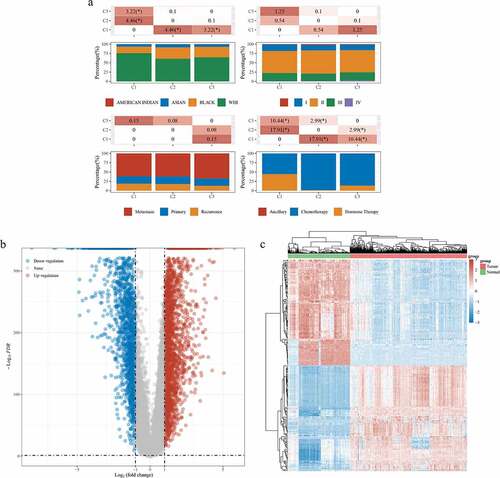
The results in ) show that there were significant differences in the races, cancer stage and treatment method among different groups of patients, which confirms the randomness of the data obtained in this study. A total of 3,937 differently expressed genes were obtained, including 2,330 up-regulated genes and 1,643 down-regulated genes (). Using the Venn 2.1.0 database, 15,247 genes in Gene Cards, 3,973 genes in TCGA and 235 genes related to SchA were compared and analyzed. Finally, 77 candidate targets of SchA against breast cancer were obtained, including 40 up-regulated targets and 37 down-regulated targets ()). The specific information of the candidate targets is shown in .
Table 2. Candidate targets of SchA against breast cancer
Figure 2. Differentially expressed targets in breast cancer and normal samples from TCGA and GTEx databases. (a) Information classification and comparison of 1480 breast cancer samples in TCGA. (b) Volcano plot of 3937 differentially expressed targets. (c) Microarray map of the top 100 differentially expressed targets
Functional enrichment analysis of candidate targets
Using the DAVID 6.8 database, 31 KEGG pathways (68, 88.3%), 130 GO BP (77, 100%) entries, 24 GO CC (77, 100%) entries and 54 GO MF (77, 100%) entries were obtained (P < 0.05). It confirmed that almost all 77 candidate targets were widely involved in the GO entries and KEGG pathways. 18 of these pathways were directly linked to breast cancer. The visualization results of the Omicshare database showed that these targets regulated serine-type endopeptidase and protein tyrosine kinase activities, thus promoting the migration and apoptosis inhibition of MDA-MB-231 cells ). These results suggested that SchA against breast cancer by regulating the targets related to cell migration and apoptosis. However, these findings need to be validated using in vitro experiments.
Construction and analysis of ‘PPI’ and ‘C-T-P’ networks
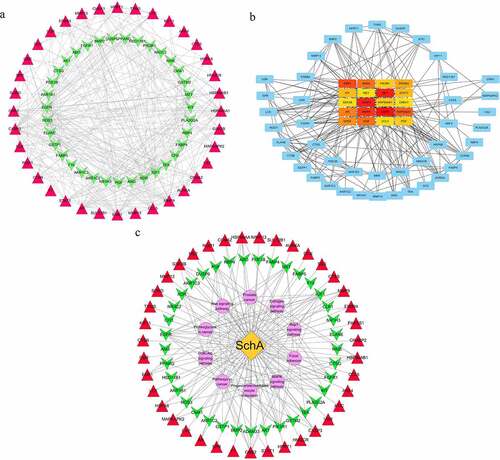
By STRING and Cytoscape 3.7.1 databases, a ‘PPI’ network with 71 nodes and 355 edges was obtained to analyze the interaction between the 77 candidate targets ()). The weak correlation between 6 of these targets and other 71 targets caused them not to be visualized in this network. Via CytoHubba analysis, the top 20 targets were further analyzed, where their colors were directly proportional to their degrees ()). In order to ensure the accuracy of core targets, a ‘C-T-P’ network with 87 nodes and 177 edges was constructed ()). The yellow node in the network represents SchA, purple nodes represent KEGG pathways, while red and green nodes represent different targets. By comprehensively comparing the degrees of the top 20 targets in ‘C-T-P’ and ‘PPI’ networks, 16 overlapping targets were obtained. Based on the median principle, the degrees of EGFR (degree = 45), MMP9 (degree = 41), PIK3R1 (degree = 36), caspase 3 (degree = 33), HSP90AA1 (degree = 32), IGF1 (degree = 30), GRB2 (degree = 37) and KDR (degree = 37) were greater than the median (degree = 25). Therefore, they were the SchA core targets.
Molecular docking and analysis
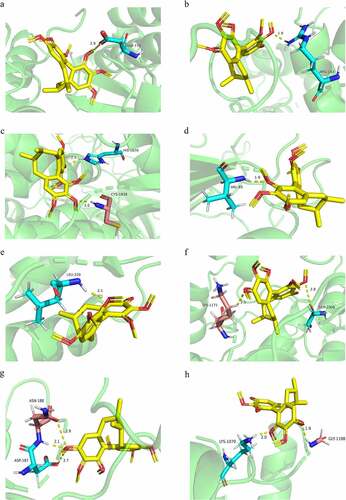
Based on the ‘PPI’ and ‘C-T-P’ networks, the core targets were screened and docked with SchA. Specific docking scores, functional groups, and amino acid residues are shown in . The docking scores of SchA with all targets were less than the threshold, which confirmed that it had a high binding affinity with them. Comprehensive analysis of the above results, EGFR, MMP9, PIK3R1, and Caspase 3 were ranked as the top 4 in each stage of bio-informatics mining. Therefore, they were more closely related to SchA and breast cancer. As is expected, tyrosine kinase receptor EGFR was involved in the proliferation, angiogenesis, invasion, metastasis and apoptosis of cancer cells. Phosphorylated EGFR significantly up-regulates the expression of PIK3R1 and MMP9, thus promoting the migration of breast cancer cells. Caspase 3, the core protein in apoptosis, and its critical interaction with SchA stipulate that it could enhance the apoptosis ability of breast cancer cells. Though these results were consistent with previous GO and KEGG enrichment analyses, they need to be verified. The PyMOL software was used to envisage the chemical formula of SchA and the proteins’ amino acid residues, visualizing them as rods with different colors. Besides, the length of all the hydrogen bonds was less than 3 nm, which confirmed the accuracy of the docking results (). However, these results still needed to be verified by molecular dynamics simulation experiments.
Table 3. Docking scores and bonds of the proteins and SchA
Schisandrin A inhibited the migration of MDA-MB-231 cells
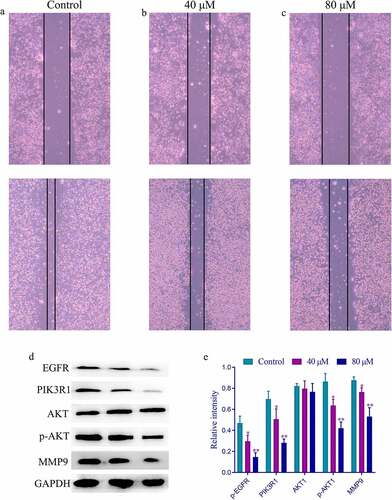
Based on network pharmacology and molecular docking results, the effect of SchA on the expression of the four hub targets needed exploration. These targets were involved in EGFR-PI3K-AKT-MMP9 pathway, widely reported to be associated with the migration and apoptosis of cancer cells. Herein, the wound healing assay was used to investigate the effect of SchA on the migration ability of MDA-MB-231 cells based on the MTT assay. Compared with the control group, the migration of MDA-MB-231 cells treated with different concentrations of SchA (40 µM and 80 µM) was significantly inhibited in a dose-dependent manner (). Western blot analysis showed a significant decrease in the expression of EGFR, PIK3R1, p-AKT, and MMP9 in the groups treated with different concentrations of SchA in comparison to the control group (*P < 0.05 or **P < 0.01). However, no significant difference was observed in the expression of the total AKT between each group. Therefore, SchA inhibited the migration of breast cancer cells by regulating the expression of these targets in the EGFR-PIK3R1-AKT-MMP9 pathway.
Figure 6. The wound healing and western blot results of different concentrations of SchA on the migration of MDA-MB-231 cells. (a) The wound healing results of the control group. (b) The wound healing results of the group treated with 40 µM SchA. (c) The wound healing results of the group treated with 80 µM SchA. (d) The western blot results of these targets. (e) The calculated gray values of these targets
Schisandrin A promoted apoptosis in MDA-MB-231 cells
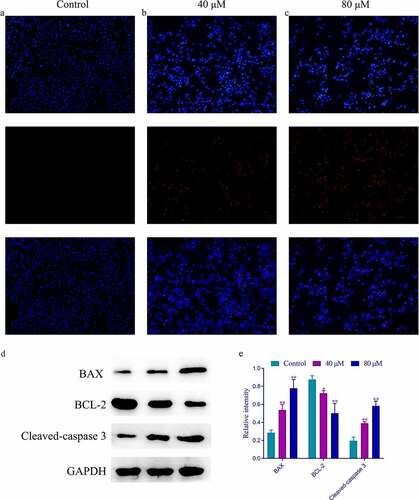
To identify whether SchA promoted apoptosis in MDA-MB-231 cells, the cells were preincubated with SchA for 24 h. The cells in each well were then stained with Hoechst 33342/PI and observed using a fluorescence microscope (magnification, ×200). Compared with the control group, apoptosis in cells treated with different concentrations of SchA (40 µM and 80 µM) was markedly increased (). BAX and BCL-2, as key downstream proteins of PIK3R1-AKT pathway, play important roles in the apoptosis of breast cancer cells. Many studies have confirmed the close relationship between these proteins and Caspase 3. Therefore, the effect of SchA on the expression of these targets was investigated through western blot assay. Compared to the control group, cleaved-caspase 3 and BAX expression increased in the groups treated with SchA in a dose-dependent manner (**P < 0.01). And a significantly decreased expression of BCL-2 was observed in the groups treated with different concentrations of SchA when compared with the control group (*P < 0.05 or **P < 0.01). Therefore, SchA promoted the apoptosis of MDA-MB-231 cells and regulated the expression of caspase 3, BAX, and BCL-2.
Figure 7. The Hoechst 33342/PI and western blot results of different concentrations of SchA on the apoptosis of MDA-MB-231 cells. (a) The Hoechst 33342/PI results of the control group. (b) The Hoechst 33342/PI results of the group treated with 40 µM SchA. (c) The Hoechst 33342/PI results of the group treated with 80 µM SchA. (d) The western blot results of BAX, BCL-2 and Cleaved-caspase 3. (e) The calculated gray values of these 3 targets
Discussion
Breast cancer has huge negative impacts on the life of women across the world because of its high incidence and mortality rates attributed to a lack of effective drugs and preventive measures. Numerous TCMs have been widely used in cancer prevention and treatment in China [Citation34]. SchA is the main active component of Schisandra chinensis (Turcz.) Baill and possesses good inhibitory effects on colon and breast cancer. However, the mechanism of SchA against breast cancer remains largely unknown. Integrated bioinformatics analysis is an important route of identifying core genes and pathways involved in disease pathogenesis [Citation35,Citation36]. Network pharmacology and molecular docking have been widely used in the research and development of new drugs in recent years [Citation37]. This study aimed to explore the mechanism of SchA against breast cancer through integrated bio-informatics analysis and in vitro experiments.
Based on ADMET parameters, SchA is a natural component with high oral bio-availability and drug-like properties and no toxicity to the liver and skin. It thus possesses the potential to be developed as a drug. Network pharmacology analysis of SchA revealed that it regulated the expression of multiple targets and affected multiple pathways against breast cancer. The primary biological function of these targets was to regulate the migration and apoptosis of cancer cells, which was consistent with the KEGG pathway analysis results. Several databases, including STRING and Cytoscape 3.7.1, were used to construct ‘PPI’ and ‘C-T-P’ networks to screen the core targets. Comprehensive analysis of the networks revealed EGFR (degree = 45), MMP9 (degree = 41), PIK3R1 (degree = 36), Caspase 3 (degree = 33), HSP90AA1 (degree = 32), IGF1 (degree = 30), GRB2 (degree = 37) and KDR (degree = 37) conformed to the median rule and were the core targets of SchA against breast cancer. These findings were further confirmed by those of molecular docking, thus strongly suggesting that EGFR, MMP9, PIK3R1 and Caspase 3 were the core targets of SchA against breast cancer.
Numerous of studies postulate that EGFR, MMP9, PIK3R1, and Caspase 3 are widely involved in regulating tumor cell migration and apoptosis [Citation38]. EGFR is a transmembrane receptor and the primary receptor of EGF [Citation39]. A synergy of EGFR and growth factors affects many downstream pathways, including the PI3K-AKT pathway, which promotes proliferation and reduces the apoptosis of breast cancer cells [Citation40]. This report is consistent with the GO enrichment analysis of EGFR. In this study, SchA docked with several amino acid residues of EGFR. Moreover, different concentrations of SchA significantly reduced the expression of EGFR in MDA-MB-231 cells, thus making EGFR a hub target of SchA against breast cancer. Heterodimer of a regulatory subunit PIK3R1 and α p110 catalytic subunit via SH2 domains, which interacts with CCDC88A/GIV 25. The interaction enables PIK3R1 to the EGFR receptor, enhancing PI3K activity and promoting the phosphorylation of AKT and MMP9. In addition, MMP9 promotes the release of growth factors, thus activating the MAPK and PI3K/AKT pathways that activate MMP9 and MMP13 by regulating the negative feedback [Citation41]. Activated MMP9 degrades the tumor basement membrane and ECM, thus promoting the formation of blood vessels and migration of tumor cells. This phenomenon is attributed to the MMP9 strong relationship between MMP9 and VEGF activation. Western blot analysis results further revealed that SchA significantly decreased the expression of PIK3R1, p-AKT, and MMP9 in a dose-dependent manner. The three proteins: PIK3R1, p-AKT, and MMP9 affected the migration of the MDA-MB-231 cells by activating the PI3K/AKT pathway. However, SchA had a good inhibitory effect against them. These results were confirmed by the wound healing assay.
Apoptosis is a process of programmed cell death that is closely related to cell morphology and biochemical changes, including endogenous and exogenous pathways [Citation42]. The apoptosis of tumor cells is inhibited. Caspase 3 is an important terminal splicing enzyme in the apoptosis process and part of the CTL cell killing mechanism. Oligomerized BAX is an important downstream protein of the PIK3R1-AKT-MMP9 pathway that forms an oligomer with BCL-2 on the mitochondrial membrane, thereby enhancing the permeability of the mitochondrial membrane and release of cytochrome C [Citation43]. Moreover, it also activates the cascade reaction of the caspase protease family, leading to the apoptosis of cancer cells. Exploring the effect of drugs on the expression of BAX, BCL-2, and cleaved-caspase 3 is thus an important way to judge whether drugs can promote apoptosis of cancer cells. In this study, Hoechst 33342/PI staining and western blot assays revealed that SchA decreased the expression of BCL-2 but increased the expression of BAX and cleaved-caspase 3, thereby promoting the apoptosis of the MDA-MB-231 cells in a dose-dependent manner.
Reckoning the study findings, SchA is a potential adjuvant treatment for breast cancer based on its ADMET parameters. Nonetheless, these results should be verified further using in vivo experiments.
Conclusion
This study reveals that SchA reduces the activation of EGFR, PIK3R1, and MMP9 and increases the expression of cleaved-caspase 3, thereby inhibiting the migration and increasing the apoptosis of MDA-MB-231 cells. Reckoning the study findings, SchA is a potential adjuvant treatment for breast cancer based on its ADMET parameters. Nonetheless, its specific effect and mechanism on breast cancer should be verified further using in vivo experiments.
Research highlights
SchA has a better inhibitory effect on MDA-MB-231 than MCF-7 cells.
SchA inhibits the migration but promotes the apoptosis of MDA-MB-231 cells.
EGFR, PIK3R1, MMP9 and Caspase 3 are the core targets of SchA against MDA-MB-231 cells.
Abbreviations
SchA: Schisandrin A; TCGA: Cancer Genome Atlas; GTEx: Genotype-Tissue Expression; GO: Gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: Protein–Protein Interaction; C-T-P: Component-Targets-Pathways; mRNAs: Messenger RNAs; TCMs: Traditional Chinese medicines; TCMSP: Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform; DAVID: Database for Annotation, Visualization and Integrated Discovery
Authors’ contributions
All authors participated in the stages from study design to submission of the paper for publication. Note that Ling Chen and Xu-Ying Huang played a more important role than other authors because they were responsible for most of the tasks.
Acknowledgements
This study was supported by the Research projects of clinical medicine in Wuhan in 2017 [WZ17B03] and [WZ17D05]. Thank you to all who had contributed to this research.
Data availability statement
As we are currently conducting experiments on this component, so we can’t disclose these data for the time being. Please understand our situation. Thank you.
Disclosure statement
All authors declare that there is no conflict of interests.
Additional information
Funding
References
- Gote V, Nookala AR, Bolla PK, et al. Drug resistance in metastatic breast cancer: tumor targeted nanomedicine to the rescue. Int J Mol Sci. 2021;22(9). DOI:10.3390/ijms22094673
- Zhang H, Yan C, Wang Y. Exosome-mediated transfer of circHIPK3 promotes trastuzumab chemoresistance in breast cancer. J Drug Target. 2021;1–39. DOI:10.1080/1061186x.2021.1906882
- Gui Y, Yang Y, Xu D, et al. Schisantherin A attenuates sepsis-induced acute kidney injury by suppressing inflammation via regulating the NRF2 pathway. Life Sci. 2020;258:118161.
- Lee K, Ahn JH, Lee KT, et al. Deoxyschizandrin, isolated from schisandra berries, induces cell cycle arrest in ovarian cancer cells and inhibits the protumoural activation of tumour-associated macrophages. Nutrients. 2018;10(1). DOI:10.3390/nu10010091
- Kong D, Zhang D, Chu X, et al. Schizandrin A enhances chemosensitivity of colon carcinoma cells to 5-fluorouracil through up-regulation of miR-195. Biomed Pharmacothe. 2018;99:176–183.
- Wan J, Jiang S, Jiang Y, et al. Data mining and expression analysis of differential lncRNA ADAMTS9-AS1 in prostate cancer. Front Genet. 2019;10:1377.
- Udhaya Kumar S, Thirumal Kumar D, Bithia R, et al. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and bioinformatics approach. Front Genet. 2020;11:734.
- Fu D, Zhang B, Yang L, et al. Development of an immune-related risk signature for predicting prognosis in lung squamous cell carcinoma. Front Genet. 2020;11:978.
- Udhaya Kumar S, Madhana Priya N, Thirumal Kumar D, et al. An integrative analysis to distinguish between emphysema (EML) and alpha-1 antitrypsin deficiency-related emphysema (ADL)-A systems biology approach. Adv Protein Chem Struct Biol. 2021;127:315–342.
- Yuan C, Wang MH, Wang F, et al. Network pharmacology and molecular docking reveal the mechanism of Scopoletin against non-small cell lung cancer. Life Sci. 2021;270:119105.
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
- Kim S, Chen J, Cheng T, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49(D1):D1388–d1395.
- Daina A, Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–1121.
- Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717.
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–w364.
- Pires DE, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42(WebServer issue):W314–319.
- Alex B, Maria-JM, Sandra O, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–d489
- Wei L, Jin Z, Yang S, et al. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34(9):1615–1617.
- Carithers LJ, Ardlie K, Barcus M, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015;13(5):311–319.
- Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):W356–w360.
- Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–d1082.
- Zhou X, Zhang XF, Guo DY, et al. Exploring the mechanism of Lingzhu San in treating febrile seizures by using network pharmacology. Comb Chem High Throughput Screen. 2020. DOI:10.2174/1386207323666200902144348
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
- Wang S, Su W, Zhong C, et al. An eight-CircRNA assessment model for predicting biochemical recurrence in prostate cancer. Front Cell Dev Biol. 2020;8:599494.
- Zhang C, Zheng Y, Li X, et al. Genome-wide mutation profiling and related risk signature for prognosis of papillary renal cell carcinoma. Ann Transl Med. 2019;7(18):427.
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–d612.
- Udhaya Kumar S, Saleem A, Thirumal Kumar D, et al. A systemic approach to explore the mechanisms of drug resistance and altered signaling cascades in extensively drug-resistant tuberculosis. Adv Protein Chem Struct Biol. 2021;127:343–364.
- Udhaya Kumar S, Thirumal Kumar D, Siva R, et al. Dysregulation of signaling pathways due to differentially expressed genes from the B-cell transcriptomes of systemic lupus erythematosus patients - a bioinformatics approach. Front Bioeng Biotechnol. 2020;8:276.
- Mishra S, Shah MI, Udhaya Kumar S, et al. Network analysis of transcriptomics data for the prediction and prioritization of membrane-associated biomarkers for idiopathic pulmonary fibrosis (IPF) by bioinformatics approach. Adv Protein Chem Struct Biol. 2021;123:241–273.
- Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242.
- Cousins KR. Computer review of ChemDraw Ultra 12.0. J Am Chem Soc. 2011;133(21):8388.
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791.
- Wei MM, Zhao SJ, Dong XM, et al. A combination index and glycoproteomics-based approach revealed synergistic anticancer effects of curcuminoids of turmeric against prostate cancer PC3 cells. J Ethnopharmacol. 2021;267:113467.
- Kumar SU, Kumar DT, Siva R, et al. Integrative bioinformatics approaches to map potential novel genes and pathways involved in ovarian cancer. Front Bioeng Biotechnol. 2019;7:391.
- Yan H, Zheng G, Qu J, et al. Identification of key candidate genes and pathways in multiple myeloma by integrated bioinformatics analysis. J Cell Physiol. 2019;234(12):23785–23797.
- Ge H, Zhang B, Li T, et al. Potential targets and the action mechanism of food-derived dipeptides on colitis: network pharmacology and bioinformatics analysis. Food Funct. 2021;12:5989–6000.
- Guo L, Shi H, Zhu L. Siteng fang reverses multidrug resistance in gastric cancer: a network pharmacology and molecular docking study. Front Oncol. 2021;11:671382.
- McGovern AJ, Barreto GE. Network pharmacology identifies IL6 as an important hub and target of tibolone for drug repurposing in traumatic brain injury. Biomed Pharmacothe. 2021;140:111769.
- Kim DY, Kim SH, Yang EK. RNA interference mediated suppression of TRPV6 inhibits the progression of prostate cancer in vitro by modulating cathepsin B and MMP9 expression. Investig Clin Urol. 2021;62:447.
- Feng X, Xue F, He G, et al. Banxia xiexin decoction inhibits the expression of PD-L1 through multi-target and multi-pathway regulation of major oncogenes in gastric cancer. Onco Targets Ther. 2021;14:3297–3307.
- Hon WC, Berndt A, Williams RL. Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene. 2012;31(32):3655–3666.
- De Marco M, Falco A, Iaccarino R, et al. An emerging role for BAG3 in gynaecological malignancies. Br J Cancer. 2021;125:789–797.