ABSTRACT
The competing endogenous RNA (ceRNA) activity of circular RNAs (circRNAs) has been implicated in the development of gastric cancer. Here, we sought to explore the ceRNA function of circRNA Jupiter microtubule associated homolog 1 (circ_HN1) in gastric tumorigenesis. Circ_HN1, microRNA (miR)-628-5p, and NT5E expression levels were quantified by qRT-PCR and western blot. Dual-luciferase reporter assays were used to assess the direct relationship between miR-628-5p and circ_HN1 or NT5E. Our data showed that circ_HN1 expression was enhanced in human gastric cancer. Depletion of circ_HN1 impeded cell proliferation, spheroid formation, invasion, and migration and promoted apoptosis in vitro, as well as diminished tumor growth in vivo. NT5E was a downstream effector of circ_HN1 function. NT5E was targeted and inhibited by miR-628-5p through the perfect complementary site in NT5E 3ʹUTR, and circ_HN1 affected NT5E expression through miR-628-5p competition. Moreover, depletion of miR-628-5p reversed the effects of circ_HN1 silencing on regulating cell functional behaviors. Our findings identify a novel ceRNA network, the circ_HN1/miR-628-5p/NT5E axis, for the oncogenic activity of circ_HN1 in gastric cancer, highlighting circ_HN1 inhibition as a promising targeted treatment against gastric cancer.
Introduction
Gastric cancer is one of the most prevalent malignancies and it ranks as the third leading cause of cancer mortality [Citation1,Citation2]. Although surgery, chemoradiotherapy, and immune therapies can control many early-stage tumors effectively, these available treatments have limited in curbing advanced-stage gastric cancer [Citation3]. Pivotal regulators of gastric carcinogenesis, including non-coding RNAs (ncRNAs) and proteins, are under intensive exploration at present [Citation4–6]. Identifying the functions of these molecules will be essential in one day developing the molecularly targeted therapies.
In recent years, the importance of the ncRNAs, including microRNAs (miRNAs) and circular RNAs (circRNAs), has become increasingly clear [Citation7,Citation8]. CircRNAs are natural RNA circles that are formed by an out-of-order arrangement of exons known as back-splicing [Citation9]. MiRNAs are small ncRNAs that function as agents of the RNA interference pathway by leading to silencing of their targets [Citation10]. Numerous studies have demonstrated that circRNAs involve the post-transcriptional RNA regulation by working as competing endogenous RNAs (ceRNAs) via miRNAs [Citation11]. Emerging evidence has also highlighted the implications of the ceRNA activity of circRNAs in human carcinogenesis [Citation7].
In a preliminary survey, when we analyzed the online dataset (GSE83521) of the Gene Expression Omnibus (GEO) database to search the dysregulated circRNAs in gastric cancer, we found that circRNA Jupiter microtubule associated homolog 1 (circ_HN1, also called circ_0045602 based on the circRNA ID of circBase database), generated by non-sequential back-splicing of exons 3–5 of HN1 pre-mRNA with a length of 260 nucleotides, is highly expressed in gastric primary tumors. The finding has been ascertained by a recent report [Citation12], where circ_HN1 promotes the malignant behaviors of gastric cancer cells via the miR-302b-3p/rho-associated coiled-coil containing protein kinase 2 (ROCK2) axis. Although the report has revealed a ceRNA determinant underlying the oncogenic activity of circ_HN1 in gastric cancer [Citation12], our understanding for its molecular basis remains incomplete.
Ecto-5ʹ-nucleotidase (NT5E, also called CD73) is a ubiquitous cell-surface protein and it has been identified as a strong oncogene in cancer biology [Citation13–15]. In gastric cancer, NT5E is a potential prognostic biomarker [Citation16,Citation17]; it is implicated in gastric tumorigenesis and gastric cancer metastasis [Citation18]. Nonetheless, it is still unclear whether NT5E is a downstream effector of circ_HN1 function in gastric tumorigenesis.
MiR-628-5p, an underexpressed miRNA in gastric cancer, has been shown to exert tumor-inhibitory activity in this disease [Citation19,Citation20]. When we used the online algorithms to help identify the ceRNA network mediated by circ_HN1, we observed the potential relationship between miR-628-5p and circ_HN1 or NT5E. Thus, we hypothesized that the miR-628-5p/NT5E axis might be responsible for the oncogenic activity of circ_HN1 in gastric cancer. In this study, we set to explore the mechanism of circ_HN1 activity in gastric cancer pathogenesis.
Materials and methods
Bioinformatics
To search the dysregulated circRNAs in gastric cancer, we used the GSE83521 dataset at https://www.ncbi.nlm.nih.gov/search/all/?term=GSE83521. To analyze the up-regulated genes in gastric cancer, we used the GSE112369 dataset at https://www.ncbi.nlm.nih.gov/search/all/?term=GSE112369. To predict miRNA-binding sites to circ_HN1 and human 3ʹUTRs, we utilized the computational program starBase at http://starbase.sysu.edu.cn/.
Cell lines
Human HGC27 (gastric cancer), MKN74 (gastric cancer) and GES-1 (nontumor gastric epithelial) cells were purchased from RIKEN Cell Bank (Tsukuba, Japan). All cells were propagated in RPMI-1640 medium (Sigma-Aldrich, Darmstadt, Germany) containing 10% newborn calf serum (HyClone, Cramlington, UK) at 37°C at 5% CO2.
Human specimens
Human specimens, including 47 gastric primary tumors and 47 adjacent normal gastric tissues, were collected at the time of the clinically indicated surgical procedures from previously untreated patients (n = 47) at the Henan Provincial People’s Hospital with informed consent. The clinical characteristics of these patients were shown in . These specimens were used to measure the expression levels of circ_HN1, NT5E and miR-628-5p. The study was approved by the Ethics Committee of Henan Provincial People’s Hospital.
Table 1. The clinical characteristics of gastric cancer patients (n = 47)
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was prepared from cultured cells and collected tissues using the RNeasy Plus Mini Kit as per the manufacturing instructions (Qiagen, Tokyo, Japan). Actinomycin D treatments were performed by incubating HGC27 and MKN74 cells with 2 mg/mL of actinomycin D (Hanhui Pharmaceuticals CO., LTD. Xian, China) for 8, 16 and 24 h at 37°C. Nuclear and cytoplasmic RNA were isolated from HGC27 and MKN74 cells using the Cytoplasmic & Nuclear RNA Purification Kit as recommended by the manufacturers (NORGEN, Thorold, ON, Canada). Complementary DNA (cDNA) was synthesized from RNA samples using random hexamers with QuantiTect reverse transcription (RT) Kit (Qiagen) or stem-loop RT primers with miRNA 1st-strand cDNA Synthesis Kit (Vazyme, Nanjing, China). qRT-PCR was performed using the MyiQ Single Color PCR System (Bio-Rad, Hemel Hemstead, UK) with the SYBR qPCR Mix (Toyobo, Osaka, Japan) and specific primers (Supplement ) designed by Primer v3.0 software (Thermo Fisher Scientific, Paisley, UK). U6 and β-actin levels were tested for normalization. For data analysis, we used the MiniOpticon System (Bio-Rad) based on the 2−ΔΔCt method [Citation21].
Construction of stable cell line
Short hairpin RNAs (shRNA) targeting circ_HN1 (sh-circ_HN1, CGCAAAACUCAUGAAUAUCAC) and the scrambled control (sh-NC, CCCUAUAGUGAAGUAAAACUC) were cloned into the lentiviral vector (GeneSeed, Guangzhou, China). Lentivirus was produced by transfecting 293 FT packaging cells (Cyagen Biosciences, Jiangsu, China) with shRNA constructs, pMDLg/pRRE and pRSV-Rev (Addgene, Teddington, UK). Virus suspension was harvested after 48 h and used to infect HGC27 and MKN74 cells. Virus-infected cell lines were selected in the growth media supplemented with 2 µg/mL puromycin (Yesen, Shanghai, China).
Plasmid and oligonucleotide transfection
Human NT5E (Accession: NM_001204813.2) coding sequence and a non-target sequence control, obtained from Abiocenter (Beijing, China), were inserted into pcDNA3.1 plasmid (Thermo Fisher Scientific) to generate appropriate expressing plasmid. MiR-628-5p mimic (5ʹ-AUGCUGACAUAUUUACUAGAGG-3ʹ), inhibitor (anti-miR-628-5p, 5ʹ-CCUCUAGUAAAUAUGUCAGCAU-3ʹ), and matched controls described by Gu et al. [Citation22] were provided by Ribobio (Guangzhou, China).
For transient transfection, we seeded 5 × 104 cells per well in 12-well dishes. Next day, cells were transfected using Lipofectamine 3000 with 200 ng of plasmid or 30 nM of oligonucleotide. We harvested the cells after 48 h transfection for further assays.
Cell colony formation, proliferation, and apoptosis assays
Colony formation experiments were carried out by plating ~100 cells per well into 6-well dishes. Following a 10-day incubation at 37°C, the number of colonies (> 50 cells) was scored using standard methods [Citation23]. Cell proliferation was gauged by the 3-(4,5-dimethylthia-zol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and 5-Ethynyl-2ʹ-Deoxyuridine (EdU) assays using the MTT Cell Proliferation Assay Kit (Biovision, Wehrheim, Germany) and Cell-Light EdU Apollo488 In Vitro Kit (Ribobio), respectively, as per the accompanying protocols and standard methods [Citation24,Citation25]. Cell apoptosis was determined using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Kit (BD Biosciences, Cowley, UK) as described elsewhere [Citation12].
Spheroid formation assay
This assay was carried out as previously described [Citation24]. Briefly, transfected HGC27 and MKN74 cells (1,000 cells/well) were plated in low adhesion 6-well dishes in complete growth media containing matrigel growth factor reduced basement membrane matrix and incubated for 14 days before quantification. Spheroids were monitored every three days and images at day 14 were taken using a microscope (Olympus, Mishama, Japan) at ×100 magnification.
In vitro migration and invasion assays
Transwell assays were performed to evaluate cell migration and invasion under standard protocols [Citation26]. For these assays, cells were resuspended in non-serum media and seeded in inserts (24-transwell, MERK Millpore, Tokyo, Japan) containing 8-µm pores with (invasion assay, 1 × 105 cells/well) or without (migration assay, 5 × 104 cells/well) matrigel (BD Biosciences). The inserts were then placed in wells containing 10% serum RPMI-1640 medium. Twenty-four hours post-seeding, the invaded or migrated cells were determined with ×100 magnification under the Olympus microscope after crystal violet (0.5%) staining.
Western blot
Protein samples were prepared using the cold RIPA buffer (Biovision) containing Protease Inhibitor Cocktails (Roche, Tokyo, Japan). Western blot analysis was performed under standard methods [Citation27], using antibodies against B cell lymphoma-2 (Bcl-2, ab692), proliferating cell nuclear antigen (PCNA, ab92552), NT5E (ab202122), Bcl-2 associated X, apoptosis regulator (Bax, ab182733) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, ab9485) from Abcam (Cambridge, UK).
Dual-luciferase reporter assay
Dual-luciferase reporter assays were performed as previously described [Citation26]. Human circ_HN1 sequence and NT5E 3ʹUTR, synthesized by Abiocenter, were inserted into the available sites of the psiCHECK-2 vector (Promega, Paris, France) to generate circ_HN1-WT and NT5E-WT, respectively. Mutations (circ_HN1-MUT and NT5E-MUT) in the binding region were done using the TaKaRa MutanBEST Kit based on the recommendations of manufacturers (TaKaRa, Dalian, China). The appropriate construct (200 ng) was co-transfected into HGC27 and MKN74 cells with miRNA mimic (30 nM). Luciferase activity was gauged after 48 h transfection using the Promega Dual-luciferase Assay System with a luminometer (Berthold, Tokyo, Japan).
RNA pull-down assay
This assay was carried out using standard methods as reported by Zhao et al. [Citation28]. Cell lysates prepared with RIPA buffer were incubated with biotinylated miR-628-5p mimic (Bio-miR-628-5p WT), mutation of biotinylated miR-628-5p in the seed region (Bio-miR-628-5p MUT), or negative control Bio-miR-NC (all from Ribobio) for 3 h at 4°C before adding streptavidin beads (Roche) for additional 3 h. Beads were harvested, and RNA was isolated from the beads to measure circ_HN1 enrichment level by qRT-PCR.
Tumor xenografts
Animal experiments were performed following a protocol approved by the Ethics Committee on Animal Care of Henan Provincial People’s Hospital and the National Animal Experiment Guidelines. For xenograft assays [Citation28], Balb/c female nude mice age-matched between 5–7 weeks (Shanghai Model Organisms, Shanghai, China) were used and divided into four groups (n = 6 per group): sh-NC, sh-circ_HN1, sh-circ_HN1+ anti-miR-NC, sh-circ_HN1+ anti-miR-628-5p. About 5 × 106 virus-infected HGC27 cells were subcutaneously injected into the right flanks of the nude mice to form xenograft mice. 10 days later, intratumor injection of PBS, anti-miR-NC or anti-miR-628-5p was performed every 5 days. Tumor growth was evaluated 10 days after cell implantation by calculating tumor volume with the (length × width2)/2 formula. All mice were requested to be euthanized on day 30 after cell implantation. The xenograft tumors were excised, weighed and subjected to expression analysis. Sections of paraffin-embedded tumors were used to measure the expression of NT5E and proliferating maker Ki67 by immunohistochemistry using NT5E (ab202122, Abcam) and Ki67 (ab15580, Abcam) antibodies, as reported elsewhere [Citation29].
Data analysis
All assays were repeated three times and performed in quintuplicate each time. Data sets were compared using Dunnett’s multiple comparisons after analysis of variance (ANOVA), Student’s t-test, or Mann-Whitney U test, where appropriate. Pearson’s correlation coefficients were used to analysis the expression correlations among circ_HN1, NT5E and miR-628-5p in gastric primary tumors. Significance was set at P < 0.05.
Results
Circ_HN1 is overexpressed in gastric cancer
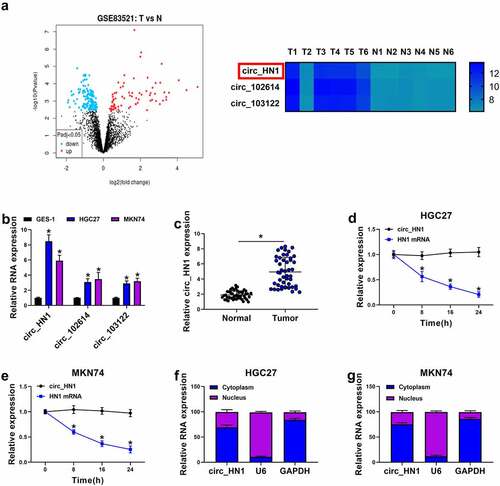
In the search of circRNAs potentially involved in gastric tumorigenesis, we analyzed the online dataset (GSE83521) of the GEO database. When we set the |log2FC| > 4, Padj < 0.05, we found 3 differently expressed circRNAs (circ_HN1, circ_102614, and circ_103122) in gastric primary tumors compared with the adjacent nontumor gastric tissues from the same patients (). Analysis of the expression levels of the 3 circRNAs in gastric cancer cells showed that circ_HN1 was the most significantly up-regulated circRNA in HGC27 and MKN74 gastric cancer cells compared with the normal GES-1 cells (). In line with the data of GSE83521 dataset, we found a striking elevation in the levels of circ_HN1 in gastric primary tumors compared with the normal controls (). To examine the stability of circ_HN1, we adopted actinomycin D assays. Incubation of actinomycin D led to reduced levels of HN1 linear mRNA, and circ_HN1 expression did not decrease in the assayed time period ( and 1e). Furthermore, subcellular localization assay revealed that circ_HN1 predominantly localized to the cytoplasm of HGC27 and MKN74 cells ( and 1g). These results together indicate that circ_HN1 is up-regulated in human gastric cancer.
Figure 1. Overexpression of circ_HN1 in gastric cancer. (a) Left: Volcano plot showing the differently expressed circRNAs in gastric cancer tissues. Right: 3 up-regulated circRNAs (circ_HN1, circ_102614, and circ_103122) in gastric cancer tissues when the |log2FC| > 4, P < 0.05. (b) qRT-PCR showing the overexpression of circ_HN1, circ_102614 and circ_103122 in HGC27 and MKN74 gastric cancer cells compared to the nontumor GES-1 cells. (c) Upregulation of circ_HN1 in 47 gastric primary tumors compared with the normal gastric tissues. (d and e) The stability of circ_HN1 analyzed by actinomycin D assay in HGC27 and MKN74 cells. (f and g) Subcellular localization assay showing the cytoplasmic localization of circ_HN1 in HGC27 and MKN74 cells. *P < 0.05
Silencing endogenous circ_HN1 hinders cell proliferation, invasion, migration, and spheroid formation and promotes apoptosis in vitro
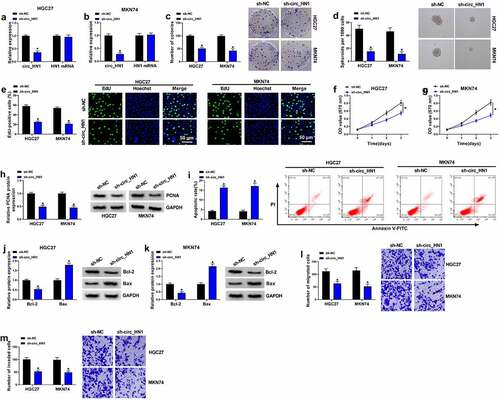
Having demonstrated the overexpression of circ_HN1 in gastric cancer, we then evaluated the functional role of circ_HN1. To do this, we generated circ_HN1-silenced cells with lentivirus expressing shRNA-circ_HN1 (sh-circ_HN1). The effectiveness of sh-circ_HN1 in knocking down circ_HN1 was verified by qRT-PCR ( and 2b). Although sh-circ_HN1 transduction led to a significant down-regulation of circ_HN1 expression, the level of HN1 linear mRNA was not reduce by sh-circ_HN1 ( and 2b). Remarkably, circ_HN1 loss of function impeded cell colony formation () and suppressed spheroid formation (), as well as reduced proliferation () in HGC27 and MKN74 cells. The reduction of the proliferating maker PCNA expression in cells transduced by sh-circ_HN1 also supported the proliferation repression caused by circ_HN1 silencing (). Knocking down circ_HN1 markedly repressed cell apoptosis compared to the control group (). Moreover, circ_HN1 depletion resulted in increased protein expression of Bax and decreased levels of Bcl-2 in the two cancer cell lines ( and 2k), validating that circ_HN1 silencing promoted cell apoptosis. Furthermore, circ_HN1-silenced HGC27 and MKN74 cells exhibited suppressed migration () and invasion () rates compared to the control group. All these findings demonstrate that circ_HN1 affects cell proliferation, invasion, migration, spheroid formation and apoptosis in vitro.
Figure 2. Effects of circ_HN1 depletion on cell migration, proliferation, invasion, spheriod formation and apoptosis in vitro. MKN74 and HGC27 cells were transduced by lentiviruses expressing sh-circ_HN1 or the scrambled control (sh-NC). (a and b) qRT-PCR showing the reduction of circ_HN1 and unvaried expression of HN1 mRNA in transduced cells. (c) Representative pictures depicting a cell colony formation assay and showing the suppressed colony formation by circ_HN1 depletion. (d) Spheriod formation. Phase contrast micrographs of day 14 in ultra-low attachment plate culture. (e) Representative images presenting a cell proliferation assay and cell proliferation by EdU assay. (f and g) MTT assay revealing cell proliferation repression caused by circ_HN1 loss of function. (h) The repression of PCNA expression in transduced cells determined by western blot. (i) Representative images showing a cell apoptosis assay performed by flow cytometry. (j and k) Western blot showing the alteration of Bcl-2 and Bax levels in circ_HN1-silencing cells. (l and m) Representative images depicting cell migration and invasion assays performed by transwell assay. *P < 0.05
NT5E is a downstream effector of circ_HN1 function
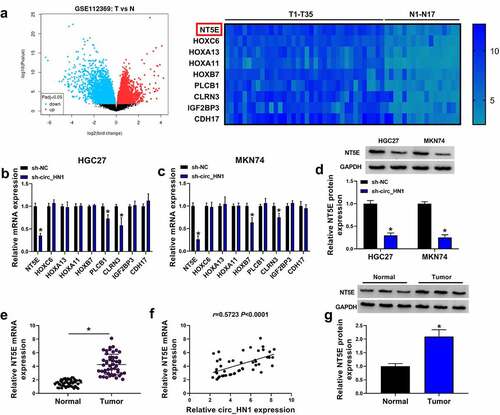
Based on the ceRNA hypothesis, to understand how circ_HN1 influenced the functional properties of gastric cancer cells, we considered the most up-regulated genes in gastric cancer to be potential effectors of circ_HN1. Using the GEO database (GSE112369), we found 9 up-regulated genes in gastric primary tumors when we set the |log2FC| > 3.5, Padj < 0.05 (). Intriguingly, in circ_HN1-silenced HGC27 and MKN74 cells, NT5E mRNA level was the most significantly underexpressed ( and 3c). We thus selected NT5E for further investigation. In agreement with the mRNA expression, NT5E protein level was down-regulated as a result of circ_HN1 depletion in the two gastric cancer cell lines (). We also used qRT-PCR to validate the overexpression of NT5E mRNA in 47 gastric primary tumors compared the adjacent nontumor gastric tissues (). Having established that circ_HN1 affected NT5E expression, we then elucidated if there was a positive correlation between NT5E and circ_HN1 levels in the gastric primary tumors. Notably, NT5E mRNA levels positively correlated with circ_HN1 expression (). Additionally, gastric primary tumors exhibited higher protein levels of NT5E compared with the controls ().
Figure 3. Overexpression of NT5E in gastric cancer. (a) Left: Volcano plot showing the deregulated genes in gastric cancer tissues. Right: 9 up-regulated genes in gastric cancer tissues when the |log2FC| > 3.5, P < 0.05. (b and c) qRT-PCR of the 9 genes expression in sh-NC- or sh-circ_HN1-infected HGC27 cells and MKN74 cells. (d) Western blot showing the reduction of NT5E protein in sh-NC- or sh-circ_HN1-transduced HGC27 cells and MKN74 cells. (e) qRT-PCR of NT5E mRNA in 47 pairs of gastric cancer tissues and the matched normal gastric tissues. (f) Correlation between circ_HN1 and NT5E mRNA expression in 47 gastric primary tumors. (g) Western blot showing the overexpression of NT5E protein in gastric cancer tissues compared to the matched normal gastric tissues. *P < 0.05
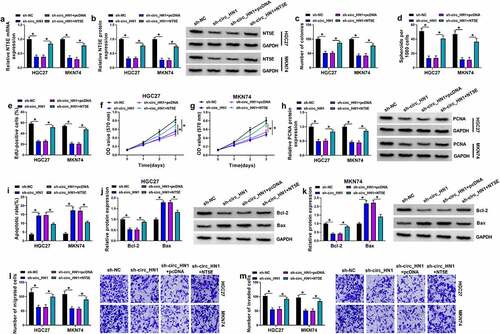
To elucidate whether NT5E was a downstream effector of circ_HN1 in the context of regulating gastric tumorigenesis, a NT5E expressing plasmid lacking the 3ʹUTR region was co-transfected and assayed for cell proliferation, spheroid formation, migration, invasion, and apoptosis. By contrast, transfection of the NT5E expressing plasmid markedly rescued sh-circ_HN1-driven NT5E expression reduction in both cell lines ( and 4b). Strikingly, restoration of NT5E reversed circ_HN1 depletion-mediated cell colony formation repression (), spheroid formation reduction (), proliferation suppression (), apoptosis enhancement (), migration inhibition (), and invasion reduction (). These findings together strongly establish the notion that the effects of circ_HN1 depletion are at least in part due to the down-regulation of NT5E.
Figure 4. The effects of circ_HN1 knockdown are mediated by NT5E. Stable sh-NC- or sh-circ_HN1-infected HGC27 and MKN74 cells were introduced with pcDNA control plasmid or NT5E overexpressing plasmid. (a) NT5E mRNA level in transfected cells. (b) NT5E protein level in transfected cells. (c) A standard colony formation assay for colony formation ability of transfected cells. (d) Spheriod formation. Phase contrast micrographs of day 14 in ultra-low attachment plate culture. Proliferation of transfected cells by EdU (e) and MTT (f and g) assay. (h) PCNA protein expression in transfected cells by western blot. (i) Flow cytometry examining cell apoptosis rate. (j and k) Western blot of the Bcl-2 and Bax expression in transfected cells. Migration (l) and invasion (m) rates of transfected cells by transwell assay. *P < 0.05
Circ_HN1 regulates NT5E expression by competitively binding to miR-628-5p
Having established that circ_HN1 regulated NT5E expression, we then undertook to identify their shared miRNAs based on the shared binding sites. Using the computational program starBase, there were 4 shared miRNAs (miR-628-5p, miR-942-5p, miR-1323, and miR-548o-3p) (). Among the 4 miRNAs, miR-628-5p was the only down-regulated miRNA in HGC27 and MKN74 gastric cancer cells compared to the nontumor GES-1 cells (). Moreover, we observed a clear elevation in the level of endogenous miR-628-5p in circ_HN1-silenced HGC27 and MKN74 cells (). The predicted data by the starBase web showed a putative binding sequence for miR-628-5p within circ_HN1 (). To validate the direct relationship between circ_HN1 and miR-628-5p, we constructed circ_HN1 luciferase reporter and transfected it into the two cancer cell lines together with miR-628-5p mimic. With this reporter and miR-628-5p mimic led to a clear down-regulation in luciferase activity ( and 5f). We then generated a mutation in the binding region and tested it by luciferase activity. However, the mutation was refractory to inhibition by miR-628-5p mimic ( and 5f). Moreover, RNA pull-down assay showed that incubation of Bio-miR-628-5p WT led to increased enrichment level of circ_HN1, and this effect was abolished by the mutation (Bio-miR-628-5p MUT) (), reinforcing the direct relationship between circ_HN1 and miR-628-5p. Using the starBase web, we also found that there existed seven nucleotides within NT5E 3ʹUTR complementarity to the seed sequence of miR-628-5p (). Using 3ʹUTR luciferase assays, we confirmed NT5E was targeted by miR-628-5p ( and 5j). Contrary to the levels of circ_HN1 and NT5E, miR-628-5p expression was strikingly reduced in gastric primary tumors compared to the normal controls (). Moreover, in gastric primary tumor tissues, miR-628-5p expression negatively correlated with circ_HN1 and NT5E levels ( and 5m). In support of these results, miR-628-5p elevation upon miR-628-5p mimic introduction, confirmed by qRT-PCR (), caused a distinct suppression in the levels of NT5E mRNA and protein ( and 5p). Conversely, knocking down miR-628-5p with anti-miR-628-5p transfection () resulted in increased expression of NT5E mRNA and protein ( and 5p). Taken together, these observations suggest that NT5E is a target of miR-628-5p and circ_HN1 acts as a ceRNA for miR-628-5p to regulate NT5E expression.
Figure 5. Circ_HN1 directly binds to miR-628-5p to up-regulate NT5E. (a) Venn diagram showing the 4 shared miRNAs of circ_HN1 and NT5E predicted by the computational program starBase. (b) qRT-PCR of the 4 shared miRNAs in HGC27, MKN74, and GES-1 cells. (c) Relative miR-628-5p expression in sh-NC- or sh-circ_HN1-transduced HGC27 and MKN74cells. (d) Predicted miR-628-5p pairing sites within circ_HN1, the mutant in the target sequence, and sequence of miR-628-5p. (e and f) Dual-luciferase reporter assays showing the validity of the miR-628-5p binding sites. (g) RNA pull-down assay performed with HGC27 and MKN74 cells using Bio-miR-NC, Bio-miR-628-5p WT or Bio-miR-628-5p MUT. (h) Predicted miR-628-5p binding sites within NT5E 3ʹUTR, the mutated seed sequence, and sequence of miR-628-5p. (i and j) Dual-luciferase reporter assays of NT5E 3ʹUTR predicted to be regulated by miR-628-5p. (k) qRT-PCR of miR-628-5p in 47 gastric cancer tissues and 47 matched normal gastric tissues. (l and m) Correlation between miR-628-5p and circ_HN1 or NT5E mRNA expression in 47 gastric primary tumors. qRT-PCR of miR-628-5p (n) and NT5E mRNA (o), and western blot of NT5E protein (p) in HGC27 and MKN74 cells transfected by anti-miR-628-5p, anti-miR-NC, miR-628-5p mimic, or miR-NC mimic. *P < 0.05
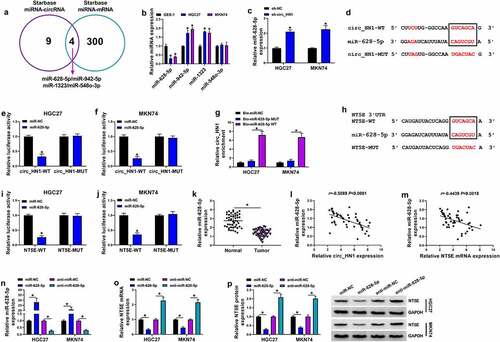
MiR-628-5p reduction reverses the impact of circ_HN1 silencing on cell functional behaviors
In order to further validate the effects of circ_HN1 depletion were mediated by miR-628-5p, we reduced miR-628-5p expression with anti-miR-628-5p introduction in circ_HN1-silenced HGC27 and MKN74 cells (). By contrast, reduced level of miR-628-5p reversed circ_HN1 depletion-mediated NT5E expression inhibition ( and 6c), reinforcing the role of circ_HN1 as a regulator of NT5E expression via miR-628-5p. Indeed, down-regulation of miR-628-5p strongly counteracted circ_HN1 depletion-mediated anti-colony formation (), anti-spheroid formation (), anti-proliferation (), pro-apoptosis (), anti-migration (), as well as anti-invasion () effects in HGC27 and MKN74 cells. In summary, these data indicate that miR-628-5p is a downstream mediator of circ_HN1 function.
Figure 6. Circ_HN1 depletion exerts regulatory effects by up-regulating miR-628-5p. Stable sh-NC- or sh-circ_HN1-infected HGC27 and MKN74 cells were transfected with anti-miR-628-5p or anti-miR-NC. (a) MiR-628-5p expression in transfected cells. (b) Relative NT5E mRNA level in transfected cells gauged by qRT-PCR. (c) Western blot showing the level of NT5E protein. (d) Colony formation using a standard colony formation assay. (e) Spheriod formation. Phase contrast micrographs of day 14 in ultra-low attachment plate culture. Proliferation capacity of transfected cells examined by EdU (f) and MTT (g and h) assays. (i) Western blot of PCNA protein level in transfected HGC27 and MKN74 cells. (j) Apoptosis of transfected HGC27 and MKN74 cells by flow cytometry. (k and l) Western blot showing Bcl-2 and Bax levels in transfected HGC27 and MKN74 cells. (m and n) Representative images showing cell migration and invasion assays performed by transwell assay. *P < 0.05
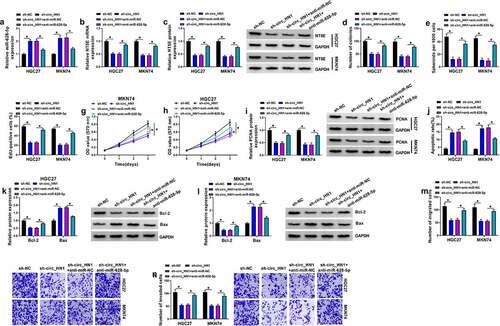
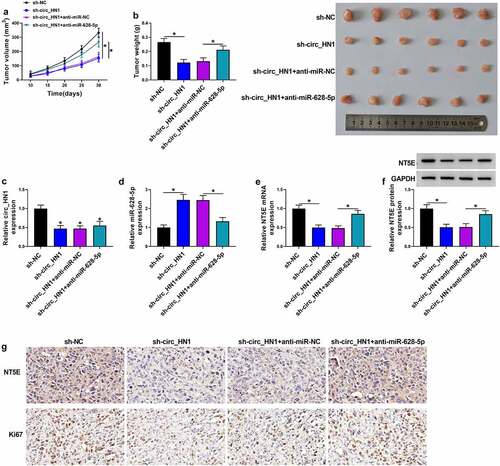
Depletion of circ_HN1 reduces tumor growth by up-regulating miR-628-5p in vivo
We further explored whether miR-628-5p was responsible for the effect of circ_HN1 on tumor growth in vivo. HGC27 cells transduced with sh-circ_HN1 formed markedly smaller xenograft tumors than the same cells transduced by the sh-NC control ( and 7b). Moreover, the introduction of anti-miR-628-5p significantly reversed the sh-circ_HN1-imposed tumor growth suppression in vivo ( and 7b). Sh-circ_HN1-transduced HGC27 tumors exhibited lower levels of circ_HN1 and NT5E and higher expression of miR-628-5p compared with the sh-NC group (). However, the introduction of anti-miR-628-5p strongly reversed sh-circ_HN1-mediated miR-628-5p elevation and NT5E down-regulation (). Additionally, circ_HN1-silenced tumors had remarkably fewer cells stained for Ki67 staining than the controls, and anti-miR-628-5p introduction reversed this impact of circ_HN1 depletion (). All these findings demonstrate that depletion of circ_HN1 diminishes tumor growth by up-regulating miR-628-5p in vivo.
Figure 7. Knocking down circ_HN1 diminishes tumor growth in vivo. (a) Graphic representation of tumor volume of mice implanted with stable sh-NC- or sh-circ_HN1-infected HGC27 cells (n = 6 per group) and performed intratumor injection of PBS, anti-miR-NC or anti-miR-628-5p. (b) Images and mean weight of the xenograft tumors derived from sh-circ_HN1-transduced or sh-NC-infected HGC27 cells with intratumor injection of PBS, anti-miR-NC or anti-miR-628-5p at the end point (n = 6 per group). qRT-PCR of circ_HN1 (c), miR-628-5p (d), NT5E mRNA (e), and western blot of NT5E protein (f) in the sh-circ_HN1-transduced or sh-NC-infected HGC27 tumors with intratumor injection of PBS, anti-miR-NC or anti-miR-628-5p at day 30 after cell implantation. (g) Representative images showing NT5E and Ki67 staining of sections from sh-circ_HN1-transduced or sh-NC-infected HGC27 tumors with intratumor injection of PBS, anti-miR-NC or anti-miR-628-5p at the end of the experiments. *P < 0.05
Discussion
The functional ceRNA crosstalk is crucial for the oncogene regulation during the tumorigenic processes [Citation11,Citation30]. Increasing experimental evidence illuminates the implications of circRNA-mediated ceRNA activity in human cancers, including gastric cancer [Citation31,Citation32]. For instance, Gao et al. uncovered that circ_0000117 contributed to gastric tumorigenesis by working as a ceRNA for miR-337-3p to induce signal transducer and activator of transcription 3 (STAT3) [Citation6]. Yang et al. ascertained that circ_0005654 promoted specificity protein 1 expression to drive gastric cancer development via miR-363 competition [Citation33]. Conversely, circ_LARP4 is a potent tumor-inhibitory circRNA in gastric carcinogenesis depending on its ceRNA activity [Citation34]. The findings described here firstly showed the suppressive effect of circ_HN1 depletion on gastric tumorigenesis, in agreement with a recent report [Citation12]. Based on these observations, we focused on the ceRNA activity of circ_HN1 in gastric cancer.
NT5E is a well-known oncogene in numerous tumors and it is a potential therapeutic target for cancer immunotherapy [Citation13,Citation35,Citation36]. Our data first demonstrated that silencing of circ_HN1 strongly down-regulated NT5E in gastric cancer cells, and suppression of NT5E phenocopied circ_HN1 depletion in impeding gastric cancer progression. CircRNAs modulate gene expression through multiple mechanisms, including miRNA sequesteration [Citation37]. Considering the positive regulation of circ_HN1 on NT5E expression and the main cytoplasmic localization of circ_HN1, we hypothesized that circ_HN1 may post-transcriptionally regulate NT5E by sponging some miRNAs.
Here, we first ascertained that miR-628-5p was a shared miRNA for circ_HN1 and NT5E. Furthermore, circ_HN1 modulated NT5E expression by competitively pairing to miR-628-5p. Several previous documents have uncovered the conflicting roles of miR-628-5p in tumor progression [Citation38–40]. These contradictory observations may be partially due to the different types of cancers in these studies, where miR-628-5p contributes to osteosarcoma progression [Citation40] and works as a strong anti-cancer agent in prostate cancer [Citation38] and ovarian cancer [Citation39]. Moreover, miR-628-5p can hinder gastric tumorigenesis by inhibiting its target mRNAs [Citation19,Citation20]. Importantly, Deng and colleagues showed that miR-628-5p was involved in a long ncRNA DLG associated protein 1 antisense RNA 1 (DLGAP1-AS1)-mediated ceRNA network in gastric cancer [Citation20]. In this report, miR-628-5p was a functional mediator of circ_HN1 in affecting gastric tumorigenesis. Additionally, the tumor xenograft assays implied the important involvement of the circ_HN1/miR-628-5p/NT5E ceRNA crosstalk in tumor growth in vivo, which should be further studied in future work. Similarly, Wang and colleagues reported that the circ_HN1/miR-302b-3p/ROCK2 ceRNA axis impacted gastric tumorigenesis [Citation12]. Xu et al. showed that NT5E possessed oncogenic activity in gastric cancer by regulating the RICS/RhoA signaling [Citation18]. Future work will build on the findings by identifying how the circ_HN1/miR-628-5p/NT5E ceRNA axis regulates gastric tumorigenesis. Additionally, when we used the online database starBase to observe the correlation between miR-628-5p/NT5E expression and overall survival of gastric cancer patients, we found that the expression of NT5E (p = 0.00019), rather than miR-628-5p (p > 0.05), was strongly correlated with the overall survival of the patients with gastric cancer; the patients with low NT5E levels had significantly longer survival time than those with high NT5E levels (Supplement ), suggesting that NT5E may be a potential prognostic biomarker of gastric cancer.
With these findings, we envision that circ_HN1 inhibitor (e.g. shRNA or lentivirus expressing shRNA) may be a potential point for the development of circRNA-based targeted therapies against gastric cancer. Based on a previous report [Citation41], intravenous injection of circ_HN1 inhibitor may be a potential method for gastric cancer treatment. Further research is warranted in this field.
Conclusion
In summary, we have defined a novel ceRNA crosstalk, the circ_HN1/miR-628-5p/NT5E axis, for the oncogenic activity of circ_HN1 in gastric cancer. Our findings provide a rationale for developing circ_HN1 as a therapeutic agent against gastric cancer.
Authors’ contributions
Mingbo Cao designed and supervised the study, Jianmin Zhang conducted the experiments, drafted the manuscript, Haihui Zhang collected and analyzed the data. Fang Wang contributed the methodology and edited the manuscript. The authors declare that all data were generated in-house and that no paper mill was used.
Ethical statement
All patients provided informed consent and the study was conducted under the approval of the Ethics Committee of Henan Provincial People’s.
Animal experiments were conducted following a protocol approved by the Ethics Committee on Animal Care of Henan Provincial People’s Hospital.
Supplemental Material
Download Zip (912 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396(10251):635–648.
- Machlowska J, Baj J, Sitarz M, et al. Gastric cancer: epidemiology, risk factors, classification, Genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21(11):4012.
- Yuan G, Ding W, Sun B, et al. Upregulated circRNA_102231 promotes gastric cancer progression and its clinical significance. Bioengineered 2021;12(1):4936–4945.
- Ning L, Zhang M, Zhu Q, et al. miR-25-3p inhibition impairs tumorigenesis and invasion in gastric cancer cells in vitro and in vivo. Bioengineered 2020;11(1):81–90.
- Gao Q, Liu Q, Chen H. Circular RNA hsa_circ_0000117 accelerates the proliferation and invasion of gastric cancer cells by regulating the microRNA-337-3p/signal transducer and activator of transcription 3 axis. Bioengineered 2021;12(1):1381–1390.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18.
- Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5(4):105.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Iwakawa HO, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25(11):651–665.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Wang D, Jiang X, Liu Y, et al. Circular RNA circ_HN1 facilitates gastric cancer progression through modulation of the miR-302b-3p/ROCK2 axis. Mol Cell Biochem. 2021;476(1):199–212.
- Gao ZW, Dong K, Zhang HZ. The roles of CD73 in cancer. Biomed Res Int. 2014;2014:460654.
- Lo Nigro C, Monteverde M, Lee S, et al. NT5E CpG island methylation is a favourable breast cancer biomarker. Br J Cancer. 2012;107(1):75–83.
- Goswami S, Walle T, Cornish AE, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med. 2020;26(1):39–46.
- Lu XX, Chen YT, Feng B, et al. Expression and clinical significance of CD73 and hypoxia-inducible factor-1α in gastric carcinoma. World J Gastroenterol. 2013;19(12):1912–1918.
- Hu S, Meng F, Yin X, et al. NT5E is associated with unfavorable prognosis and regulates cell proliferation and motility in gastric cancer. Biosci Rep. 2019;39(5):BSR20190101.
- Xu Z, Gu C, Yao X, et al. CD73 promotes tumor metastasis by modulating RICS/RhoA signaling and EMT in gastric cancer. Cell Death Dis. 2020;11(3):202.
- Chen Y, Wu Y, Yu S, et al. Deficiency of microRNA-628-5p promotes the progression of gastric cancer by upregulating PIN1. Cell Death Dis. 2020;11(7):559.
- Deng J, Zhang Q, Lu L, et al. Long noncoding RNA DLGAP1-AS1 promotes the aggressive behavior of gastric cancer by acting as a ceRNA for microRNA-628-5p and raising astrocyte elevated gene 1 expression. Cancer Manag Res. 2020;12:2947–2960.
- Jaca A, Govender P, Locketz M, et al. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J Clin Pathol. 2017;70(4):331–356.
- Gu E, Pan W, Chen K, et al. LncRNA H19 Regulates Lipopolysaccharide (LPS)-induced apoptosis and inflammation of BV2 microglia cells through targeting miR-325-3p/NEUROD4 Axis. J Mol Neurosci. 2021;71(6):1256–1265.
- Zhao F, Jia Z, Feng Y, et al. Circular RNA circ_0079593 enhances malignant melanoma progression by the regulation of the miR-573/ABHD2 axis. J Dermatol Sci. 2021;102(1):7–15.
- Parashar D, Geethadevi A, McAllister D, et al. Targeted biologic inhibition of both tumor cell-intrinsic and intercellular CLPTM1L/CRR9-mediated chemotherapeutic drug resistance. NPJ Precis Oncol. 2021;5(1):16.
- Cai S, Zhu Q, Guo C, et al. MLL1 promotes myogenesis by epigenetically regulating Myf5. Cell Prolif. 2020;53(2):e12744.
- Chen C, Gupta P, Parashar D, et al. ERBB3-induced furin promotes the progression and metastasis of ovarian cancer via the IGF1R/STAT3 signaling axis. Oncogene. 2020;39(14):2921–2933.
- Kanda T, Sugihara T, Takata T, et al. Low-density lipoprotein receptor expression is involved in the beneficial effect of photodynamic therapy using talaporfin sodium on gastric cancer cells. Oncol Lett. 2019;17:3261–3266.
- Zhao X, Dong W, Luo G, et al. Silencing of hsa_circ_0009035 suppresses cervical cancer progression and enhances radiosensitivity through MicroRNA 889-3p-dependent regulation of HOXB7. Mol Cell Biol. 2021;41(6):e0063120.
- Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293.
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121.
- Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661–1680.
- Guan YJ, Ma JY, Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19(1):183.
- Yang C, Han S. The circular RNA circ0005654 interacts with specificity protein 1 via microRNA-363 sequestration to promote gastric cancer progression. Bioengineered. 2021;12(1):6305–6317.
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151.
- Ghalamfarsa G, Kazemi MH, Raoofi Mohseni S, et al. CD73 as a potential opportunity for cancer immunotherapy. Expert Opin Ther Targets. 2019;23(2):127–142.
- Chen S, Wainwright DA, Wu JD, et al. CD73: an emerging checkpoint for cancer immunotherapy. Immunotherapy. 2019;11(11):983–997.
- Braicu C, Zimta AA, Gulei D, et al. Comprehensive analysis of circular RNAs in pathological states: biogenesis, cellular regulation, and therapeutic relevance. Cell Mol Life Sci. 2019;76(8):1559–1577.
- Chen J, Hao P, Zheng T, et al. miR-628 reduces prostate cancer proliferation and invasion via the FGFR2 signaling pathway. Exp Ther Med. 2019;18:1005–1012.
- Li M, Qian Z, Ma X, et al. MiR-628-5p decreases the tumorigenicity of epithelial ovarian cancer cells by targeting at FGFR2. Biochem Biophys Res Commun. 2018;495(2):2085–2091.
- Wang JY, Wang JQ, Lu SB. miR-628-5p promotes growth and migration of osteosarcoma by targeting IFI44L. Biochem Cell Biol. 2020;98(2):99–105.
- Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–347.
