ABSTRACT
Hepatocellular carcinoma (HCC) is one of the most prevalent malignant neoplasms with high relapse and mortality rate. It is of great importance to identify novel and effective molecular markers to predict prognosis for the treatment of HCC. The Ewing sarcoma breakpoint region 1 (EWSR1) gene is well known to fuse with various partner genes and involved in promoting the development of multiple sarcomas, especially the Ewing sarcoma family of tumors. Nevertheless, seldom studies have focused on the role of EWSR1 in cancers of epithelial origin, let alone in HCC. In the current study, the transcriptional and clinical data of EWSR1 in HCC patients were obtained from TCGA and GEO databases, as well as 124 cases from the department of Pathology of Sichuan Jianyang People’s Hospital. Kaplan-Meier and Cox regression analysis were used to assess patient prognosis. EWSR1 mRNA levels were significantly upregulated in HCC tissues than in normal liver tissues (P < 0.001). The TCGA database analysis showed upregulation of EWSR1 was associated with histological grade, pathologic T stage and death, in addition to that, the T staging, N staging, TNM staging, Ki67, AFP expression were extremely higher in the EWSR1 over-expression group in our cohort. Univariate and multivariate Cox hazard regression analysis results revealed that EWSR1 was an independent prognostic factor for OS in HCC, and bioinformatics analysis showed RNA splicing process represented the major function and pathway. In conclusion, our data showed EWSR1 could serve as a novel promising prognostic biomarker for HCC patients.
Abbreviations: AFP, Alpha-fetoprotein; CCL14, C-C motif chemokine ligand 14; CK19, Cytokeratin 19; CI, coefficient interval; COL1A1, Collagen 1A1; DFS, Disease-free Survival; EWSR1, Ewing Sarcoma breakpoint region 1/EWS RNA binding protein 1; FLI1, Friend leukemia virus integration 1; GEO, Gene Expression Omnibus; GO, Gene Ontology; HCC, Hepatocellular carcinoma; HR, Hazard ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; mRNA, messenger Ribonucleic Acid; N, nodule; OS, Overall survival; PPI, Protein-Protein Interaction analysis; RNA, Ribonucleic Acid; SD, Standard Deviation; TCGA, The Cancer Genome Atlas; T, tumor; TNM, tumor-nodule-metastasis.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide, with increasing incidence and mortality rates, particularly in China [Citation1]. In recent years, due to improvements in imaging and serological examination technology, including computed tomography, ultrasound, and alpha-fetoprotein (AFP) and des-gamma-carboxy prothrombin assays, the detection rate of early-stage HCC has increased. However, the mortality rate of HCC in China remains high, ranking as the second most lethal cancer in the country according to the GLOBCAN 2018 online database [Citation2]. This is attributed the complex and heterogeneous nature of HCC, which involve multiple stages and factors. Although several tissue prognostic biomarkers, such as C-C motif chemokine ligand 14 (CCL14), cytokeratin 19 (CK19), CD133, and CD90, have been investigated, most of them can be located in the cytoplasm as well as on the cell membrane, which may lead to confusing results and nonspecific background staining. Therefore, substantial effort is still needed to identify novel biomarkers, especially nuclei-labeled markers, that can help improve the accuracy of prognosis predictions.
EWSR1 (Ewing sarcoma breakpoint region 1/EWS RNA binding protein 1) was initially identified as a translocation-generated fusion gene between EWSR1 and FLI1 in Ewing’s sarcoma and neuroectodermal tumors [Citation3]. Recently, EWSR1 has been considered as a ‘hybrid’ gene involved in multiple mesenchymal tumor translocations, with evidence showing that it could be translocated and fused with many partner genes, including EWSR1-FLI1 and EWSR1-ERG in Ewing’s sarcoma [Citation4,Citation5], EWSR1-WT1 in desmoplastic small round cell tumors [Citation6], EWSR1-DDIT3 in myxoid liposarcoma [Citation7], EWSR1-CREB in angiomatoid fibrous histiocytoma [Citation8] and EWSR1-ATF1 in clear-cell sarcoma-like tumors of the gastrointestinal tract. In addition, EWSR1 acts as a multifunctional RNA/DNA binding protein and is involved in various cellular processes, such as transcription regulation and RNA splicing [Citation9]. Nevertheless, few studies have focused on the biological role of EWSR1 in epithelial tumors, especially in HCC.
The aim of this study was to investigate the expression and clinical significance of EWSR1 in HCC based on publicly available data and our cohorts. The biological function and pathway of the enrichments were explored through bioinformatics analysis, thereby providing valuable information on whether EWSR1 could be used as a candidate predictor of HCC clinical outcomes and an effective biomarker in routine clinical practice.
Materials and methods
TCGA and GEO datasets
EWSR1 RNA-sequencing and detailed clinicopathological data profiles in HCC (n = 371) and healthy liver tissues (n = 50) from 1995 to 2015 were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/projects/TCGA-LIHC) [Citation10]. Clinicopathological parameters were obtained for 369 cases, and a few of parameters with incomplete information were marked as Not reported. Gene expression profiles were obtained using R version 3.6.1 software (R Foundation for Statistical Computing, Vienna, Austria) from the E-MTAB-6695 cohort from October 2006 to November 2015, comprising seven Gene Expression Omnibus (GEO) datasets (accession no. GSE40873, GSE41804, GSE45436, GSE6222, GSE62232, GSE75271, and GSE29721) [Citation11].
Patients and tissue samples
A total of 124 paraffin-embedded HCC tissue specimens and adjacent healthy liver tissues, collected in 2013–2019 from patients naïve to preoperative radiotherapy or chemotherapy, were obtained from the Department of Pathology of The People’s Hospital of Jianyang City, China. The clinicopathological characteristics of all patients were obtained and the pathological diagnosis of each tissue specimen was confirmed by at least two pathologists. Follow-up data were acquired from the medical records and by telephone calls. Tumor differentiation was evaluated according to the World Health Organization Classification of Tumors of the Digestive System (5th Edition) [Citation12]. Disease-free survival (DFS) was measured from the date of surgery until disease relapse or metastasis, and overall survival (OS) was measured from the date of surgery until patient death. All patients provided written informed consent during hospitalization, and the Ethics Committee of The People’s Hospital of Jianyang City approved the study (No.2021017).
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissue sections (4 μm) were deparaffinized in xylene and rehydrated by serial ethanol washes. The slides were treated with 3% H2O2 for 15 min to quench the endogenous peroxidase. Antigen retrieval was performed by incubating the slides with 0.01 M citrate buffer (pH 6.0) at 100°C for 10 min. Standard immunohistochemical protocol was then implemented using a Ventana Benchmark XT autostainer (Ventana Medical Systems, Tucson, AZ, USA) as described previously [Citation13]. Negative control slides omitting the primary antibodies were used for all assays.
Evaluation and scoring
A semi-quantitative scoring system considering both intensity of staining and the extent of tumor cells was applied for EWSR1 (nuclear staining) and AFP (cytoplasmic staining). The staining intensity was scored as follows: 0, negative; 1, weak positive; 2, intermediate positive; and 3, strong positive. The scores of the extent of immunostaining signals were assessed according to the percentage of HCC cells that showed positive staining in each microscopic field (0–100). A final score ranging from 5 to 300, with a median score of 100, was achieved by multiplying the scores of intensity and extent. All cases were divided into two groups: low (score 0 − 100) and high (100 − 300) expression [Citation14,Citation15]. Ki67 status was evaluated based on the percentage of positive nuclear-stained tumor cells: low expression (≤ 25%) and high expression (> 25%) [Citation16].
PPI and KEGG/GO biological process enrichment
Correlation analysis was performed using the Pearson correlation test. Differentially expressed genes (DEGs) in protein-protein interaction analysis (PPI) with a connectivity degree > 20 and with a combination score > 0.75 were represented using Cytoscape (https://cytoscape.org) [Citation17]. Gene ontology (GO) biological process [Citation18] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses [Citation19] were conducted.
Statistical analysis
A chi-square test was applied to evaluate associations between the immunoreactivity of EWSR1 and other markers, as well as with the clinicopathological characteristics of HCC patients. Kaplan-Meier survival analysis was performed to estimate the prognostic relevance of EWSR1, and the survival difference between groups was assessed using the log-rank test. Univariate and multivariate Cox regression analyses were performed to evaluate the differences in all possible risk factors for death. SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was used for all statistical analyses. For all tests, P < 0.05 was considered to indicate statistical significance.
Results
In this study we explored the prognostic potential and role of EWSR1 in HCC. Based on the publicly available data, as well as in primary tissue samples, we demonstrate that EWSR1 is overexpressed in HCC, and is associated with high histological tumor grade and short overall patient survival. Moreover, EWSR1 was found to be an independent factor for poor prognosis in patients with HCC by univariate and multivariate cox regression analyses. GO and KEGG enrichments results also showed that EWSR1 was most likely to contribute for RNA splicing and DNA replication processes in HCC. These results, taken together, suggest that the EWSR1 may represent a valuable prognostic marker for HCC outcome in the clinical practice.
EWSR1 is upregulated in HCC patients
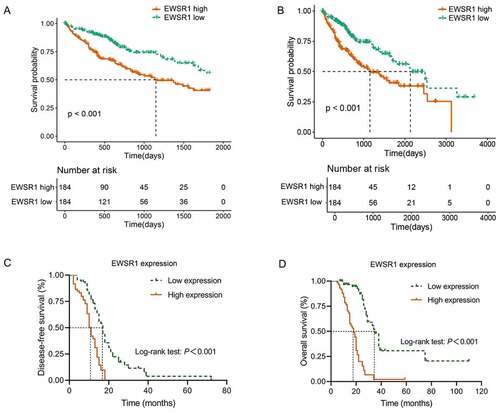
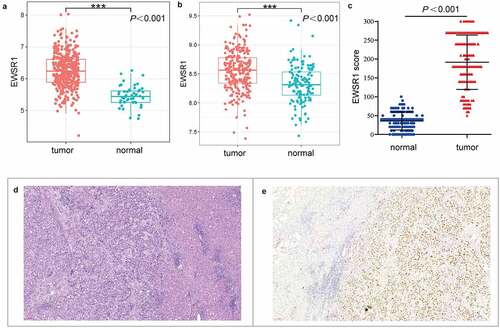
In this study, we first analyzed ESWR1 expression in the TCGA and E-MTAB-6695 cohorts. As shown in , EWSR1 levels were significantly upregulated in HCC tissues compared with healthy liver tissues (P < 0.001, )). In addition, immunohistochemical analysis of EWSR1 in our cohort showed low expression in all the healthy liver samples (124/124) whereas it was overexpressed in 81.45% (101/124) of the HCC tissues, suggesting that EWSR1 is upregulated in HCC (P< 0.001, )), which is consistent with data from TCGA and GEO databases.
Figure 1. EWSR1 expression between tumor and non-tumor liver tissues in HCC patients in TCGA (a) and GEO (b) datasets, and our cohort (c). Hematoxylin and eosin staining of HCC (left) and paratumoral, healthy (right) tissues (d). Positive immunohistochemical staining of EWSR1 in HCC (right), negative in paratumoral normal (left) tissue (e). Scale: 100 μm. Abbreviations: GEO, Gene Expression Omnibus; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas
Overexpression of EWSR1 correlates with clinicopathological features of HCC
To explore the role of upregulated EWSR1 in HCC progression, the association between EWSR1 expression and patient clinicopathological features was evaluated using data from the TCGA database and our cohort. Analysis of TCGA data showed that overexpression of EWSR1 was associated with higher histological grade (P < 0.001), pathologic T stage (P = 0.026) and mortality (P = 0.007) (). In our cohort, the degree of tumor differentiation was lower in patients with high expression of EWSR1 as compared with those in the EWSR1 low-expression group (). Moreover, the Tumor (T) staging (P = 0.004), Nodule (N) staging (P = 0.008), and Tumor-Nodule-Metastasis (TNM) staging (P = 0.001), was well as AFP (P < 0.001) and Ki67 (P < 0.001) expression rates, and mortality (P < 0.001) were markedly higher in the EWSR1 overexpression group (). No significant differences in sex, age, tumor size, capsular invasion, vascular tumor thrombus, cirrhosis, recurrence rate, or other indicators were observed between the two groups ().
Table 1. Correlation between EWSR1 expression and clinicopathological parameters of HCC patients in the TCGA cohort
Table 2. Correlation between EWSR1 expression and clinicopathological parameters of HCC patients in our cohort
Upregulation of EWSR1 correlates with poor prognosis in HCC
To evaluate the prognostic significance of EWSR1, we examined the association between EWSR1 expression and OS in HCC patients. Kaplan-Meier analyses indicated that overexpression of EWSR1 was associated with poor OS (P < 0.01, ) in the TCGA dataset, and the DFS and OS curve of our cohort also showed that HCC patients with low EWSR1 expression had better clinical outcomes than those with high expression (P < 0.01, ). We then performed a Cox regression model to evaluate the independent risk factors for disease-free survival (DFS) and OS based on our cohort. The mean DFS and OS were of 17 (95% confidence interval [CI]: 14.78–19.23) and 35 months (95% CI: 30.74–39.26), respectively, in the EWSR1 low expression group; and 11 (95% CI: 9.76–12.24) and 18 months (95% CI: 14.20–21.80), respectively, in the EWSR1 high expression group. Univariate Cox regression analysis showed that T stage, N stage, TNM stage, AFP overexpression, and EWSR1 overexpression were significantly associated with DFS (P < 0.05). Multivariate Cox regression analysis further confirmed that AFP and EWSR1 overexpression were independent risk factors for DFS (P < 0.05, ). In addition, univariate Cox regression analysis showed that tumor size, differentiation degree, T stage, N stage, TNM stage, AFP overexpression, Ki67 expression, and EWSR1 overexpression were each significantly associated with OS (P < 0.05). Following multivariate analysis confirmed that T stage, N stage, AFP and EWSR1 overexpression were independent risk factors for OS (P < 0.05, ).
Table 3. Univariate and multivariate Cox regression analyses of DFS in HCC patients
Table 4. Univariate and multivariate Cox regression analyses of OS in HCC patients
Bioinformatic analysis of EWSR1 in HCC
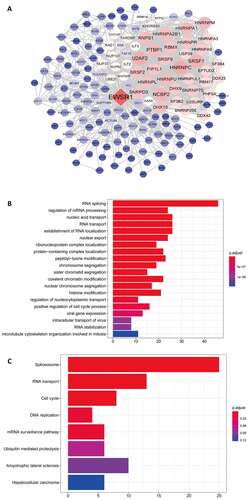
Using data from TCGA, we performed the Pearson correlation test on EWSR1 and other differential genes (with a threshold of 0.7), and defined the PPI network of EWSR1 using Cytospace ()). Subsequently, the identified genes of the PPI with adjusted P < 0.05 were further analyzed by GO and KEGG pathway analyses. GO functional enrichment analysis revealed significantly enriched biological processes, including RNA splicing, mRNA processing, nucleic acid and RNA transport, and establishment of RNA localization and nuclear export ()). KEGG pathway analysis further showed that the spliceosome, RNA transport, cell cycle, and DNA replication were the most enriched ()), suggesting that EWSR1 plays a significant role in the RNA splicing and DNA replication processes.
Discussion
The EWSR1 gene is a member of the TET (also known as FET) family [Citation20], including Fused in sarcoma/Translocated in Liposarcoma (FUS/TLS) and TATA-box binding Associated Factor 15 (TAF15) [Citation21]. As mentioned before, EWSR1 is well-known to be able to fuse with a range of genes in the mesenchymal neoplasms, and a few series of epithelial tumors as myoepithelial[Citation22], primary clear cell carcinoma of the thymus [Citation23] and the Xp11 translocation renal cell carcinoma [Citation24]. Recently, EWSR1 rearrangement is reported as a frequent event in papillary microcarcinoma and related to classic and small-cell morphology [Citation25]. More than that, EWSR1 can participate in a variety of cell processes by regulating gene expression, mitotic cell division, RNA splicing, cell signaling and DNA repair and transcription [Citation9]. However, the role of EWSR1 protein expression in tumor, especially in epithelial cancers is rarely reported.
In the present study, EWSR1 was found to be significantly upregulated in HCC tissues compared with the healthy counterparts, as determined by the bioinformatic analysis of GEO and TCGA datasets, was well by immunohistochemical assessment in primary samples. Hence, EWSR1 may be considered a novel biomarker to distinguish HCC from healthy liver. Noteworthy, the expression of EWSR1 was found to be closely related to aggressive factors and mortality, further illustrating that EWSR1 expression is positively correlated with poor tumor differentiation and advanced clinical stage. EWSR1 overexpression was reported to be a poor prognostic predictor in multiple myeloma [Citation26], however, to date, no investigations on the role of EWSR1 in HCC were available. Analysis of high-throughput RNA sequencing data publicly available showed that increased EWSR1 expression was associated with shortened OS after liver resection in this work, and this observation was further confirmed in our cohort, in which EWSR1 overexpression was correlated with significantly shorter PFS and OS after a 5-year follow-up period. In addition, analysis using the Cox proportional hazards regression models showed that overexpression of EWSR1 is an independent predictor of PFS and OS in HCC patients. Therefore, these findings support the use of EWSR1 as a novel and valuable biomarker for predicting poor prognosis in HCC.
To our knowledge, several prognostic molecular markers for HCC have been revealed in the recent years. In Gu’s study, CCL14 is not only a potential prognostic biomarker but also correlated with tumor immune cells infiltration in HCC [Citation27]. CK19, a traditional bile duct marker, has been increasingly reported to be associated with aggressive behaviors and poor outcomes in HCC. Furthermore, as TGFβR1 may be a targeted therapeutic factor for CK19 positive HCC, it is supported to include CK19 positive hepatocellular carcinoma as a special subtype [Citation28]. Ma et al reveal Collagen 1A1 (COL1A1) can be used as a survival advantage predicator and downregulation of COL1A1 could suppress the oncogenicity and epithelial-to-mesenchymal transition process of HCC cells [Citation29]. AFP, a serum marker for HCC detection, is also frequently elevated in HCC, recenty study show AFP acetylation can promote HCC progress by blocking binding to the phosphatase PTEN and the pro-apoptotic protein caspase-3 [Citation30]. In our samples, AFP was overexpressed in HCC tissues and acted as an independent prognostic factor. However, due to the high heterogeneity of HCC, a panel of markers should be used to comprehensively evaluate the prognosis assessment. As most of the markers reported are located in the cytoplasm or on the cell membrane, which may lead to confusing results and nonspecific background staining. EWSR1, as a marker for nuclear staining, has the unique advantage of being able to evaluate results more accurately and reduce false negatives and false positives. Comparison of specificity and sensitivity of EWSR1 with other markers will be carried out in the future.
In the past decades, studies on EWSR1 have mainly focused on the role of fusion proteins formed by EWSR1 translocation in tumorigenesis and cancer development [Citation31], whereas the biological function of EWSR1 itself remains poorly understood. EWSR1 is known to be involved in mitotic progression by promoting microtubule acetylation in the mitotic spindle and inhibiting the activity of HDAC6 [Citation32]. We performed a comprehensive bioinformatics analysis of the TCGA dataset to reveal the underlying mechanism of EWSR1 in HCC. In accordance with the previously reported role of EWSR1, herein we found that EWSR1 and is related/partner genes were mainly involved in RNA splicing, mRNA processing, nucleic acid transport, and RNA transport processes. Moreover, the spliceosome was the most enriched pathway, as determined by KEGG pathway analysis. Taken together, these results confirm that EWSR1 may contribute for HCC pathological mechanisms by partaking in RNA splicing and DNA replication.
Conclusion
To our knowledge, this is the first study to investigate the biological function of EWSR1 in an epithelial cancer. Our results suggest that high EWSR1 expression is an independent predictor of shorter DFS and OS in patients with HCC, by regulating RNA splicing and DNA replication. Taken together, these results indicate that EWSR1 may represent as a novel biomarker of poor prognosis in patients with HCC.
Research highlights
EWSR1 expression is upregulated in HCC tissues compared with paratumor tissues.
EWSR1 overexpression is associated with high histological grade and short OS.
EWSR1 is an independent factor for poor prognosis in patients with HCC.
RNA splicing processing represents the major function and pathway of EWSR1 in HCC.
Disclosure statement
The authors have declared that no competing interest exists.
Additional information
Funding
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 global cancer statistics? Cancer Commun (Lond). 2019;39(1):22.
- Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165.
- Grunewald TGP, Cidre-Aranaz F, Surdez D, et al. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5.
- Chen S, Deniz K, Sung YS, et al. Ewing sarcoma with ERG gene rearrangements: a molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer. 2016;55:340–349.
- Thway K, Noujaim J, Zaidi S, et al. Desmoplastic small round cell tumor: pathology, genetics, and potential therapeutic strategies. Int J Surg Pathol. 2016;24(8):672–684.
- Baranov E, Black MA, Fletcher CDM, et al. Nuclear expression of DDIT3 distinguishes high-grade myxoid liposarcoma from other round cell sarcomas. Mod Pathol. 2021;34(7):1367–1372.
- Bale TA, Oviedo A, Kozakewich H, et al. Intracranial myxoid mesenchymal tumors with EWSR1-CREB family gene fusions: myxoid variant of angiomatoid fibrous histiocytoma or novel entity? Brain Pathol. 2018;28:183–191.
- Lee J, Nguyen PT, Shim HS, et al. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim Biophys Acta Mol Basis Dis. 2019;1865(7):1938–1945.
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68–77.
- Bin Lim S, Chua MLK, Yeong JPS, et al. Pan-cancer analysis connects tumor matrisome to immune response. NPJ Precis Oncol. 2019;3(1):15.
- Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76(2):182–188.
- Zhang X, Gu G, Song L, et al. ID4 promotes breast cancer chemotherapy resistance via CBF1-MRP1 pathway. J Cancer. 2020;11(13):3846–3857.
- Zhang H, Ma RR, Wang XJ, et al. KIF26B, a novel oncogene, promotes proliferation and metastasis by activating the VEGF pathway in gastric cancer. Oncogene. 2017;36(40):5609–5619.
- Lv BB, Ma RR, Chen X, et al. E2F1-activated SPIN1 promotes tumor growth via a MDM2-p21-E2F1 feedback loop in gastric cancer. Mol Oncol. 2020;14(10):2629–2645.
- Jones A, Kroneman TN, Blahnik AJ, et al. Ki-67 “hot spot” digital analysis is useful in the distinction of hepatic adenomas and well-differentiated hepatocellular carcinomas. Virchows Arch. 2021;478(2):201–207.
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11.
- Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56.
- Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
- Hoell JI, Larsson E, Runge S, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18(12):1428–1431.
- Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1(2):82–92.
- Thway K, Fisher C. Myoepithelial tumor of soft tissue: histology and genetics of an evolving entity. Adv Anat Pathol. 2014;21(6):411–419.
- Porubsky S, Rudolph B, Ruckert JC, et al. EWSR1 translocation in primary hyalinising clear cell carcinoma of the thymus. Histopathology. 2019;75:431–436.
- Fukuda H, Kato I, Furuya M, et al. A novel partner of TFE3 in the Xp11 translocation renal cell carcinoma: clinicopathological analyses and detection of EWSR1-TFE3 fusion. Virchows Arch. 2019;474(3):389–393.
- Kovacevic B, Caramelo A, Skuletic V, et al. EWSR1 rearrangement in papillary thyroid microcarcinoma is related to classic morphology and the presence of small-cell phenotype. Bosn J Basic Med Sci. 2021. DOI:10.17305/bjbms.2021.6181
- Nishiyama D, Chinen Y, Isa R, et al. EWSR1 overexpression is a pro-oncogenic event in multiple myeloma. Int J Hematol. 2021;113(3):381–394.
- Gu Y, Li X, Bi Y, et al. CCL14 is a prognostic biomarker and correlates with immune infiltrates in hepatocellular carcinoma. Aging (Albany NY). 2020;12(1):784–807.
- Zhuo JY, Lu D, Tan WY, et al. CK19-positive hepatocellular carcinoma is a characteristic subtype. J Cancer. 2020;11:5069–5077.
- Ma HP, Chang HL, Bamodu OA, et al. Collagen 1A1 (COL1A1) is a reliable biomarker and putative therapeutic target for hepatocellular carcinogenesis and metastasis. Cancers (Basel). 2019;11(6):786.
- Xue J, Cao Z, Cheng Y, et al. Acetylation of alpha-fetoprotein promotes hepatocellular carcinoma progression. Cancer Lett. 2020;471:12–26.
- Cantile M, Marra L, Franco R, et al. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med Oncol. 2013;30(1):412.
- Wang YL, Chen H, Zhan YQ, et al. EWSR1 regulates mitosis by dynamically influencing microtubule acetylation. Cell Cycle. 2016;15(16):2202–2215.