ABSTRACT
Chronic exposure to high concentrations of circulating palmitic acid and stearic acid leads to impaired β cell function, which accelerates the development of type 2 diabetes. However, differences in the mechanisms underlying this process between these two saturated fatty acids remain largely unknown. In this study, we screened for potential circular RNAs (circRNAs) and their associated regulatory pathways in palmitic acid- and stearic acid-induced mouse β-TC6 cell dysfunction. CircRNA high-throughput sequencing, gene ontology enrichment and Kyoto Encyclopedia of Genes and Genomes analysis were performed and co-expression and competing endogenous RNAs (ceRNA) networks were constructed. We identified that four circRNAs that were differentially expressed specifically in β cells exposed to palmitic acid, whereas four circRNAs were differentially expressed specifically in β cells exposed to stearic acid. Seven circRNAs were differentially co-expressed in palmitic acid- and stearic acid-treated β cells. In pathway exploration, we identified the core protein Solute carrier family 2 member 2 (SLc2a2), which is mainly involved in insulin resistance, maturity onset diabetes of the young and type 2 diabetes. The expressions of key circRNAs in β-TC6 cells were validated by Real time quantitative PCR, with a consistent result in high-throughput sequencing. The findings aid our understanding of the mechanisms governing the difference between palmitic acid- and stearic acid-induced β cell dysfunction and provide potential therapeutic targets for developing treatments against long-term high fat diet-induced β cell injury.
Abbreviations: Acvr1c: Activin A receptor, type 1C; CeRNA, Competing endogenous RNAs; circRNA, circular RNA; DEcircRNA: Differentially Expressed circular RNA; DEmiRNA: Differentially Expressed microRNA; DEmRNA: Differentially Expressed mRNA; GO: Gene Ontology; HPDHigh Palmitic acid Diet; HSD: High Stearic acid Diet; KEGG: Kyoto Encyclopedia of Genes and Genomes; miRNA: microRNA; ncRNAs: non-coding RNAs; qPCR: Real time quantitative PCRS; FAs: Saturated Fatty Acids; SLc2a2: Solute carrier family 2 member 2; T2D: Type 2 Diabetes
Introduction
Long-term consumption of a high fat diet can lead to a significant increase in the circulating concentration of saturated fatty acids (SFAs) [Citation1,Citation2]. Chronic exposure to high levels of SFAs results in β cell dysfunction, which is a leading cause of the development of type 2 diabetes (T2D) [Citation3,Citation4]. Both palmitic acid (C16:0) and stearic acid (C18:0) are the main constituents of SFAs in food or serum and accumulating evidence suggests that these two SFAs exert their functions in different ways in many similar biological processes [Citation5,Citation6]. Our previous study also indicated that the destructive effect of palmitic acid and stearic acid on pancreatic β cells differ significantly; however, the causes of this difference remain unresolved.
In recent years, non-coding RNAs (ncRNAs) have received increasing attention as important regulators in various biological processes and in the regulation of a series of human metabolic diseases, such as obesity, atherosclerosis and T2D [Citation7–9]. There is strong evidence suggesting that different types of ncRNAs, including long non-coding RNAs and microRNAs (miRNAs), play a critical role in SFA-induced β cell dysfunction [Citation10–12]. However, the role of circular RNAs (circRNAs) in inducing such dysfunction has not been elucidated. CircRNAs generally exist in eukaryotic transcription groups in large quantities, are usually generated from precursor mRNAs with a closed loop and lack poly-adenylated and cap structures [Citation13]. These RNAs are expressed in cell-type specific manners and conserved across species. Growing evidence indicates that circRNAs function via multiple mechanisms, including controlling their parent gene [Citation14–16] interacting with RNA-binding proteins or miRNAs [Citation17–19] and competing with the splicing of linear transcripts [Citation20]. Currently, circRNAs are considered to be novel regulators of β cell activities [Citation21], however, the role of circRNAs in β cell dysfunction induced by palmitic acid and stearic acid is poorly understood.
In the present study, we aimed to explore the differences of circRNAs and their related signaling pathways in SFAs-induced β cell dysfunction between palmitic acid and stearic acid. High-throughput sequencing and bioinformatics analysis were used to investigate the expression profile of circRNAs and screen for differentially expressed (DE) circRNAs in palmitic acid- and stearic acid-treated β-TC6 cells, respectively. We then performed gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Furthermore, we compared and predicted the novel circRNA-miRNA-mRNA competing endogenous RNAs (ceRNA) regulatory mechanisms between palmitic acid- and stearic acid-induced β cell dysfunction. This study enriches our understanding of the mechanisms governing the difference between palmitic acid- and stearic acid-induced β cell dysfunction and provides new targets and therapeutic strategies for T2D prevention.
Material and methods
Chemicals
Stock solutions of palmitic acid (P0500, Sigma-Aldrich, St. Louis, MO, USA) and stearic acid (S4751, Sigma-Aldrich) were prepared as described previously [Citation22]. Palmitic acid or stearic acid (0.1 g) was dissolved in 20 ml ethanol and then saponified with 244 μl sodium hydroxide (1.6 mol/l). After drying, the sodium salt was re-suspended in saline to 65.5 ml at 80°C for 4 h. Then, 65.5 ml of 20% (w/v) bovine serum albumin was added and stirred at 50°C for 2 h until the mixture was dissolved completely. Finally, this solution was sterilized and aliquoted for storage at −20°C.
Cell culture
Mouse β-TC6 cells were obtained from the Cell Library of Chinese Academy of Sciences (Shanghai, China), and mouse AML12 cells and C2C12 cells were purchased from the Global Bioresource Center (Manassas, VA, USA). The β-TC6 cells and C2C12 cells were incubated in Dulbecco’s Modified Eagle’s Medium supplemented with 15% (v/v) fetal bovine serum (Gibco, Waltham, MA, USA), 50 μg/l streptomycin and 50 IU/l penicillin (Gibco). The AML12 cells were cultured in Dulbecco’s Modified Eagle’s Medium with 10% (v/v) fetal bovine serum (Gibco), 50 μg/l streptomycin and 50 IU/l penicillin (Gibco), 1% (v/v) Insulin-Transferrin-Selenium (Gibco) and 40 ng/ml dexamethasone (Sigma). All cells were treated with palmitic acid or stearic acid at 400 μmol/l for 24 h.
circRNA sequencing
An Illumina HiSeq system was used to perform circRNA sequencing by GENEWIZ (Suzhou, China). Total RNA of nine samples were extracted by the TRIzol Reagent (Invitrogen, Carlsbad, CA, USA)/RNeasy Mini Kit (Qiagen, Hilden, Germany) with ribosomal RNA (rRNA) depletion and quantified and qualified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1% (w/v) agarose gels. One microgram total RNA with an RIN value above seven was used for the following library preparation. Following the manufacturer’s protocol (NEBNext®UltraTMDirectional RNA Library Prep Kit for Illumina®, New England Biolabs, Inc. Ipswich, MA, USA), next-generation sequencing library preparation was constructed. First-strand cDNA was synthesized by ProtoScript II Reverse Transcriptase with random primers and actinomycin D. Second-strand cDNA was synthesized using Second Strand Synthesized Enzyme Mix (includes dACG-TP/dNTP). The double-stranded cDNA was purified by using AxyPrep Mag PCR Clean-up (Axygen) and subsequently treated with End Prep Enzyme Mix to repair both ends and add a dA-tail in one reaction. This was followed by T-A ligation to add adaptors to both ends. Size selection of the adaptor-ligated DNA was then performed using AxyPrep Mag PCR Clean-up, and fragments of ~360 bp (with the approximate insert size of 300 bp) were recovered. The dUTP-marked second strand was digested with Uracil-Specific Excision Reagent (USER) enzyme (New England Biolabs, Ipswich, MA, USA). Each sample was amplified by PCR for 11 cycles using P5 and P7 primers with both primers carrying sequences that anneal with the flow cell to perform bridge PCR, and the P7 primer carrying a six-base index facilitated multiplexing. The PCR products were purified by AxyPrep Mag PCR Clean-up, validated using an Agilent 2100 Bioanalyzer and quantified by Qubit 2.0 Fluorometers (Invitrogen). Sequencing was implemented using a 2 × 150 paired-end configuration and image analysis and base calling were conducted by the Hiseq Control Software (HCS) +OLB+GAPipeline-1.6 (Illumina) on the Hiseq instrument.
Mapping and identification of circRNAs
Reference genome sequences and gene model annotation files of relative species were downloaded from genome websites, such as UCSC, NCBI and ENSEMBLE. Then, bwa (0.7.12-r1039) was used to index reference genome sequences and alignments were performed for clean data. CIRI (V2.0) software was used to identify circRNAs.
Differential expression analysis of circRNAs
Junction reads at back-splicing loci of circRNAs were used to calculate the expression levels of circRNAs. SRPBM (spliced reads per billion mapping) was used to normalize the reads. Differential expression analysis used the DESeq Bioconductor package (DESeq2 v1.6.3 and DESeq v1.18.0), a model based on the negative binomial distribution, with |log2FC| > 1 and P-value <0.05 set as thresholds.
Prediction of circRNA-miRNA and miRNA-mRNA interactions
We predicted interactions between circRNAs and miRNAs by combining Targetscan 5.0 and miRanda 3.3a software with a perfect seed match. Full-length sequences of circRNA and miRNA were selected. The intersection of the results was set at a TargetScan score ≤ 50 and miRanda_Energy ≤ −10. miRNA-mRNA interactions were identified from both TargetScan and miRwalk database, as well as high-throughput sequencing (log2fold ≥ 1 or log2fold ≤ −1) in this study and our previous study [Citation23]. Finally, candidate DEmiRNAs and DEmRNAs were selected through cross-prediction results from miRwalk database and high-throughput sequencing (P ≤ 0.05).
Functional enrichment analyses
Potential biological functions of RNAs with different levels of expression were predicted by functional enrichment, including GO terms and pathway analysis [Citation23]. In GO terms, a series of enriched genes (with a significant P-value less than 0.05) were annotated using GO-TermFinder for identification. We used scripts in the KEGG database to enrich significantly differentially expressed genes and perform pathway analysis.
Construction of DEcircRNA-DEmiRNA-DEmRNA co-expression and ceRNA networks
We used TargetScanMouse (version 7.2) and miRwalk (version 2.0) to predict the interaction between DEcircRNA-DEmiRNA and DEmiRNA-DEmRNA based on sequence complementarity, conserved target sites, and formed free energy information, as described previously [Citation24–27]. DEcircRNA-DEmiRNA-DEmRNA regulatory axes were included in the network. A ceRNA network was constructed when circRNA and mRNA share the same miRNA, and circRNA is positively related to mRNA (P < 0.05). Cytoscape (version 3.7.2) was used to visualize the results and build DEcircRNA-DEmiRNA-DEmRNA co-expression and ceRNA networks.
Animal experiments
Seven-week male C57BL/6 mice from Beijing Vital River Laboratory Animal Technology Company (Beijing, China) were randomly divided into normal diet, high stearic acid diet (HSD) and high palmitic acid diet group (n = 5 each group). Mice were fed at constant temperature (20°C) in a constant humidity (60% relative humidity) laboratory with no food or water restrictions. After 12 weeks feeding, mice islets were collected [Citation22] for circRNA measurement. All animal procedures were approved by the Institutional Animal Protection and Use Committee of Harbin Medical University and were in accordance with the guidelines of the Animal Experimentation Center of Harbin Medical University. The composition for the diets used in this study is the same with that in our previous study [Citation28].
Real-time quantitative (q) PCR and sanger sequencing
Total RNA was extracted from mouse pancreatic islets and cell lines using TRIzol reagent (Invitrogen), and the expression level of β-actin was used as an internal control. qPCR was performed with the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Additionally, all primers were synthesized (Supplementary Table 1, https://figshare.com/s/da74d93a44f6f595b683) and PCR products were sequenced by Sangon Biotech Co., Ltd (Shanghai, China) (Supplementary Figure 1, https://figshare.com/s/1e654918249b55ccabfa).
Statistical analysis
All data were reported as mean ± standard deviation and analyzed using SPSS 21.0 (IBM Inc., Armonk, NY, USA). Comparisons between two groups were analyzed by Student’s t-test and multiple groups using one-way ANOVA, followed by the Student–Newman–Keuls test. Statistical significance was set at P < 0.05.
Data and resource availability`
The dataset generated and analyzed in this study is available from the corresponding authors upon reasonable request.
Results
In this study, a comparison of DEcircRNAs and their signaling pathways in β cell dysfunction induced by palmitic acid and stearic acid were performed using CircRNA high-throughput sequencing and bioinformatics analysis. We constructed DEcircRNA-DEmiRNA-DEmRNA co-expression and ceRNA networks to explore the potential signaling pathways. Moreover, the expressions of the candidate circRNAs (4 specific to palmitic acid group; 4 specific to stearic acid group; 7 common to both palmitic and stearic acid groups) were verified by qPCR in β-TC6 cells, mice islets, AML 12 and C2C12 cells. These findings provide alternative preventive targets for high fat diet-induced β cell dysfunction in the development of T2D.
Identification of circRNAs in palmitic acid- and stearic acid-treated β-TC6 cells
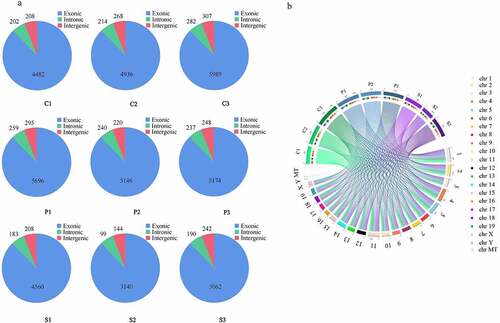
A total of 19,022 circRNAs were identified in the nine samples, of which 820 were annotated and 18,202 were novel unannotated circRNAs, including 16,747 exonic circRNAs (88.04%), 1205 intronic circRNAs (6.33%) and 1070 intergenic circRNAs (5.63%) ()). The circRNAs transcripts were distributed across all chromosomes ()).
Analysis of DEcircRNAs in palmitic acid- and stearic acid-treated β-TC6 cells
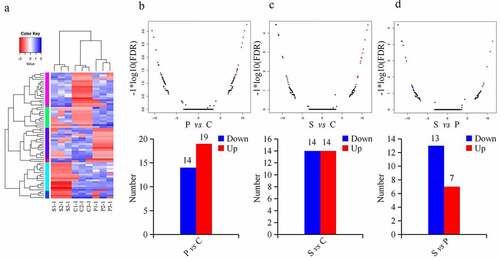
Hierarchical clustering analysis showed that the expression of circRNAs from the three groups were distinct ()). Compared with the control group, there were 33 circRNAs differentially expressed in the palmitic acid group, including 19 upregulated and 14 downregulated circRNAs (), Supplementary Table 2, https://figshare.com/s/da74d93a44f6f595b683). In stearic acid-treated β-TC6 cells, 14 circRNAs were upregulated and 14 circRNAs were downregulated (), Supplementary Table 3, https://figshare.com/s/da74d93a44f6f595b683). Compared with palmitic acid group, seven upregulated and 13 downregulated circRNAs were exclusively expressed in the stearic acid group (), Supplementary Table 4, https://figshare.com/s/da74d93a44f6f595b683).
Figure 2. DEcircRNAs in β-TC6 cells analyzed by pairwise comparison. (a) Hierarchical clustering analysis of circRNAs in the three groups. (b–d) Volcano plots and histograms comparing DEcircRNAs in the palmitic acid and control group, stearic acid and control group, and stearic acid group and palmitic acid group. Red and blue points represent upregulated and downregulated circRNAs, respectively. P vs C, palmitic acid versus control group; S vs C, stearic acid versus control group; S vs P, stearic acid versus palmitic acid group
Comparative analysis of DEcircRNAs between each group
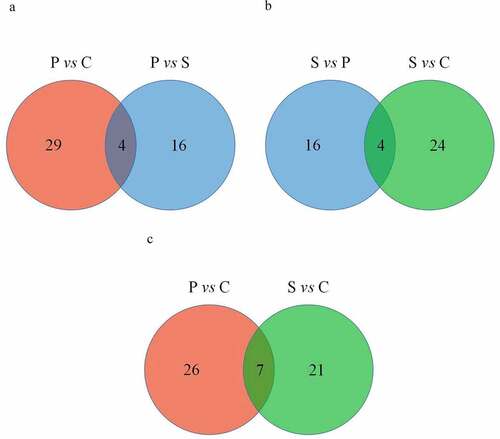
Venn diagram analysis showed that four DEcircRNAs were selected specifically in the palmitic acid group: chr14_16277799_16283473_+ was upregulated by 8.74 log2fold change, whereas chr10_43393871_43413547_+, chr2_155432816_155440785_− and chr5_117500713_117505639_+ were downregulated by −9.09, −8.38 and −10.53 log2fold change, respectively (), ). Four circRNAs, chr11_31055458_31061586_+, chr6_31139729_31181149_−, chr6_145032783_145039635_− and chr13_100763550_100772781_−, were exclusively differentially expressed in the stearic acid-treated β-TC6 cells, of which these circRNAs were downregulated by −10.99, −10.91, −9.86 and −8.82 log2fold change, respectively (), ). Additionally, seven circRNAs were differentially expressed in both palmitic acid and stearic acid groups ()). These circRNAs were chr14_31030844_31036059_+, chr15_72999972_73031643_−, chr19_28530873_28540408_−, chr1_133001283_133018884_−, chr2_34204780_34213442_−, chr6_31163894_31,197,851_− and chr9_121727316_121731565_+ ().
Table 1. The differential expressed circRNAs specific to palmitic acid-induced β-TC6 cells compared both with stearic acid and control group
Table 2. The differential expressed circRNAs specific to stearic acid-induced β-TC6 cells compared both with palmitic acid and control group
Table 3. The common differentially expressed circRNAs in stearic acid and palmitic acid group
Figure 3. Comparative analysis of DEcircRNAs between each group using a Venn diagram. (a–b) The number of circRNAs specific to the palmitic acid group and stearic acid group. (c) The number of commonly expressed circRNAs in both the palmitic acid and stearic acid groups. P vs C, palmitic acid versus control group; S vs C, stearic acid versus control group; S vs P, stearic acid versus palmitic acid group
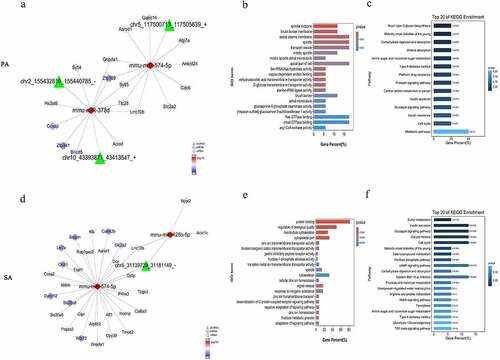
Prediction of the specific interactions among circRNAs, miRNAs and mRNAs in palmitic acid-treated β-TC6 cells
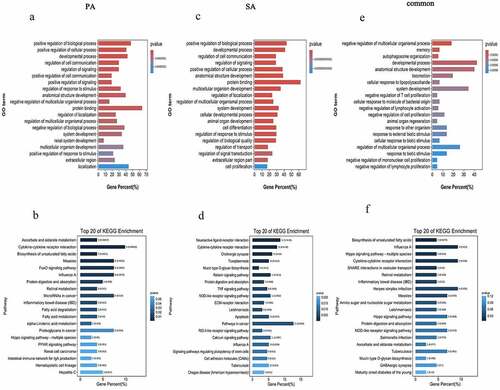
Based on the miRanda (3.3a) database, there are 92 miRNAs (Supplementary Table 5, https://figshare.com/s/da74d93a44f6f595b683) that have possible relationships with the four circRNAs specific to the palmitic acid group. Combined with the sequencing results, mmu-miR-378d and mmu-miR-574-5p were predicted to target chr2_155432816_155440785_−, chr10_43393871_43413547_+ and chr5_117500713_117505639_+. These two miRNAs target 192 DEmRNAs (168 upregulated and 24 downregulated) (Supplementary Table 6, https://figshare.com/s/da74d93a44f6f595b683), and these genes are closely related to protein binding, positive regulation of biological processes, localization, developmental processes, and positive regulation of cellular processes according to GO enrichment analysis (), Supplementary Table 7, https://figshare.com/s/da74d93a44f6f595b683). Moreover, KEGG pathway analysis showed that these 192 DEmRNAs are involved in biological regulatory pathways such as cytokine-cytokine receptor interactions, the FoxO (Forkhead box, sub-group O) signaling pathway and miRNAs involved in cancer, measles, and influenza A (), Supplementary Table 8, https://figshare.com/s/da74d93a44f6f595b683).
Figure 4. GO and KEGG pathway analyses of dysregulated mRNAs in the DEcircRNA-DEmiRNA-DEmRNA triple network. Top 20 enrichment GO terms of DEmRNAs in the palmitic acid group (a), stearic acid group (c) and the common to both palmitic acid and stearic acid group (e). The length of the bar represents the number of DEmRNAs. The color of the bar is related to the P-value with smaller P-values closer to red. Top 20 KEGG pathways of DEmRNAs in the palmitic acid group (b), stearic acid group (d) and the common to both palmitic acid and stearic acid group (f). The color of the bar is related to the P-value with smaller P-values closer to dark blue. PA, palmitic acid group; SA, stearic acid group
Prediction of the specific relationships among circRNAs, miRNAs and mRNAs in stearic acid-treated β-TC6 cells
Based on the miRanda (3.3a) database, we obtained 145 miRNAs (Supplementary Table 9, https://figshare.com/s/da74d93a44f6f595b683) that were closely related to the four circRNAs specific to the stearic acid group. After combining with High-Seq results, downregulated mmu-miR-770-5p and mmu-miR-323-5p and upregulated mmu-miR-574-5p and mmu-miR-125b-5p were predicted to have relationships with these four circRNAs. Additionally, 456 DEmRNAs were selected, 378 upregulated and 78 downregulated (Supplementary Table 10, https://figshare.com/s/da74d93a44f6f595b683), and GO enrichment analysis revealed the top 20 significantly enriched GO terms and showed that these 456 DEmRNAs are related to protein binding, positive regulation of biological processes, developmental processes, anatomical structure development and positive regulation of cellular processes. (), Supplementary Table 11, https://figshare.com/s/da74d93a44f6f595b683). KEGG enrichment demonstrated that these DEmRNAs are major involved in cancer-related pathways and are also widely involved in neuroactive ligand-receptor interactions, cytokine-cytokine receptor interactions and the NOD-like receptor signaling pathway. These genes also participate in protein digestion and absorption, the TNF signaling pathway, apoptosis, and cell adhesion molecules (), Supplementary Table 12, https://figshare.com/s/da74d93a44f6f595b683).
Searching for the relationship among circRNAs, miRNAs and mRNAs in both the palmitic acid and stearic acid groups
195 miRNAs were predicted to be associated with the seven circRNAs identified to be differentially co-expressed in both the palmitic acid and stearic acid groups (Supplementary Table 13, https://figshare.com/s/da74d93a44f6f595b683). Among them, mmu-miR-574-5p was found to be associated with both chr6_31163894_31197851_− and 88 DEmRNAs (83 upregulated and 5 downregulated) (Supplementary Table 14, https://figshare.com/s/da74d93a44f6f595b683), of which these enriched GO terms have been implicated in many biological modulation activities such as developmental processes, anatomical structure development and system development (), Supplementary Table 15, https://figshare.com/s/da74d93a44f6f595b683). KEGG enrichment revealed that these DEmRNAs are associated with the process of influenza A, cytokine-cytokine receptor interactions and herpes simplex infection (), Supplementary Table 16, https://figshare.com/s/da74d93a44f6f595b683).
Construction of DEcircRNA-DEmiRNA-DEmRNA networks
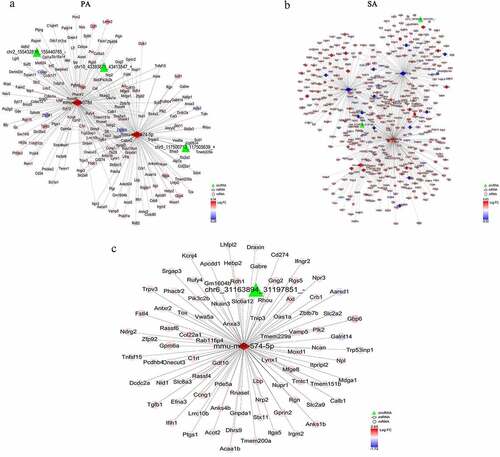
We constructed three co-expression networks for the candidate circRNAs, miRNAs and mRNAs identified to be specific to the palmitic acid, stearic acid and the group common to both SFAs. The co-expression network specific to the palmitic acid group was composed of three DEcircRNAs, two DEmiRNA and 192 DEmRNAs nodes ()). The co-expression network specific to the stearic acid group included two DEcircRNAs, four DEmiRNAs and 456 DEmRNAs ()), whereas the co-expression network specific to both palmitic acid and stearic acid included chr6_31163894-31197851, mmu-miR-574-5p and 88 DEmRNAs nodes ()).
Figure 5. The DEcircRNAs-DEmiRNAs-DEmRNAs regulatory network. (a) View of the DEcircRNAs-DEmiRNAs-DEmRNAs network specific to the palmitic acid group. The network consists of 261 edges with three DEcircRNAs, two DEmiRNAs and 192 DEmRNAs. (b) In the stearic acid group, the preliminary DEcircRNAs-DEmiRNAs-DEmRNAs network consists of two circRNAs, four miRNAs, 456 genes and 644 edges. (c) DEcircRNAs-DEmiRNAs-DEmRNAs co-expression regulatory network in the common to both palmitic acid and stearic acid group. The red and blue nodes represent upregulated and downregulated DEmRNAs, respectively. Edges denote their relationship
Construction of a sub-network of ceRNA
Sub-networks of ceRNA specific to the palmitic acid and stearic acid groups were created. In the palmitic acid group, chr2_155432816_155440785_− and chr10_43393871_43413547_+ were found to compete for mmu-miR-378d, whereas chr5_117500713_117505639_+ may act as a sponge of mmu-miR-574-5p (), ). The targeted mRNAs () play a critical role in mucin type O-glycan biosynthesis, maturity onset diabetes of the young, carbohydrate digestion and absorption, mineral absorption, amino sugar and nucleotide sugar metabolism and type 2 diabetes mellitus pathways (), Supplementary Table 17–18, https://figshare.com/s/da74d93a44f6f595b683). In the stearic acid group, the sub-network comprising chr6_31139729_31181149_−, mmu-miR-574-5p, mmu-miR-125b-5p and 31 DEmRNAs, including 32 DEcircRNA-DEmiRNA-DEmRNA competing pairs (), ). GO enrichment indicated that these 31 DEmRNAs participate mainly in protein binding functions, biological quality regulation and the microtubule cytoskeleton (), Supplementary Table 19, https://figshare.com/s/da74d93a44f6f595b683). These DEmRNAs were identified to be widely associated with sulfur metabolism, insulin secretion, glucagon signaling, cell cycle and maturity onset diabetes of the young pathways (), Supplementary Table 20, https://figshare.com/s/da74d93a44f6f595b683). No ceRNA network was generated for the circRNAs identified in both the palmitic acid and stearic acid group. In particular, for insulin secretion, glucagon signaling, maturity onset diabetes of the young and type 2 diabetes mellitus pathways, as the target gene of mmu-miR-574-5p, Solute carrier family 2 member 2 (SLc2a2, ENSMUST00000029240) was selected, which was found to positively correlate with chr6_31139729_31181149_− and specific to the stearic acid group. The chr6_31139729_31181149_−/mmu-miR-574-5p/Camk2b (ENSMUST00000109813) axis was chosen from the stearic acid group. In the palmitic acid group, SLc2a2 was selected to form a competing pair with chr5_117500713_117505639_+, which was enriched in the maturity onset diabetes of the young, type 2 diabetes mellitus and nutrient absorption pathways.
Table 4. Screening results of DEcircRNA-DEmRNA competing pairs in palmitic acid group
Table 5. Screening results of DEcircRNA-DEmRNA competing pairs in stearic acid group
Validation of key circRNAs in β-TC6 cells by qPCR
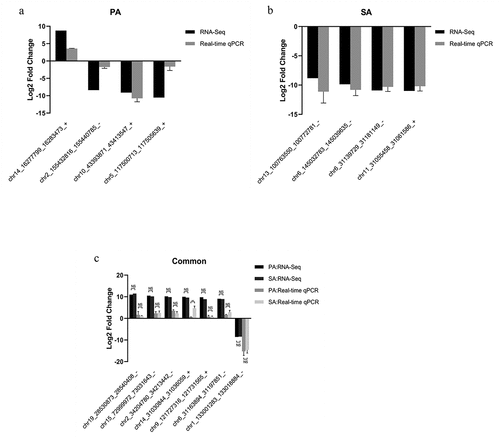
As shown in ), expressions of chr2_155432816_155440785_−, chr10_43393871_43413547_+ and chr5_117500713_117505639_+ decreased significantly, whereas chr14_16277799_16283473_+ increased in the palmitic acid group when compared with those of the control group. As for circRNAs specific to the stearic acid group, chr11_31055458_31061586_+, chr6_31139729_31181149_−, chr6_145032783_145039635_− and chr13_100763550_100772781_− were significantly downregulated in stearic acid-treated β-TC6 cells ()). Additionally, the overexpression levels of chr14_31030844_31036059_+, chr15_72999972_73031643_−, chr19_28530873_28540408_−, chr2_34204780_34213442_−, chr6_31163894_31197851_− and chr9_121727316_121731565_+ were significantly higher in both palmitic acid and stearic acid treated β-TC6 cells when compared with those of the control group ()). In contrast, chr1_133001283_133018884_− was downregulated when compared with that of the control group. These observations are consistent with the RNA sequencing results. In addition, the similar results were observed in islets from HPD/HSD mice, as well as palmitic/stearic acid-treated AML 12 cells and C2C12 cells (Supplementary Figure 2A-2C, https://figshare.com/s/567b5b966e8427e47d02). However, among them, there is significance of the change in the expressions of chr19_28530873_28540408_−, chr14_31030844_31036059_+, chr15_72999972_73031643_− and chr1_133001283_133018884_− between HPD and HSD group (Supplementary Figure 2A, https://figshare.com/s/567b5b966e8427e47d02). In AML 12 cells, chr15_72999972_73031643_− level is significant lower in palmitic acid group than that in stearic acid group (Supplementary Figure 2B, https://figshare.com/s/567b5b966e8427e47d02). In C2C12 cells, no significance was observed only in the change of chr14_31030844_31036059_+ and chr19_28530873_28540408_− expressions (Supplementary Figure 2C, https://figshare.com/s/567b5b966e8427e47d02).
Figure 7. Validation of candidate circRNAs in β-TC6 cells by qPCR. (a) Expression of four DEcircRNAs specific to the palmitic acid group. (b) Expression of four DEcircRNAs specific to the stearic acid group. (c) Levels of the seven DEcircRNAs common to both the palmitic and stearic acid groups. **P < 0.01. n = 3 independent cell cultures per group. Ctrl, Control group; PA, palmitic acid group; SA, stearic acid group; NS, no significance
Discussion
Chronic exposure to high levels of SFAs, including palmitic acid and stearic acid, leads to impaired insulin secretion by β cells, which accelerates the development of T2D. Although accumulating evidence indicates non-coding RNAs play critical roles in this process, the function of circRNAs in β cell damage has received nominal attention. Here, we identified four circRNAs specific to palmitic acid, four circRNAs specific to stearic acid and seven circRNAs common to both SFAs. We then constructed and compared the co-expression and ceRNA networks between palmitic acid- and stearic acid-treated β-TC6 cells.
The four novel circRNAs specific to the stearic acid group belonged to the exonic type. qPCR results showed that these four circRNAs were significantly downregulated, which was consistent with the sequencing results. Among them, chr13_100763550_100772781_− was found to be downregulated the most. circRNAs specific to the palmitic acid group also belonged to the exonic type, with chr14_16277799_16283473_+ showing the greatest level of upregulation and chr10_43393871_43413547_+ showing the greatest level of downregulation in β-TC6 cells. Because circRNAs are stable [Citation29], these candidate circRNAs can be used as specific markers for detecting elevated levels of stearic acid or palmitic acid in serum. Despite differences in the digestion and action of palmitic acid and stearic acid in β cells, common circRNAs associated with β cell dysfunction induced by palmitic acid and stearic acid were identified, with chr1_133001283_133018884_− the most significantly differentially expressed circRNA found. However, the expressions of chr15_72999972_73031643_− and chr14_31030844_31036059_+ showed the most significant change in mice islets from HPD and HSD group, respectively. This difference is probably due to the compensatory effect in vivo. This observation indicates that these molecules may be potential targets for ameliorating the harmful effects of high fat diets.
To explore the potential mechanism of circRNAs on SFAs-impaired β cell function, DEmiRNAs of DEcircRNAs and DEmRNAs of DEmiRNAs downstream in the regulatory network were further analyzed. Initially, we generated a ceRNA network specific to palmitic acid and identified three circRNAs (chr2_155432816_155440785_−, chr5_117500713_117505639_+ and chr10_43393871_43413547_+), two miRNAs (mmu-miR-378d and mmu-miR-574-5p) and 17 mRNAs that were closely related to nutrient metabolism and T2D, especially the mmu-miR-378d/Gnpda1 (Glucosamine-6-Phosphate Isomerase 1) axis for chr2_155432816_155440785_− and chr10_43393871_43413547_+, which may be involved in glucose metabolism [Citation30]. Furthermore, clustering of the GO terms and KEGG pathways showed that these 17 mRNAs were mainly enriched in the development of T2D. For the stearic acid group, the constructed ceRNA network included mmu-miR-125b-5p, mmu-miR-574-5p and chr6_31139729_31181149_−. We found that the chr6_31139729_31181149_−/mmu-miR-125b-5p/Activin A receptor, type 1 C (Acvr1c) regulatory axis played a critical role in cytokine-cytokine receptor interactions and the TGF-β (Transforming growth factor-β) signaling pathway. Expression of Acvr1c, a metastasis suppressor [Citation31], was observed to decrease in stearic acid-induced β-TC6 cells.
Additionally, mmu-miR-574-5p/SLc2a2 is the common axis for chr5_117500713_117505639_+ (Palmitic acid) and chr6_31139729_31181149_− (Stearic acid). SLc2a2 is a major glucose transporter in rodent islet β cells [Citation32], indicating that glucose transport signaling pathways are involved in palmitic acid- and stearic acid-induced β cell dysfunction.
There are still some limitations to our study. First, the four circRNAs have not been reported previously. More experiments are required to verify the effects of these circRNAs on SFA-induced β cell dysfunction. Second, extrapolation of the data obtained from the mouse cell line to human probably generates a number of uncertainties. Further experiments should be performed to verify these issues.
Conclusion
In summary, this is the first report to compare differences in dysregulated circRNAs and their related co-expression networks in β cell dysfunction induced by palmitic acid and stearic acid. Among them, chr2_155432816_155440785_− and chr10_43393871_43413547_+/mmu-miR-378d/Gnpda1 axis may be involved in glucose metabolism specific to palmitic acid group. While in stearic acid group, chr6_31139729_31181149_−/mmu-miR-125b-5p/Acvr1c regulatory axis plays a critical role in cytokine-cytokine receptor interactions and the TGF-β signaling pathway. Additionally, mmu-miR-574-5p/SLc2a2 is the common axis closely related to glucose transport signaling in palmitic acid- and stearic acid-induced β cell dysfunction. The findings of this study provide potential targets for prevention and treatment of SFA-impaired insulin secretion and enrich our understanding of the mechanisms underlying this process between palmitic acid and stearic acid.
Supplemental Material
Download ()Acknowledgements
H.L. conceived and designed the experiments. Y.Z., Q.Z., S.S., L.D., X.L., Y.W., Q.L. and Z.T. performed the experiments. Y.Z., Q.Z., S.S. analyzed the data. H.L. and Y.Z wrote the manuscript. H.L. and C.S. reviewed and edited the manuscript. We thank Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Lee HS, Nam Y, Chung YH, et al. Beneficial effects of phosphatidylcholine on high-fat diet-induced obesity, hyperlipidemia and fatty liver in mice. Life Sci. 2014;118(1):7–14.
- Tung YT, Chen HL, Wu HS, et al. Kefir peptides prevent hyperlipidemia and obesity in high-fat-diet-induced obese rats via lipid metabolism. Modulation.Molecular nutrition & food research. 2018;62(3).
- Cnop M, Abdulkarim B, Bottu G, et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63(6):1978–1993.
- Eguchi K, Manabe I, Oishi-Tanaka Y, et al. Saturated fatty acid and TLR signaling link β cell dysfunction and islet inflammation. Cell Metab. 2012;15(4):518–533.
- Acosta-Montaño P, Rodríguez-Velázquez E, Ibarra-López E, et al. Fatty acid and lipopolysaccharide effect on beta cells proteostasis and its impact on insulin secretion. Cells. 2019;8(8):884.
- Gouk SW, Cheng SF, Ong AS, et al. Stearic acids at sn-1, 3 positions of TAG are more efficient at limiting fat deposition than palmitic and oleic acids in C57BL/6 mice. Br J Nutr. 2014;111(7):1174–1180.
- Lorente-Cebrián S, González-Muniesa P, Milagro FI, et al. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: emerging roles as biomarkers and therapeutic targets. Clinical science (London, England). 2019;133(1):23–40
- Poller W, Dimmeler S, Heymans S, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. 2018;39(29):2704–2716.
- Sathishkumar C, Prabu P, Mohan V, et al. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018;12(1):41.
- Håversen L, Danielsson KN, Fogelstrand L, et al. Induction of proinflammatory cytokines by long-chain saturated fatty acids in human macrophages. Atherosclerosis. 2009;202(2):382–393.
- Mononen N, Lyytikäinen LP, Seppälä I, et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci Rep. 2019;9(1):8887.
- Yang WM, Jeong HJ, Park SW, et al. Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Mol Nutr Food Res. 2015;59(11):2303–2314.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7.
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806.
- Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol. 2019;234(11):19130–19140.
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858.
- Shi Y, Guo Z, Fang N, et al. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed Pharmacothe. 2019;117:109151.
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842.
- Brozzi F, Regazzi R. Circular RNAs as novel regulators of β-Cell functions under physiological and pathological conditions. Int J Mol Sci. 2021;22(4):1503.
- Guo R, Yu Y, Zhang Y, et al. Overexpression of miR-297b-5p protects against stearic acid-induced pancreatic β-cell apoptosis by targeting LATS2. Am J Physiol Endocrinol Metab. 2020;318(3):E430–E439.
- Yu Y, Guo R, Zhang Y, et al. miRNA-mRNA profile and regulatory network in stearic acid-treated β-cell dysfunction. J Endocrinol. 2020;246(1):13–27.
- Xiong DD, Dang YW, Lin P, et al. circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220.
- Zhang J, Liu Y, Shi G. The circRNA-miRNA-mRNA regulatory network in systemic lupus erythematosus. Clin Rheumatol. 2021;40(1):331-339.
- Zhang F, Zhang R, Zhang X, et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY). 2018;10(9):2266–2283.
- Miao L, Yin RX, Zhang QH, et al. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Sci Rep. 2019;9(1):18314.
- Lu H, Hao L, Li S, et al. Elevated circulating stearic acid leads to a major lipotoxic effect on mouse pancreatic beta cells in hyperlipidaemia via a miR-34a-5p-mediated PERK/p53-dependent pathway. Diabetologia. 2016;56(6):1247–1257.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461.
- Arreola R, Valderrama B, Morante ML, et al. Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study. FEBS Lett. 2003;551(1):63–70.
- Michael IP, Saghafinia S, Hanahan D. A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis. Proc Natl Acad Sci U S A. 2019;116(48):24184–24195.
- Thorens B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia. 2015;58(2):221–232.