 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Rapid, inexpensive, and laboratory-free diagnostic of viral pathogens is highly critical in controlling viral pandemics. In recent years, nanopore-based sensors have been employed to detect, identify, and classify virus particles. By tracing ionic current containing target molecules across nano-scale pores, nanopore sensors can recognize the target molecules at the single-molecule level. In the case of viruses, they enable discrimination of individual viruses and obtaining important information on the physical and chemical properties of viral particles. Despite classical benchtop virus detection methods, such as amplification techniques (e.g., PCR) or immunological assays (e.g., ELISA), that are mainly laboratory-based, expensive and time-consuming, nanopore-based sensing methods can enable low-cost and real-time point-of-care (PoC) and point-of-need (PoN) monitoring of target viruses. This review discusses the limitations of classical virus detection methods in PoN virus monitoring and then provides a comprehensive overview of nanopore sensing technology and its emerging applications in quantifying virus particles and classifying virus sub-types. Afterward, it discusses the recent progress in the field of nanopore sensing, including integrating nanopore sensors with microfabrication technology, microfluidics and artificial intelligence, which have been demonstrated to be promising in developing the next generation of low-cost and portable biosensors for the sensitive recognition of viruses and emerging pathogens.
1. Introduction
As one of the major causes of infectious disease, viruses continue causing global pandemics and constituting significant challenges and concerns in global public health [Citation1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been rapidly spreading worldwide since December 2019, when it first was discovered in Hubei, China [Citation2]. This pandemic once again pointed out the need for developing versatile point-of-need (PoN) monitoring and point-of-care (PoC) diagnostic devices for performing accurate, fast, cost-effective, and laboratory-free tests upon the emergence of a new class of pathogen [Citation3,Citation4]. In this paper, PoN devices refer to the ones used to monitor non-patient samples such as air and water, while PoC devices refer to technologies used to test human body fluids. Implementation of PoN and PoC techniques is very critical in controlling epidemics and pandemics by quickly identifying and isolating the infected environments and people and providing early treatment [Citation5]. An ideal detection technique should satisfy the criteria of specificity, sensitivity, simplicity, speed, and cost [Citation6].
Traditional approaches to diagnose viral infections are 1) direct detection of viral particles in the clinical samples, 2) isolation and identification of virus cultured cells, and 3) human antibody detection in the patient’s serum. However, these methods are laboratory-based and require costly equipment and trained personnel to be performed. In recent decades, direct detection methods have advanced significantly. For example, Polymerase Chain Reaction (PCR)-based techniques can provide a definitive answer in less than 24 h; however, they have limitations such as high cost and cross-contamination [Citation7]. The process of producing recombinant proteins and antibodies in immunological assays is also very complex [Citation8]. A summary of the approaches to diagnosing viral infections and their major strengths and limitations are given in . Although the limit of detection (LOD) is a key characteristic for method comparison in terms of the performance and utility of viral detection methods in various clinical settings, due to variability in the reference standard material and methods of quantitation used during the evaluation of limits of detection for different viruses, it has been challenging to compare performance among these detection methods. In addition, some immunological assays have been developed based on the immune response and immunity to specific pathogens by measuring antibodies or antigens. In this case, LOD cannot be determined for method comparison [Citation9]. The limitations and challenges associated with the traditional viral detection approaches point out the need for developing reliable, rapid, low-cost, and portable diagnostic techniques to quantify viral pathogens [Citation8].
Table 1. Various viral diagnostic methods and their advantages and disadvantages
Nanopore sensing is an emerging technology that enables real-time and label-free recognition and characterization of different molecules down to the single-molecule level[Citation10]. Nanopores are structures at the molecular scale and can be assembled through genetic engineering by using protein ion channels, or they can be synthetically fabricated on a solid substrate by taking advantage of recent progress in nanotechnology and nanofabrication [Citation11,Citation12]. Applying an electrical signal across a nanopore causes ions to flow through it and generate an ionic current. When a target molecule diffuses or passes through the nanopore, the ionic current would be obstructed. When the nanopore size is in order of the analyte size, the change in various properties of the ionic current becomes very prominent. Analyzing the ionic current trace provides important information regarding the target analyte in real-time response [Citation10]. Due to the electrical nature of this technology, nanopore-based sensors are extremely sensitive and reliable. Additionally, they are rapid, cost-effective, and capable of being miniaturized, which are essential requirements in developing PON sensors [Citation13].
The idea of employing the nanopore sensing technique in single-molecule detection was first proposed in the early 1990s when the possibility of sequencing a single strand of DNA molecule by threading it through a nanopore and recording the discrete levels of conductance blockade for each of its bases was discussed [Citation14]. Since then, nanopore sensing as a novel analytical technique has been applied for sequencing nucleic acids [Citation15], analyzing peptides and proteins [Citation16,Citation17], detecting metal ions [Citation18,Citation19] and small molecules [Citation20], characterizing particles for drug delivery systems (e.g., emulsions and liposomes) [Citation21,Citation22], polymeric and inorganic nanoparticles [Citation23,Citation24], extracellular vesicles [Citation25], and quantifying biomacromolecules, including bacteria and viruses [Citation26–28]. This method enables the analysis and investigation of the single molecule’s characteristics that cannot be studied within ensemble systems [Citation15]. In addition, complicated or time-consuming procedures for sample preparation are not needed in this method. However, due to limitations in the size of available pores (mainly protein nanopores that are much smaller than the size of virus particles) and fabrication techniques for the development of synthetic pores with customized properties, there have been limited reports on the application of this method for detection of virus particles until recent years.
Significant advancements in nano/microfabrication methods have enabled the fabrication of robust and low-cost solid-state nanopores with tunable geometry, size, and surface chemistry. Therefore, there has been rising interest in utilizing nanopore sensing platforms for efficient and rapid bioanalytical applications, including detecting single-virus [Citation26,Citation29], monitoring the virus-antibody interactions [Citation30], identifying virus subtypes [Citation27,Citation31], monitoring virus inactivation for evaluation of various inactivation methods [Citation32], and precise characterization of individual viruses such as mass [Citation33], zeta-potential [Citation34], and mechanical properties [Citation35] measurements.
Although there are multiple review papers on nanopore sensing technology, they mainly discuss the kinetics of nanopore sensing and review the applications of this method in sequencing DNA molecules, detecting proteins and small molecules [Citation12,Citation36–38]. There is also a review on the quantification of viruses using nanopore sensors which discusses the advancements in this field till 2016 [Citation39]. Meanwhile, this review discusses the fundamentals of nanopores and nanopore sensing methods and provides their up-to-date applications in viral sensing. Furthermore, the integration of nanopore sensors with microfluidic platforms and artificial intelligence (AI) to develop the next generation of PoC virus recognition sensors with automated analysis (i.e., via smartphones) is discussed.
2. Nanopore sensors
A nanopore sensor consists of a pore in the scale of a few to hundreds of nanometers formed within an insulating membrane, which separates two electrolyte chambers [Citation12]. Applying an electrical signal across this insulating membrane causes ions to flow across the nanopore and generates an ionic current. When a target molecule diffuses or passes through the nanopore, it blocks the ions’ flow, as demonstrated in . If the size of the nanopore is in order of the analyte’s size, this current blockade becomes very prominent and can be used to sense and characterize the analyte of interest. By statistical analysis of the blockade event, such as its shape, amplitude, duration, and frequency, one can obtain the target molecule’s different properties, such as its size, geometry, weight, surface charge, and interaction with other molecules, at the single-molecule level and in real-time responses [Citation10].
Figure 1. A schematic view representing the fundamentals of the nanopore sensing method. a) The flow of Ions across the nanopore as a reason for an applied electric potential across cell membrane. Passage of a protein (of interest) partially blocks the ionic flow which results in a drop in the pore’s ionic current. Once the protein is translocated, the ion flow is restored. b) A representative graph of monitoring the ionic current over time shows a single molecule (protein) translocation. By processing the data, the nanopore data and characteristics are extracted from the current-time response
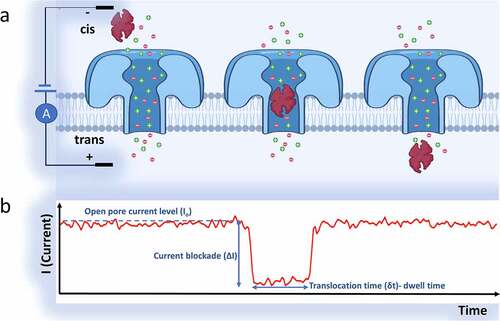
Figure 2. a) A biological nanopore (Aerolysin protein, dia. ~1–1.7 nm) inserted in a lipid bilayer for in vitro detection applications. b) A solid-state nanopore which is fabricated within a Si3N4 membrane layer. The nanopore’s diameter can be tuned finely (usually ranges more than 2 nm). The solid-state nanopore’s shape can be a cone, cylinder, or an hourglass, based on the fabrication techniques. The membrane thickness in cylindrical nanopores defines the depth of the nanopore (usually ranges between 10–200 nm). Using semiconductor processing methods or surface chemistry, the solid-state silicon nitride nanopores can be easily modified
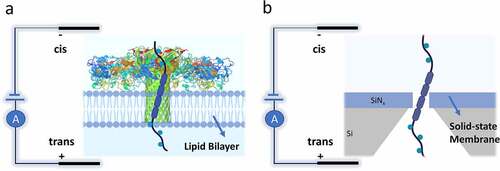
Based on the type of nanopore and membrane layer, nanopore sensors can be classified into two main groups of biological and solid-state nanopores [Citation36], as demonstrated in . In biological nanopores, the pore consists of a protein pore inserted in a lipid bilayer, while in solid-state type, it is a nano aperture fabricated in a thin solid membrane.
At first, the nanopore term referred to the biological ion channels, consisting of the self-assembled membrane proteins to shape a nanopore. Structures, Properties, and analyte types of frequently used biological nanopores are summarized in . Protein pores are responsible for almost all physiological processes, such as transducing electrical signals (ion transportation) in the muscles and nervous system through the cell membrane and energy storage [Citation11]. The biological nanopores are usually embedded in the lipid bilayers, such as cyclodextrin nanotubes [Citation45] and metal-organic polyhedral [Citation46]. They can be prepared in reproducible manners with the same physical and chemical properties. Since the pioneering demonstration of detecting nucleotide using alpha-hemolysin [Citation47], scientists have been developing and engineering various biological nanopores to use in many applications within the field of nano-biotechnology, especially for the detection, discrimination, and manipulation of single molecules [Citation11]; some examples include metal ions [Citation48], nucleotide [Citation49], small-molecule [Citation50], and protein [Citation51]. The idea of using nanopores in biosensing applications was first proposed in the early 1990s by Branton et al. [Citation52], where they demonstrated the possibility of sequencing the entire DNA strand by different levels of conductance blockade for each base of DNA strands. Shortly after, the revolutionary work done by Kasianowicz et al. [Citation47] demonstrated precise characterization of DNA and RNA molecules translocation across a biological nanopore, more specifically the α-hemolysin protein.
Table 2. Structure, Properties, and analyte types of frequently used biological ion channels (nanopores)
Similar to the biological pores, solid-state nanopores are apertures in the scale of a nanometer. They are mainly fabricated within various organic and inorganic synthetic thin films (usually SiNx or SiO2 membranes) of several nanometers to a few micrometers thickness. Solid-state nanopores are being implemented by leveraging nano/microfabrication technology.
The main advantages of biological nanopores over their synthetic counterparts are low detection noise, biocompatibility, and excellent measurement stability due to their precise diameter attributed to their amino acid sequence. Although the dimensions and structures of biological nanopores are predetermined by nature, different attempts have been made to engineer and alternate their pore sizes [Citation53]. Also, site-directed mutagenesis has been employed to precisely alter the surface functionalities of biological nanopores [Citation54]. However, the flexibility of the biological nanopores in tuning their properties is limited and requires intensive engineering efforts [Citation55].
One of the main challenges of using biological nanopores in sensing applications is the complicated designing process of a new recombinant protein. Moreover, after the design, the synthesis process has limitations in the produced protein (a few mg) [Citation11]. Although the Oxford Nanopore developed ultra-fast DNA sequencing that uses α-HL channel proves that this produced amount is sufficient for applying single-molecule sensing [Citation64], this channel cannot be used to create a high-density nanopore membrane with a large surface.
Another limitation of biological nanopores is the instability of the lipid bilayers [Citation11]. Although most of the biological nanopores show acceptable tolerance in different experimental conditions, the high sensitivity of their lipid bilayer to their environments, such as temperature, pH, ionic forces, and other external conditions, limits their performance and use [Citation11]. The instability of lipid bilayers limits the application of biological nanopores to specific experimental conditions and limits the mass production of biological nanopore sensors [Citation36]. It has been reported that tethering lipid bilayer on solid supports can improve its stability [Citation65]. Furthermore, the glass nanopore approach presents smaller bilayer capacitance and lower noise. Other reported methods for this purpose include using precast gels to protect the planar bilayer and polymerization of phospholipid bilayer membranes [Citation66]. Bayley et al. [Citation67] used a planar phospholipid bilayer with an embedded single biological α-HL nanopore and inserted it between two agarose gel layers. Analyte molecules could access the encapsulated nanopore through the thin porous agarose layer and could be detected. The resulted protein nanopore chips could be stored, transported, and used repeatedly. Despite protein pores, solid-state nanopores’ shape, dimensions, and surface properties can be tuned for various applications. They also have more chemical, thermal, and mechanical stability and can be integrated within nanofluidic and microfluidic systems. Moreover, the synthetic nanopores are easy to use and can be upscaled at large surface areas. Other specific properties of biological nanopores, such as the gating property induced by an external stimulus and perm-selectivity, can be mimicked by synthetic nanopores. This category of nanopores has been of great interest for various applications such as osmotic energy conversion, tissue regeneration, drug delivery, and biosensors [Citation68,Citation69]. Recent progress in the design, fabrication, and engineering of solid-state nanopores has made nanopore sensing technology a versatile method by providing low cost, high throughput, and reliable biosensors at the scale of single-molecule [Citation70].
Since the virus particles are usually larger than the diameter of biological pores, mainly solid-state nanopores have been employed to quantify the whole virus particles. However, since biological nanopores have recently been applied in sensing viruses’ RNA promoters [Citation29], we have discussed them in this paper. It is worth mentioning that there are some other platforms of nanopore detecting, including hybrid nanopores [Citation67] and plasmonic nanopores [Citation71], which leverage the combination of biological and synthetic nanopores’ properties or use additional electromagnetic fields to alternate and modify the nanopore sensor’s performance. However, they are beyond the scope of this review since they have not been employed in viral sensing applications.
2.1. Mechanism, theory, and fundamentals
2.1.1. Resistive pulse sensing method
In both biological and solid-state nanopore sensors, a nanopore inserted membrane that acts as a conduit separates the two liquid reservoirs containing electrolytes, such as NaCl or KCl [Citation37]. Within each of the reservoirs, there is a non-polarizable electrode by which the electrical potential is applied. The generated steady-state flow of ions is called the open-pore ionic current. In a cylindrical synthetic nanopore and KCl solution with high ionic strength (more than 100 mM), the generated open-pore ionic current can be approximately calculated using EquationEquation 1(1)
(1) [Citation37].
Where V is the applied voltage, μCl- and μK+ are electrophoretic mobilities of Cl− and K+, nKCl, e, d, and l are the number density of KCl, elementary charge, nanopore diameter, and nanopore length (usually equal to the membrane’s thickness), respectively. The ions are translocated across the pore by applying this electrical potential, resulting from the interplay between diffusion, electroosmotic and electrophoretic effects [Citation72].
Since the nanopore sensing technique is very sensitive, the effect of external noise on the sensor’s response cannot be neglected. Therefore, the nanopore sensing experiments are mostly performed in a Faraday cage [Citation73,Citation74]. The open-pore ionic current is stable during a typical experiment of the nanopore. The translocation of an analyte interrupts this stable current (called ionic current blockade) and causes a change in the current response over time. The sensor output (ionic current) in nanopore sensors is transferred to an amplifier and a data acquisition (DAQ) system. By processing the ionic current-time response data, different characteristics of the nanopore and analyte can be obtained. This method of sensing is called the resistive pulse sensing (RPS) method [Citation75]. presents the blockade of the open pore current upon translocation of the target analyte. The most critical parameters that are deduced from the ionic current response for characterization of the analyte translocation events are:1) The duration of blockade or the molecule’s residence time inside the nanopore (tdwell), 2) The blockade current’s amplitude (or the difference between the amplitude of the blockade and open-pore currents (ΔI), 3) The duration between the translocation events (τ), 4) The capture rate (translocation event numbers/unit time). All these factors are directly related to the geometry and structure of nanopore, applied bias (based on EquationEquation 1(1)
(1) ), concentration as well as chemical and physical properties of the translocated analyte, and thus can be used to extract the analyte’s characteristics [Citation11]. This method accurately identifies and recognizes analytes at a single-molecule level and wide range from small molecules to molecular complexes in different applications.
2.1.2. Temporal resolution and noise
As mentioned previously, for sensing applications, the dimensions and structure of the nanopore need to be comparable with the analyte of interest. The reason for that is to increase the signal-to-noise ratio by maximizing the change in the ionic current when a translocation is happening [Citation12]. Characterization of a translocation event is typically performed by measuring the blockade and the open pore current. The ratio of these two currents’ mean is defined as the fractional blockade current (EquationEquation 2(2)
(2) ) and used to report the translocation event.
In EquationEquation 2(2)
(2) , IB is fractional blockade current, where ib and io are blockade and open-pore currents, respectively. EquationEquation 3
(3)
(3) presents another way to quantify and report the translocation event with fractional event amplitude.
In this equation, is the difference in blockade current and open-pore current. Since, based on EquationEquation 3
(3)
(3) , fractional event amplitude is 1-IB, it is usually used interchangeably with fractional blockade current (EquationEquation 1
(1)
(1) ). The fractional event amplitude depends on the nanopore blockage by the molecule of interest or the target analyte. It can be approximated to the blocked volume of a sensing nanopore over its whole sensing volume [Citation12].
By assuming a cylindrical nanopore, the open pore current’s mean value can be quantified using EquationEquation 4(4)
(4) .
Where R is the total resistance of nanopore, and σ is bulk conductivity. The total resistance of the nanopore can be obtained from EquationEquation 5(5)
(5) .
The term (4 l/σπd2) is related to the geometry of the nanopore (cylindrical in this case). The second term in the resistance, (1/σd), is called access resistance and comes from the convergence of the ionic current from the bulk solution into the vicinity of the nanopore. Based on EquationEquation 5(5)
(5) , the narrowest section of a nanopore experiences the highest potential drop (resistance) upon translocation of an analyte. Therefore, this region is considered as the sensing part of the nanopore. For instance, while the entire length of an a-hemolysin nanopore is 10 nm, the sensing part is estimated to be about half of its length (̴ 5 nm) [Citation14]. Solid-state nanopores sculpted with electron beams usually have a double-conical shape. Therefore, the nanopore’s effective length for sensing is significantly smaller than the film’s nominal thickness. Estimating these measurements can be done experimentally by characterizing a known analyte’s translocation, such as DNA, through the nanopore [Citation76].
For resolving the molecular structures of analytes, such as the primary or secondary structure of a protein, a narrow sensing part is preferred [Citation77]. However, the main disadvantage of a narrow sensing part is its shallow mechanical stability. Another drawback is that the tdwell in narrow nanopores is short, and therefore not all the translocation events can be resolved within the inadequate temporal bandwidth of the experiment. Adequate temporal bandwidth is required to capture the signal related to nanopore sensors. For example, if the tdwell of a translocation event is 1 µs, then the bandwidth of this system would be ̴ 1 MHz. Since a system’s overall noise increases speedily at the higher frequencies, working in high bandwidth can cause difficulties, complicate the related measurements, and obscure the translocation signals. The often-used range of frequency and acquisition bandwidths in nanopore sensing applications have been reported to be 10–100 kHz, and it can only capture a fraction of translocation events [Citation12].
The overall noise in the nanopore sensing systems originates from two primary sources. The first source of the noise is the electrical circuit, including the internal circuit of the amplifier, transfer of the charge at the electrodes, and electrical filters. The second source of noise in the nanopore sensing systems originates from the physical properties of the nanopore sensor, such as the membrane material and its composition, capacitance, and charge [Citation78]. In nanopore sensing applications, the temporal resolution is defined by the ratio of the fractional event amplitude and the total electrical noise, as indicated in EquationEquation 6(6)
(6) .
In this equation, in is the overall electrical noise of the system. In general, in can be approximated by the open pore current’s RMS value (io). In the temporal resolutions smaller than ̴ 2, the differentiation of the translocation events from the background noises is usually unfeasible. Therefore, the temporal resolutions smaller than ̴ 2 are usually considered to be unpractical.
2.2. Materials and Procedures to Design Synthetic Nanopores
The selection of the materials and methods highly depends on the application and the characteristics of the target analyte. In general, the materials used to fabricate solid-state nanopores are categorized into six main groups based on their main properties, as summarized in , while the most used material is Si3N4 due to its well-established fabrication methods in the semiconductor and microelectronics industry.
Table 3. Most common materials for fabrication of solid-state nanopores
The nanopores based on semiconductor materials such as silicon have been extensively investigated in applications such as protein identification [Citation79], DNA sequencing [Citation80], and molecule translocation physics [Citation81]. Polymers and polymeric films have also been widely studied in designing various shapes of synthetic nanopores, mainly with a track-etch approach, which is compatible with multi pore membranes and, therefore, suitable for large-scale applications. Common polymers used for this purpose are polycarbonate (PC), polyimide (PI), and polyethylene terephthalate (PET) [Citation82].
Composite materials have been of great interest in synthetic nanopores since they can combine the different materials and benefit from their advantages. For example, nanopores made based on metal-polymer composites have enhanced stability and structural and chemical properties[Citation83].
Incorporating microfabrication technology from the semiconductor industry into the biosensing platform during the last two decades has received significant attention due to enabling the miniaturization of the biosensing components, such as microsensors, microfluidics, and miniaturized signal processing units [Citation98]. Miniaturization of the biosensing components enables the fabrication of scalable and portable devices and provides the ability to control and sense the analytes within the nano-scale range of analytes, even at the single-molecule level.
Different techniques can be used to fabricate solid-state nanopores based on the selected material and required pore size for maximizing the signal-to-noise ratio. P. Yu et al [Citation99]. used a controlled track-etching method and fabricated nanopores of 20 nm diameter on a polyethylene terephthalate (PET) membrane [Citation82]. Direct drilling method using a highly focused electron, ion, and Helium ion beam (through a transmission electron microscope (TEM) or ion microscopes) is another common technique for the solid-state nanopores fabrication that can achieve reliable pore sizes of sub-10 nm [Citation100]. The current TEM-based drilling methods enable the nanopores fabrication with diameters down to a nanometer [Citation101]. However, beam-based drilling methods are low throughput and costly. Other methods of fabricating solid-state nanopores include laser ablation (using glass or silver) and controlled dielectric breakdown (CDB). CDB is considered a low-cost and simple method of fabricating nanopores with an obtainable pore size of sub-5 nm [Citation102]. In this method, a single nanopore is formed in a thin insulating membrane (~50 nm) due to applying a high electric field across it [Citation103]. Lithographic processes in combination with various etching techniques are also widely used in nanopore fabrication due to their well-established procedures in microelectronics. Although this method enables the mass fabrication of nanopores with various arrangements and pore sizes, it is hard to achieve reliable pore sizes of sub-10 nm due to spatial lithography resolution and the multistep fabrication process. This method usually requires post pore shrinking approaches such as oxidation or deposition to achieve the desired pore sizes [Citation104]. summarizes the methods based on the materials used to design the solid-state nanopores. An up-to-date review of in-situ fabrication methods of nanopores can be found in ref [Citation105].
Table 4. Fabrication methods of solid-state nanopores
It has been proven that high-aspect-ratio nanopores could improve the detection by slowing down the translocation event (increasing tdwell, see section 2.1). In a recent attempt, Nguyen et al. [Citation118] developed a temporal voltage variation method that optimizes etching parameters to fabricate high aspect ratio nanopores (with a dia. around 12 nm) on silicon using electrochemical breakdown etching. Their developed process is fast, low-cost, and capable of batch fabrication. It can be employed in single-molecule sensing applications to detect DNA, protein markers, exosomes, and viruses [Citation118].
2.2.1. Integration of solid-state nanopores with microfluidics
Microfluidic-based detection platforms and microfluidics integrated biosensors have been employed extensively in implementing Lab-on-a-chip (LOC) and PoC or PoN devices [Citation119]. Microfluidics technology enables portable, cost-effective, and prompt sensors with reduced reaction times by manipulating very tiny amounts of samples within channels and chambers on a scale of a few to hundreds of micrometers [Citation120].
Integrating required equipment for performing nanopore experiments, such as electrolyte chambers, tubing, electrodes, electrical connections, and external circuitry, is challenging. Also, there is a need to purify some samples, such as protein or nucleic acids. Therefore, there has been a significant interest in integrating nanopores with microfluidic chips to simplify the sensing process and expand their utility.
At first, the role of microfluidic system in nanopore sensing was to provide a structural platform. However, the recent advancements in microfluidics have resulted in improved performance of microfluidic-based nanopore sensors [Citation121]. Integration of solid-state nanopores with microfluidics allows analyzing biomolecules while benefiting from sample processing capabilities of microfluidic. Micro-scale electrolyte chambers and flow channels reduce the sample loss and required sample volume, thus improving detection efficiency. Also, minimizing the required fluidic tubing and electrode increases the scalability. Integrated microelectrodes with fixed positions provide precise measurement by reducing the noise of the system and the chance of air exposure [Citation121]. Microfluidics also enables obtaining a high throughput nanopore sensing platform by designing nanopore arrays and performing parallel sensing operations on-chip, leading to a high-throughput sensing platform at the single-molecule level. For example, Jain et al. [Citation122] developed an integrated microfluidic-based nanopore sensor. They used polydimethylsiloxane (PDMS) as their structural material to fabricate the microfluidic device and integrated it with a nanopore fabricated by electron beam sculpturing. They could successfully detect the translocation of single DNA molecules using the integrated sensor. Yanagi et al. [Citation123] showed the integration of solid-state nanopore arrays with a microfluidic chip comprising an acrylic flow cell and utilized it to detect the translocation of DNA. Tahvildari et al. [Citation124] fabricated solid-state nanopore arrays using the CDB method and integrated them with a PDMS microfluidic chip. More recent work used microvalves based on PDMS to control the access of biological samples to the nanopore [Citation125]. Shuo et al [Citation126]. further integrated a microfluidic-nanopore sensor with optical sensing and developed a dual-mode optical-electrical sensing device to detect the H1N1 influenza A virus. In their electro-optofluidic sensor, the integrated solid-state nanopore was responsible for the controlled delivery of single fluorescently labeled influenza viruses virus particles for further optical analysis, as demonstrated in . Performing optical and electrical characterization in parallel provides additional characterization data and enables parallel detection in high throughput sensors with many channels and optical signals without a crosstalk challenge, which is not the case for electrical signals. Recently, Varongchayakul et al. [Citation127] developed a stand-alone microfluidic-based nanopore sensor to characterize DNA translocation. demonstrates their highly integrated microfluidic–nanopore sensor consisted of different sections for temperature control, sample preparation and processing units. They performed simultaneous optical and electrical detection and could successfully detect DNA molecules.
Figure 3. Nanopore sensors integrated with microfluidic and optofluidic devices. a) A solid-state nanopore (silicon dioxide) integrated with optofluidic device for single nanoparticles’ simultaneous electro-optical analysis. Liquid-core and solid-core waveguides on a silicon chip are shown in blue and Orange, respectively. Electrodes and particles are in metallic reservoirs. The inset demonstrates a magnified schematic view of particle translocation through the nanopore. The red zone demonstrates the optical excitation volume. Adapted from ref [Citation106]. with permission. b) A schematic diagram of the microfluidic integrated nanopore biosensor with on-chip purification/ bioassay and multilayer fluidic channels. i) Assembly of microfluidic chip. ii) Top view. iii) Cross‐section of the device zoomed‐in at nanopore chambers. iv) The assembled microfluidic chip. Inset is the nanopore’s TEM image. Reproduced from ref [Citation107]. with permission of John Wiley & Sons, Ltd
![Figure 3. Nanopore sensors integrated with microfluidic and optofluidic devices. a) A solid-state nanopore (silicon dioxide) integrated with optofluidic device for single nanoparticles’ simultaneous electro-optical analysis. Liquid-core and solid-core waveguides on a silicon chip are shown in blue and Orange, respectively. Electrodes and particles are in metallic reservoirs. The inset demonstrates a magnified schematic view of particle translocation through the nanopore. The red zone demonstrates the optical excitation volume. Adapted from ref [Citation106]. with permission. b) A schematic diagram of the microfluidic integrated nanopore biosensor with on-chip purification/ bioassay and multilayer fluidic channels. i) Assembly of microfluidic chip. ii) Top view. iii) Cross‐section of the device zoomed‐in at nanopore chambers. iv) The assembled microfluidic chip. Inset is the nanopore’s TEM image. Reproduced from ref [Citation107]. with permission of John Wiley & Sons, Ltd](/cms/asset/ff069f50-29ef-4d73-bfd4-4c00e5dd7ef0/kbie_a_1995991_f0003_oc.jpg)
The microfluidic-nanopore devices in the reported works could advance the nanopore sensing technology by providing an integrated and automated platform for different stages such as preparation, handling and introduction of samples, isolation or purification of analytes and adaptability to fabrication at the commercial stage. Combining single-molecule solid-state nanopore sensors with microfluidic technology indicates that the integrated sensors offer further impetus for novel designs and set the stage for implementing this technology into PoC and PoN applications.
2.2.2. Surface modification of solid-state nanopores
One of the important factors that enable the application of nanopore sensors in the characterization and analysis of various molecules is their amenability to surface modification [Citation80]. Introducing point-mutations onto the surface of nanopores enables controlling the interactions between the engineered nanopore surface with the target analyte [Citation128], which enables the design and probe of the chemical interactions at the single-molecule level [Citation129]. In general, a variety of surface modifications are applied to nanopores to increase their stability, change their diameter, minimize their clogging, manipulate their surface charges, reduce nonspecific interactions, increase the analytes’ residence time in the pore, enable interactions with target analytes, and reduce the current recording noise across nanopores [Citation130]. For example, increasing the analyte residence time (tdwell) enables the extraction of more information from each translocation event (see section 2.1). Also, decreasing the pore diameter and altering its shape can increase the signal-to-noise ratio. Furthermore, regulating the on-rate kinetics of target analytes to the nanopore enhances the detector’s sensitivity and reduces the detection limit for nanopore application.
Although biological nanopores can be genetically engineered and functionalized at specific sites [Citation131] and are naturally resistant to pore-clogging [Citation132], because of their small diameter (from 0.4 nm- 3.6 nm), they can only be used for the characterization of analytes with small dimensions, such as RNA, DNA, unfolded proteins, peptides, organic molecules, and ions. The development of solid-state nanopores with large-scale tunable diameters enables the detection and characterization of macromolecules such as proteins, viruses, and bacteria. However, one of the significant challenges of using solid-state nanopores is pore-clogging upon nonspecific interactions of analytes with the pore surface [Citation133]. Hydrophobic interactions [Citation134], van der Waals forces [Citation135], and electrostatic attractions [Citation136] are the main factors that contribute to these nonspecific interactions. Surface modifications can help to decrease the intensity of nonspecific interactions, thus enabling unperturbed translocation [Citation137].
A wide range of functionalities can be introduced to solid-state nanopores by modifications of their surface. The main methods for surface modification of solid-state nanopores, along with their primary applications, are summarized in . Some practical examples include single protein receptors immobilized via a self-assembled monolayer to probe the single-molecule binding kinetics, ssDNA immobilization to differentiate and determine target sequences, or methods to decelerate the biomolecules translocation time, such as atomic layer deposition of ZnO and piranha (mixture of H2O2 and H2SO4) cleaning. Using piranha solution also helps remove the organic compounds and contaminants from the surface of the nanopore and improves its wettability.
Table 5. The main methods for surface modification of solid-state nanopores and their primary applications
Tuning the surface charge density of nanopores is another critical factor in designing nanopore sensors. Surface modifications provide a way to alter the surface charges on the surface of nanopore [Citation138], which in turn adjusts the screening length of the formed electrical double layer (EDL) on the pore surface and thus can be utilized to manipulate the electroosmotic flow resulted from the electrophoretic passage of ions in the EDL. For instance, Arima et al. [Citation27] exploited electroosmotic flow resulted from a negative native charge of Si3N4 nanopore surface to filter contaminants from entering nanopore while sensing individual Influenza viruses driven with electrophoretic forces. It is crucial to keep the electroosmotic flow constant to analyze translocation time series from the flow of analyte molecules through the pore.
Vapor-deposition techniques allow the deposition of single-molecule layers on the nanopore walls. Deposition on membranes containing nanopores can also increase their stability against slow etching in the electrolyte solution during sensing [Citation139]. Surfactants (surface-active agents), which consist of both hydrophilic and hydrophobic residues, can adsorb on nanopore surfaces and change their surface properties based on the specifications of the designed translocation event. For example, surfactants such as cetyl trimethyl ammonium bromide (CTAB) and Tween 20 are used in coatings nanopores to reduce biomolecules’ interactions with the pore wall [Citation140]. Surfactants are usually used to lower surface tension and have additional roles such as killing bacteria, altering surface charge density, inhibiting corrosion, and foaming [Citation141].
Surface modifications using self-assembly-monolayers (SAMs) based on organosilanes and organothiols are also among the most frequently used techniques to alter the surface chemistry of nanopore sensors. The monolayers generated in this method can form over large surfaces, and the preparation of SAMs does not require specific equipment. They can provide surface groups that can covalently link to analyte molecules, interact with them or repel molecules [Citation142]. In nanopore-based sensing, they are mainly applied for minimizing nonspecific interactions [Citation143], manipulating surface charge [Citation144], sensing specific analytes [Citation145] and adding different functionalities such as preferential transport [Citation146], gating properties [Citation147], and enhancing the plasmonic nanopores signal [Citation148]. Silanes have both inorganic and organic moieties and can covalently bond to surfaces of various substrates such as iron oxide, aluminum oxide, and quartz. Silanization without polymerization can generate thin coatings, reducing nonspecific adhesion or increasing hydrophobicity. Silanization can also functionalize pore walls by allowing the attachment of polymer brushes, peptides, carboxylic acid, cysteines, spiropyran moieties, aldehydes, nucleoporins, dendrimers, and DNA to the attached chemical groups to the silane molecule [Citation130]. Thiol terminated self-assembly monolayers are very popular in the surface functionalization of gold-modified nanopores or the nanopores made in gold substrates due to their versatile tail groups, and easy deposition.
Layer-by-layer self-assembly (LBL) method is another technique for surface modification of nanopores that uses alternating deposition of oppositely charged polyions on the surface. LBL method provides precise deposition of bilayers in thicknesses less than 1 nm and is useful when adjusting the diameter of a nanopore. Surface modifications using the LBL method are mainly used to tailor surface functionality, manipulate nanopore size, or incorporate other molecules for specific recognition of target analytes [Citation155].
Other surface modifications that have been used for modifying the surface of solid-state nanopores include biochemical modification using proteins, lipids, and nucleic acids, covalent modification methods such as plasma-induced graft polymerization, hydrosilylation, spin-coating, and direct crosslinking of functional groups such as spiropyrans or DNA to the pore wall’s surface [Citation130]. Fluid lipid coatings have been shown to provide significant advantages in sensing proteins and macromolecules [Citation129]. For example, they are shown to minimize or prevent nonspecific protein adsorption to the nanopore walls, which in turn eliminates clogging. Furthermore, by incorporating receptor molecules or lipid-anchored ligand to these coatings, they can interact with target analytes with the continued translocation capability due to the lipid coating’s fluidic nature [Citation137]. Lipid anchors are shown to decrease the translocation speed of the anchored analytes due to anchor drag in the viscous lipid coating. Furthermore, phosphatidylcholine head groups and the zwitterionic nature of lipids minimize or nearly eliminates the electroosmotic flow [Citation156], as well as nonspecific interactions with various proteins [Citation157].
3. Nanopore sensors for detection and classification of viruses
Nanopore sensing as a novel analytical technique has been used in different applications, such as sequencing nucleic acids, analyzing peptides and proteins [Citation16,Citation17,Citation47,Citation158], detecting metal ions and small molecules, and biomacromolecules, including bacteria and viruses [Citation26–28,Citation159].
Debolis et al. [Citation160] reported the first application of nanopore sensors in virology in 1977. They used the RPS technique to measure the size of type C Oncornaviruses and Bacteriophage T2 particles using Nuclepore membranes. Later in 1978, Feuer et al. [Citation161] employed this technique to measure the spike lengths on viruses. Though, due to limitations in the size of available pores (mainly protein nanopores) and fabrication techniques, there have been almost no reports on nanopore sensing applications in virology since then until recent years.
During recent years, significant advancements in nano/microfabrication methods have enabled the fabrication of low-cost and robust solid-state nanopores with tunable geometry, size, and surface chemistry [Citation11]. Therefore, there has been rising interest in utilizing nanopore sensing platforms for efficient and rapid bioanalytical applications, especially in single-virus detection. In this regard, Uram et al. [Citation30] used a solid-state nanopore fabricated with a femtosecond-pulsed laser on a borosilicate cover glass to detect Paramecium Bursaria Chlorella Virus (PBCV-1). Their label-free nanopore sensor detected the single virus particles and probed immunoprecipitation of a virus with a specific antibody. They could also monitor the virus-antibody interactions and assembly of antibodies onto virus particles, and determine the number of attached antibodies, without need for immobilization or modification of the antibody or virus and only by resistive pulse responses upon translocation of viruses with and without antibody through their sub-micron pore as shown in .
Figure 4. Characterization of antibody binding to virus particles. A) Detection of single viruses with monitoring resistive pulses (transient current reduction) upon translocation of virions through nanopore (dotted line is mean of current spikes. B) Detection of virus-antibody conjugates. Antibody addition cause volume increase of translocating particles which increase the peak amplitude upon passing through the pore. Reproduced from ref [Citation20]. with permission of John Wiley & Sons, Ltd
![Figure 4. Characterization of antibody binding to virus particles. A) Detection of single viruses with monitoring resistive pulses (transient current reduction) upon translocation of virions through nanopore (dotted line is mean of current spikes. B) Detection of virus-antibody conjugates. Antibody addition cause volume increase of translocating particles which increase the peak amplitude upon passing through the pore. Reproduced from ref [Citation20]. with permission of John Wiley & Sons, Ltd](/cms/asset/05ded224-4f2c-46f5-841b-9726e45779a5/kbie_a_1995991_f0004_b.gif)
Figure 5. a) Detection of Single-influenza-virus using a Si3N4 nanopore. i) Nanopore measurements schematic. Applied bias voltage (Vb) causes electrophoretic translocation of influenza virions in chorioallantoic fluid to pass through the nanopore. ii) Resistive pulse in cross-membrane ionic current upon translocation of single virus. iii) Ionic current traces for fluids containing different Influenza subtypes. Adapted with permission from ref [Citation17]. Copyright {2018} American Chemical Society. b)Detection of Single-influenza-virus using a Au/Si3N4 nanopore modified with peptide probes. i) The interactions between virus and surface peptides. ii) Enlarged views of the resistive pulses acquired in a influenza A(H1N1) containing buffer solution with Si3N4 (Orange) and P2 (ASHRVGSTYIAC) modified nanopores (red). The residence time (tdwell) of viruses in the peptide-modified pore pore increases due to temporal trapping of the virions on the wall surface The inset shows the molecular structure of P2. Adapted with permission from ref [Citation21]. Copyright {2018} American Chemical Society
![Figure 5. a) Detection of Single-influenza-virus using a Si3N4 nanopore. i) Nanopore measurements schematic. Applied bias voltage (Vb) causes electrophoretic translocation of influenza virions in chorioallantoic fluid to pass through the nanopore. ii) Resistive pulse in cross-membrane ionic current upon translocation of single virus. iii) Ionic current traces for fluids containing different Influenza subtypes. Adapted with permission from ref [Citation17]. Copyright {2018} American Chemical Society. b)Detection of Single-influenza-virus using a Au/Si3N4 nanopore modified with peptide probes. i) The interactions between virus and surface peptides. ii) Enlarged views of the resistive pulses acquired in a influenza A(H1N1) containing buffer solution with Si3N4 (Orange) and P2 (ASHRVGSTYIAC) modified nanopores (red). The residence time (tdwell) of viruses in the peptide-modified pore pore increases due to temporal trapping of the virions on the wall surface The inset shows the molecular structure of P2. Adapted with permission from ref [Citation21]. Copyright {2018} American Chemical Society](/cms/asset/1c4d8337-3a31-4cf7-a420-5889658f4604/kbie_a_1995991_f0005_oc.jpg)
In another work, Zhou et al [Citation162]. developed a solid-state nanopore sensor to characterize hepatitis B virus (HBV) capsids. They fabricated nanopores (∼40 nm in diameter) on poly (ethylene terephthalate) membranes using the track-etching method. They modified the surface of their nanopore with triethylene glycol to control its charge and reduce the virus capsid adsorption to the pore. Later, Arjmandi et al. [Citation33] fabricated nanopores of 20–500 nm diameter using chemical wet etching of SiO2 membranes and utilized them for precise mass and zeta-potential measurements of different nanoparticles as well as HIV and EBV viruses. McMullen et al. [Citation163] used soli-state nanopores on Si3N4 to detect and characterize filamentous virus (fd) and rod-shaped viruses. Later, Darvish et al. [Citation26] demonstrated label-free recognition of HIV-1 virus particles using the RPS technique. They could capture a single HIV-1 virion above their Si3N4 nanopore and squeeze it through the pore by applying an electric potential, which is crucial in gaining a fundamental understanding of biological phenomena such as membrane fusion.
Recently, Miyagawa et al. [Citation164] combined the nanopore sensing procedure with a specialized virus detection process. They first captured target viruses from a complex sample with their specific probes on an Au electrode. Next, they released the captured virions by electrochemical method and transferred them toward a quartz nanopore by electrophoretic force. Using this method, they could selectively detect Influenza viruses at a resolution of one particle. Although solid-state nanopores are mainly used for the detection and characterization of whole-virus particles, biological nanopores are still being used to detect viruses based on their RNA promoters. For instance, in a recent work Influenza, A virus (IAV) RNA promoters were detected using α-HL nanopore at the single-molecule level [Citation29].
One of the most important advantages of nanopore sensing technology is its capability in discriminating different allotypes of a virus species. Recently, Arima et al. [Citation27] utilized solid-state nanopores combined with artificial intelligence (AI) for selective and label-free identification of Influenza virus subtypes (influenza types A, B, and influenza A subtypes) with high accuracy. The sensor consisted of a 300 nm diameter nanopore fabricated on a Si3N4 membrane (50 nm thickness), using chemical wet etching. Their results showed identifications of Influenza subtypes with 68% accuracy at the single-virus level. The sensor could discriminate different allotypes of influenza virus at the single-virus level by analyzing ionic signal patterns through AI pattern recognition, as demonstrated in .
In a later work [Citation31], they utilized molecular mechanisms to improve their sensor performance. They sputtered Au (45 nm thickness) on the surface of the fabricated nanopore and decorated its surface with synthetic peptides with a weak affinity toward the Influenza A virus, enabling specific perturbation of viral envelopes (). Since Influenza A and B have almost the same size, the magnitude of the ionic current blockade was not sufficient to distinguish different types as it mainly reflects the size of the target. However, machine learning revealed that tdwell (translocation time), which directly reflects the electrophoretic mobility of target molecules (see section 2.1), was significantly different for different allotypes. Different translocation times of virus subtypes can be attributed to their distinct surface charge densities and intermolecular interactions with surface peptides.
Mechanical characterization of single virus particles has been performed recently using the nanopore sensing method. Darvish et al. [Citation35] used a 300 nm diameter nanopore fabricated by chemical wet etching on Si3N4 membrane to characterize the mechanical properties of HIV-1 virions. Since the membrane rigidity of a virus can be a measure of its infectivity, they used the nanopore sensing method to determine the infectivity of target viruses. Their result indicated that mature viruses with softer membranes undergo electrodeformation in the nanopore and yield smaller peak amplitudes in current traces. On the other hand, immature viruses with a rigid membrane yielded larger current peak amplitudes. Also, they showed that the recognition of virus particles was not possible when the applied bias voltage was under a specific value since the electrophoretic forces that drive charged viruses through the nanopore were reduced.
In the most recent work, Taniguchi et al. [Citation165] applied the combination of nanopore sensing and artificial intelligence for high-sensitive coronavirus detection without the need for RNA extraction, which is a main drawback of the current RT-PCR method [Citation166]. The sensor consisted of a nanopore with a 300 nm diameter on a Si3N4 membrane, a high-speed, portable, precise current measuring instrument, and machine learning software on a server. demonstrates their developed sensor was able to detect SARS-CoV-2 with 96% specificity and 90% sensitivity in saliva specimens in only 5 minutes. The sensor also accurately identified four coronavirus types with similar sizes: SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-229E.
Figure 6. Solid-state nanopore sensor for detection of coronavirus. a) Structure of the fabricated nanopore in a Si3N4 membrane. Cis and trans channels contain the specimen and buffer and are connected to Ag/AgCl electrodes for signal transduction. b) The photographic image of the developed nanopore chip. Adapted with permission from ref [Citation145]. Copyright {2021} Springer nature
![Figure 6. Solid-state nanopore sensor for detection of coronavirus. a) Structure of the fabricated nanopore in a Si3N4 membrane. Cis and trans channels contain the specimen and buffer and are connected to Ag/AgCl electrodes for signal transduction. b) The photographic image of the developed nanopore chip. Adapted with permission from ref [Citation145]. Copyright {2021} Springer nature](/cms/asset/0eb00f60-893c-4c82-bac4-e248025145a2/kbie_a_1995991_f0006_oc.jpg)
Other than identification, characterization and typing, virus inactivation has been reported by using nanopore sensors. Studying the effect of viral therapy methods is very important for preparing safe biotherapeutics. Conventional methods of monitoring the changes caused by an inactivation technique in virions are based on plaque-forming assays and thus are very time-consuming due to the required time for bacterial growth and plaque-forming (24–48 hours).
Recently, Nazari et al. [Citation32] developed a rapid method of assaying virus inactivation in only a few minutes using a solid-state nanopore sensor. The sensor consisted of a nanopore with a diameter of 38 nm on the Si3N4 membrane. By analyzing ionic current traces of virus translocation before and after applying an optical virus inactivation method (using femtosecond laser radiation), they could study the effect of this inactivation method on various physical properties (fragmentation, agglomeration, etc.) of ΦX174 bacteriophage, which is widely used as a surrogate for human enteric viruses. Observing a decrease in the fractional current blockade and dwell time after laser treatment concluded that virus perforation upon the applied laser treatment. includes a summary of nanopore sensing applications in the detection of viruses.
It is worth mentioning that other than single pore sensors and RPS, nanoporous membranes such as alumina membranes have also been used to detect viruses [Citation167–169]. However, since their detection methodology is based on electrochemical impedance spectroscopy (EIS), they are not discussed in this paper. However, we included them in the last three rows of .
Table 6. Summary of nanopore sensors used for detection of viruses
3.1. Combination of machine learning with nanopore sensors
Although the nanopore sensing technique is a versatile method of analyzing single molecules, extracting the required data from the time-series signals is very time-consuming. During recent years, nanopore sensors have been integrated with machine learning and artificial intelligence for fully automated data extraction and thus digital recognition and classification of analytes [Citation177]. Utilizing a convolutional artificial neural network (CNN) to analyze nanopore data in a recent work revealed that deep learning could improve the accuracy in classifying translocation events and increase the number of events that can be analyzed by a factor of five [Citation178]. In the case of virus targets, the supervised machine learning method has shown to be effective in classifying virus sub-types [Citation177]. In this technique, a classification model (classifier) learns feature parameters from signal data and attributes them as a class to a virus sub-type. Although there is a challenge of variation in size, shape, and surface charge of viruses even within the same sub-type, defining the appropriate boundary conditions can enable precise discrimination. Amongst the various models for classification such as SVM, naïve Bayes, and k-nearest neighbor models, rotation forest and random forest models that randomly extract the training data from the dataset have shown promising results in previous works [Citation177]. One of the main advantages of employing machine learning in virus classification is using physical properties of virus particles instead of biological aspects, which can be useful in identification of new strains without any antibody or marker. Machine learning approach combined with nanopore sensing can open a new window of interest in developing rapid digital diagnostic devices that can be integrated with smartphones and enable portable and on-site diagnostic systems.
4. Summary and future outlook
Infectious viruses have been causing severe emerging and reemerging diseases during the recent decades. Viruses can spread very fast, create ongoing pandemics, and harm lots of lives. Since viruses are capable of rapid mutation during their replication, they are most likely to cause the emergence of new variants and infectious diseases [Citation179]. Viruses also cause environmental contamination, and their environmental hazards have been studied in food and environmental virology. The resulting environmental contamination is associated with consequent hazards of viral reintroduction into animal and human populations [Citation180]. Therefore, developing rapid, portable, low-cost and sensitive sensors for monitoring and detecting viruses in clinical and environmental samples has become essential for public health organizations [Citation181].
Single-virus detection and identification using nanopores is an emerging yet promising technology and, in many regards, is still in its early stages. The main strength of this electrical sensing method is enabling sensitive, label-free, and real-time detection of target viruses down to the scale of the single-virion level. Furthermore, driven by advances in micro/nanofabrication and nanotechnology, this method provides a significantly low-cost and portable platform for detecting a wide range of viral contaminants and helps gain a fundamental understanding of the physical and chemical properties of single viruses, their chemical interactions and bioanalysis that will advance the knowledge in virology. Nanopore sensing will likely be implemented quickly for commercial applications in various fields, such as biomedicine and biotechnology for drug development applications, molecular diagnostics, environmental quality control, etc.
Current trends imply that several obstacles remain and need to be overcome. In the case of using biological nanopores, the most critical challenge is the instability of the lipid bilayer and its capacitive behavior. However, significant efforts have been made to improve its stability, such as tethering lipid bilayer on solid supports [Citation65], using precast gels [Citation182], and polymerization of phospholipid bilayer membranes [Citation183]. Since most nanopore sensors rely on weak non-covalent interactions to analyze and detect the target analyte, the target analytes can interfere with matrix components in complex biofluids. Although biological nanopores can be genetically engineered and functionalized at specific positions, and solid-state nanopores are functionalized with different selectivity and functional groups, there is a lack of investigation in employing nanopore sensors in complex biological samples. Other than utilizing protein-ligand interactions [Citation184], the development of nanopore sensors consisted of nanopore arrays modified with various non-covalent bonding sites can enhance the selectivity and specificity of the sensor [Citation185]. Nanopore arrays can produce collective diagnostic patterns that can serve as an analyte signature, and the discrimination of analytes in complex samples can be performed with enhanced resolution compared to single-pore configuration.
Solid-state nanopores have attracted great interest in whole-virus detection and identification since they are robust and can be fabricated with required pore sizes, suitable for relatively large virus particles that vary in size and morphology. Further improvements in fabricating nanopores with reproducible pore size will enhance the resolution, reproducibility, and sensitivity of measurements in the characterization of small viruses. Also, advancements in introducing coatings or surface modifications within nanopores can minimize pore-clogging and nonspecific adsorptions. Integration of nanopores and their required equipment (tubings, reservoirs, electrodes, and electrical circuitry, including A/D converter, amplifier, etc.) in microfluidic devices has resulted in the development of miniaturized, portable, and low-cost sensors. Taking further advantages of microfluidic technology can advance the current nanopore-based sensors and provide an automated, portable, and affordable virus recognition platform that is an advantageous substitute for traditional bench assays.
Another challenge in the nanopore sensing method is the time-consuming process of analyzing the generated signals by analyte and extracting their characteristics, especially in nanopore arrays with simultaneous translocation events. However, the recent attempts to integrate nanopore sensors with state-of-the-art artificial intelligence are reported to be very efficient in automated, accurate, and rapid processing of data signals and recognition of specific patterns, thus enabling nanopore sensing technology to be mastered and utilized [Citation186]. Recently, using PCR-negative/-positive SARS-COV-2 specimens’ data as the training dataset has enabled detecting both negative and positive specimens with high throughput and sensitivity. Modifying the training data enables artificial intelligent nanopore sensors to be versatile virus recognition devices. For example, machine learning of influenza A virus (H1N1) and SARS-CoV-2 that pose similar symptoms indicated a very high discrimination factor [Citation165]. Using the collected clinical specimens from the infected patients of different viruses as training data enables the development of highly accurate diagnostic sensors with excellent discrimination capability.
The compatibility of the RPS method in nanopore sensors with non-electrical sensing methods has been of great interest in recent years. For example, simultaneous RPS with optical analysis was used for accurate measurement and obtaining ‘double-checked’ results. Combination of the RPS method with optical spectrometry, such as plasmonic resonance, Raman scattering, and fluorescence, not only can increase accuracy and precision and provide advanced information in the analysis of individual molecules [Citation126,Citation187], but also enables achieving high throughput analysis with furthest advancements of optical signal-to-noise ratio.
Nanopore sensors have reached a significant stage of advancement and shown a great capacity for recognizing individual entities, and there are commercial sensors released to the market that utilize this technology for sequencing applications [Citation188]. The application of this versatile method to detect, identify, and classify viruses in recent years has resulted in promising performance. Analyzing specifications of a greater diversity of virions such as their surface charge, concentration, and size and determining the kinetics of virus-antibody binding and total to infectious particle ratio would be of particular interest. Taking strict steps is critical to avoid false-negative and false-positive results in diagnostic applications. Detection of new and unknown virus species and variants could be achieved using the nanopore sensing technology, while it is inherently difficult using the traditional detection methods as there is a need for information regarding the membrane proteins or genome before designing antibodies or PCR primers, respectively. Additionally, to isolate and obtain the source protein or RNA/DNA from the viruses, the host cells must be found. Nanopore sensing technology can detect unknown virions based on their electrical properties or signature without requiring biochemical information [Citation39]. Further advances in this multidisciplinary field can lead to the substitution of nanopore-based sensors for traditional biochemical assays. This method can be helpful for point-of-care testing at very early stages of infection that can prevent disease outspread by enabling clinical treatments before symptom onsets. From a long-term perspective, we believe that nanopore-based sensing or RPS technology will take on a more dominant role in detecting and quantifying viruses. It can lead to the development of wearable and mobile recognition devices for detecting and monitoring infectious virions, which could enhance public health and safety.
Conclusion
In conclusion, nanopore sensing technology has grown rapidly during the last few decades. Dramatic advances in microfabrication and nanotechnology, and integration of microfluidics and artificial intelligence have enabled significant applications of this electrical-based technique in bio-detection, including quantification of viral particles in a sensitive, real-time, and efficient manner. This method has shown to be very promising in viral particle quantification by overcoming certain limitations found in traditional detection techniques. Thus, this method is expected to be developed for use at point-of-care and point-of-need, particularly during pandemics, in the near future.
Acknowledgements
The authors are sincerely thankful to the Mitacs-Accelerate Canada, Natural Sciences and Engineering Research Council of Canada Discovery Grant and James and Joanne Love Chair in Environmental Engineering for their financial supports to PR and SB.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Tarim EA, Karakuzu B, Oksuz C, et al. Microfluidic-based virus detection methods for respiratory diseases. Emergent Mater. 2021;2:143–168.
- Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 Apr;14(4):3822–3835.
- Adigal SS, Rayaroth NV, John RV, et al. A review on human body fluids for the diagnosis of viral infections: scope for rapid detection of COVID-19. Expert Rev Mol Diagn. 2021 Jan;21(1):31–42.
- Diao M, Lang L, Feng J, et al. Molecular detections of coronavirus: current and emerging methodologies. Expert Rev Anti Infect Ther. 2021 Jul;1–12. DOI:10.1080/14787210.2021.1949986
- Wu K, Saha R, Su D, et al. Magnetic-nanosensor-based virus and pathogen detection strategies before and during COVID-19. ACS Appl Nano Mater. 2020 Oct;3(10):9560–9580.
- Gray MC, Su W-Y, Van Hal SJ. Improving influenza virus detection. Expert Opin Med Diagn. 2012;6(1):75–87.
- Prasad S, Potdar V, Cherian S, et al. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020;151(2–3):241.
- Sheikhzadeh E, Eissa S, Ismail A, et al. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020;220(May):121392.
- Kumar P. Methods for rapid virus identification and quantification. Mater Methods. 2013;3:207.
- Cao C, Long YT. Biological nanopores: confined spaces for electrochemical single-molecule analysis. Acc Chem Res. 2018;51(2):331–341.
- Lepoitevin M, Ma T, Bechelany M, et al. Functionalization of single solid state nanopores to mimic biological ion channels: a review. Adv Colloid Interface Sci. 2017;250:195–213.
- Varongchayakul N, Song J, Meller A, et al. Single-molecule protein sensing in a nanopore: a tutorial. Chem Soc Rev. 2018;47(23):8512–8524.
- Xu J, Merlier F, Avalle B, et al. Molecularly imprinted polymer nanoparticles as potential synthetic antibodies for immunoprotection against HIV. ACS Appl Mater Interfaces. 2019;11(10):9824–9831.
- Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26(10):1146–1153.
- Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6(10):615–624.
- Heerema SJ, Vicarelli L, Pud S, et al. Probing DNA translocations with inplane current signals in a graphene nanoribbon with a nanopore. ACS Nano. 2018;12(3):2623–2633.
- Tsutsui M, Yoshida T, Yokota K, et al. Discriminating single-bacterial shape using low-aspect-ratio pores. Sci. Rep. 2017;7(1):1–9.
- Wang L, Yao F, Kang X. Nanopore single-molecule analysis of metal ion-chelator chemical reaction. Anal Chem. 2017;89(15):7958–7965.
- Roozbahani GM, Chen X, Zhang Y, et al. Nanopore detection of metal ions: current status and future directions. Small Methods. 2020;4(10):2000266.
- Cai S, Sze JYY, Ivanov AP, et al. Small molecule electro-optical binding assay using nanopores. Nat Commun. 2019;10(1):1–9.
- Somerville JA, Willmott GR, Eldridge J, et al. Size and charge characterisation of a submicrometre oil-in-water emulsion using resistive pulse sensing with tunable pores. J Colloid Interface Sci. 2013;394:243–251.
- Stoloff DH, Wanunu M. Recent trends in nanopores for biotechnology. Curr Opin Biotechnol. 2013;24(4):699–704.
- Willmott GR, Fisk MG, Eldridge J. Magnetic microbead transport during resistive pulse sensing. Biomicrofluidics. 2013;7(6):64106.
- Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38(8):2360–2384.
- Maas SLN, Broekman MLD, De Vrij J Tunable resistive pulse sensing for the characterization of extracellular vesicles. In: Exosomes and microvesicles, pp. 21-33. Humana Press, New York, NY, 2017.
- Darvish A, Goyal G, Kim M. Sensing, capturing, and interrogation of single virus particles with solid state nanopores. Adv Glob Heal through Sens Technol. 2015;9490(May 2015):94900M.
- Arima A, Tsutsui M, Harlisa IH, et al. Selective detections of single-viruses using solid-state nanopores. Sci. Rep. 2018;8(1):16305.
- Apetrei A, Ciuca A, Lee J, et al. A protein nanopore-based approach for bacteria sensing. Nanoscale Res Lett. 2016;11(1):1–12.
- Oh S, Lee MK, Chi SW. Single-molecule-based detection of conserved influenza A Virus RNA promoter using a protein nanopore. ACS Sens. 2019;4(11):2849–2853.
- Uram JD, Ke K, Hunt AJ, et al. Submicrometer pore‐based characterization and quantification of antibody–virus interactions. Small. 2006;2(8‐9):967–972.
- Arima A, Harlisa IH, Yoshida T, et al. Identifying single viruses using biorecognition solid-state nanopores. J. Am. Chem. Soc. 2018;140(48):16834–16841.
- Nazari M, Li X, Alibakhshi MA, et al. Femtosecond photonic viral inactivation probed using solid-state nanopores. Nano Futur. 2018;2(4). DOI:10.1088/2399-1984/aadf9d
- Arjmandi N, Van Roy W, Lagae L. Measuring mass of nanoparticles and viruses in liquids with nanometer-scale pores. Anal Chem. 2014;86(10):4637–4641.
- Arjmandi N, Van Roy W, Lagae L, et al. Measuring the electric charge and zeta potential of nanometer-sized objects using pyramidal-shaped nanopores. Anal Chem. 2012;84(20):8490–8496.
- Darvish A, Lee JS, Peng B, et al. Mechanical characterization of HIV-1 with a solid-state nanopore sensor. Electrophoresis. 2019;40(5):776–783.
- Shi W, Friedman AK, Baker LA. Nanopore Sensing. Anal Chem. 2017;89(1):157–188.
- Miles BN, Ivanov AP, Wilson KA, et al. Single molecule sensing with solid-state nanopores: novel materials, methods, and applications. Chem Soc Rev. 2013;42(1):15–28.
- Kono N, Arakawa K. Nanopore sequencing: review of potential applications in functional genomics. Dev Growth Differ. 2019 Jun;61(5):316–326.
- Yang L, Yamamoto T. Quantification of virus particles using nanopore-based resistive-pulse sensing techniques. Front Microbiol. 2016;7:1500.
- Burrell CJ, Howard CR, Murphy FA. Laboratory diagnosis of virus diseases. Fenner White’s Med Virol. 2017;135.
- Morales-Narváez E, Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274.
- Hong KH, Lee SW, Kim TS, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann. Lab. Med. 2020;40(5):351–360.
- Grant BD, Anderson CE, Williford JR, et al. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92(16):11305–11309.
- Yuce M, Filiztekin E, Zkaya KG. COVID-19 diagnosis-A review of current methods. Biosens. Bioelectron. 2020;172:112752.
- Mamad-Hemouch H, Ramoul H, Abou Taha M, et al. Biomimetic nanotubes based on cyclodextrins for ion-channel applications. Nano Letters. 2015;15(11):7748–7754.
- Kawano R, Osaki T, Sasaki H, et al. A polymer‐based nanopore‐integrated microfluidic device for generating stable bilayer lipid membranes. Small. 2010;6(19):2100–2104.
- Kasianowicz JJ, Brandin E, Branton D, et al. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci. 1996;93(24):13770–13773.
- Braha O, Gu L-Q, Zhou L, et al. Simultaneous stochastic sensing of divalent metal ions. Nat Biotechnol. 2000;18(9):1005–1007.
- Cao C, Ying Y-L, Hu Z-L, et al. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat Nanotechnol. 2016;11(8):713–718.
- Ettedgui J, Kasianowicz JJ, Balijepalli A. Single molecule discrimination of heteropolytungstates and their isomers in solution with a nanometer-scale pore. J Am Chem Soc. 2016;138(23):7228–7231.
- Fahie M, Chisholm C, Chen M. Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore. ACS Nano. 2015;9(2):1089–1098.
- Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nanoscience and technology: A collection of reviews from Nature Journals, pp.261-268.
- Ayub M, Stoddart D, Bayley H. Nucleobase recognition by truncated α-hemolysin pores. ACS Nano. 2015;9(8):7895–7903.
- Wang Y-Q, Cao C, Ying Y-L, et al. Rationally designed sensing selectivity and sensitivity of an aerolysin nanopore via site-directed mutagenesis. ACS Sens. 2018;3(4):779–783.
- Naveed H, Liang J. Engineering biological nanopores with enhanced properties. Biophys J. 2012;102(3):188a–189a.
- Ying YL, Cao C, Long YT. Single molecule analysis by biological nanopore sensors. Analyst. 2014;139(16):3826–3835.
- Tanaka Y, Hirano N, Kaneko J, et al. 2 Methyl 2, 4 pentanediol induces spontaneous assembly of staphylococcal hemolysin into heptameric pore structure. Protein Sci. 2011;20(2):448–456.
- Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science. 2004;303(5661):1189–1192.
- Iacovache I, De Carlo S, Cirauqui N, et al. Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat Commun. 2016;7(1):12062.
- Guasch A, Pous J, Ibarra B, et al. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle11Edited by R. Huber. J. Mol. Biol. 2002;315(4):663–676.
- Mueller M, Grauschopf U, Maier T, et al. The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature. 2009;459(7247):726–730.
- Subbarao GV, Van Den Berg B. Crystal structure of the monomeric porin OmpG. J Mol Biol. 2006;360(4):750–759.
- Otwinowski Z, Schevitz RW, Zhang R-G, et al. Crystal structure of trp represser/operator complex at atomic resolution. Nature. 1988;335(6188):321–329.
- Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34(5):518–524.
- White RJ, Ervin EN, Yang T, et al. Single ion-channel recordings using glass nanopore membranes. J. Am. Chem. Soc. 2007 Sep;129(38):11766–11775.
- Bello J, Kim Y-R, Kim SM, et al. Lipid bilayer membrane technologies: a review on single-molecule studies of DNA sequencing by using membrane nanopores. Microchim Acta. 2017;184(7):1883–1897.
- Hall AR, Scott A, Rotem D, et al. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat Nanotechnol. 2010;5(12):874–877.
- De La Escosura-Muñiz A, Merkoçi A. Nanochannels preparation and application in biosensing. ACS Nano. 2012;6(9):7556–7583.
- Zhang Z, Sui X, Li P, et al. Ultrathin and ion-selective janus membranes for high-performance osmotic energy conversion. J. Am. Chem. Soc. 2017 Jul;139(26):8905–8914.
- Lee K, Park K-B, Kim H-J, et al. Recent progress in solid state nanopores. Adv. Mater. 2018;30(42):1704680.
- Spitzberg JD, Zrehen A, Van Kooten XF, et al. Plasmonic‐nanopore biosensors for superior single‐molecule detection. Adv Mater. 2019;31(23):1900422.
- Firnkes M, Pedone D, Knezevic J, et al. Electrically facilitated translocations of proteins through silicon nitride nanopores: conjoint and competitive action of diffusion, electrophoresis, and electroosmosis. Nano Lett. 2010;10(6):2162–2167.
- Spitzberg JD, Van Kooten XF, Bercovici M, et al. Microfluidic device for coupling isotachophoretic sample focusing with nanopore single-molecule sensing. Nanoscale. 2020;12(34):17805–17811.
- Roelen Z, Bustamante JA, Carlsen A, et al. Instrumentation for low noise nanopore-based ionic current recording under laser illumination. Rev Sci Instrum. 2018;89(1):15007.
- Pan R, Hu K, Jiang D, et al. Electrochemical resistive-pulse sensing. J Am Chem Soc. 2019;141(50):19555–19559. ACS Publications.
- Li J, Stein D, McMullan C, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412(6843):166–169.
- Liu S, Lu B, Zhao Q, et al. Boron nitride nanopores: highly sensitive DNA single‐molecule detectors. Adv. Mater. 2013;25(33):4549–4554.
- Freedman KJ, Haq SR, Fletcher MR, et al. Nonequilibrium capture rates induce protein accumulation and enhanced adsorption to solid-state nanopores. ACS Nano. 2014;8(12):12238–12249.
- Yusko EC, Bruhn BR, Eggenberger O, et al. Real-time shape approximation and 5-D fingerprinting of single proteins. Nature nanotechnology 2017;12(4):360-367.
- Wanunu M, Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007 Jun;7(6):1580–1585.
- Balme S, Lepoitevin M, Dumée LF, et al. Diffusion dynamics of latex nanoparticles coated with ssDNA across a single nanopore. Soft Matter. 2017;13(2):496–502.
- Apel PY, Blonskaya IV, Dmitriev SN, et al. Surfactant-controlled etching of ion track nanopores and its practical applications in membrane technology. Radiat. Meas. 2008;43:S552–S559.
- Pevarnik M, Healy K, Davenport M, et al. A hydrophobic entrance enhances ion current rectification and induces dewetting in asymmetric nanopores. Analyst. 2012;137(13):2944–2950.
- Storm AJ, Chen JH, Ling XS, et al. Fabrication of solid-state nanopores with single-nanometre precision. Nat Mater. 2003;2(8):537–540.
- Venkatesan BM, Shah AB, Zuo JM, et al. DNA sensing using nanocrystalline surface enhanced Al2O3 nanopore sensors. Adv Funct Mater. 2010;20(8):1266–1275.
- Larkin J, Henley R, Bell DC, et al. Slow DNA transport through nanopores in hafnium oxide membranes. ACS Nano. 2013;7(11):10121–10128.
- Apel P. Track etching technique in membrane technology. Radiat Meas. 2001;34(1–6):559–566.
- Garaj S, Hubbard W, Reina A, et al. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467(7312):190–193.
- Jain T, Rasera BC, Guerrero RJS, et al. Heterogeneous sub-continuum ionic transport in statistically isolated graphene nanopores. Nat. Nanotechnol. 2015;10(12):1053–1057.
- Feng L, Cao M, Ma X, et al. Superparamagnetic high-surface-area Fe3O4 nanoparticles as adsorbents for arsenic removal. J Hazard Mater. 2012;217–218:439–446.
- Feng J, Liu K, Graf M, et al. Electrochemical reaction in single layer MoS2: nanopores opened atom by atom. Nano Letters. 2015;15(5):3431–3438.
- White HS, Bund A. Ion current rectification at nanopores in glass membranes. Langmuir. 2008 Mar;24(5):2212–2218.
- Steinbock LJ, Krishnan S, Bulushev RD, et al. Probing the size of proteins with glass nanopores. Nanoscale. 2014;6(23):14380–14387.
- Wang G, Bohaty AK, Zharov I, et al. Photon gated transport at the glass nanopore electrode. J Am Chem Soc. 2006 Oct;128(41):13553–13558.
- Fornasiero F, In JB, Kim S, et al. pH-Tunable ion selectivity in carbon nanotube pores. Langmuir. 2010 Sep;26(18):14848–14853.
- Geng J, Kim K, Zhang J, et al. Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature. 2014;514(7524):612–615.
- Siria A, Poncharal P, Biance A-L, et al. Giant osmotic energy conversion measured in a single transmembrane boron nitride nanotube. Nature. 2013;494(7438):455–458.
- Létant SE. Functional nanostructured platforms for chemical and biological sensing. Micro Nanotechnologies Sp Appl. 2006;6223(May 2006):62230B.
- Petrov YV, Ubyivovk EV, Baraban AP. Fabrication of nanopores in silicon nitride membrane by means of wet etching enhanced by focused helium ion beam irradiation. AIP Conf Proc. 2019;2064(1):30012.
- Gierak J, Madouri A, Biance AL, et al. Sub-5 nm FIB direct patterning of nanodevices. Microelectron. Eng. 2007;84(5–8):779–783.
- Kennedy E, Dong Z, Tennant C, et al. Reading the primary structure of a protein with 0.07 nm 3 resolution using a subnanometre-diameter pore. Nat Nanotechnol. 2016;11(11):968–976.
- Chung NX, Gatty HK, Lu X, et al. Optimized electrochemical breakdown etching using temporal voltage variation for formation of nanopores in a silicon membrane. Sens Actuators B Chem. 2021;331(December 2020):1–9.
- Waugh M, Briggs K, Gunn D, et al. Solid-state nanopore fabrication by automated controlled breakdown. Nat. Protoc. 2020;15(1):122–143.
- Chen Q, Wang Y, Deng T, et al. Fabrication of nanopores and nanoslits with feature sizes down to 5 nm by wet etching method. Nanotechnology. 2018;29(8):85301.
- Fried JP, Swett JL, Nadappuram BP, et al. In situ solid-state nanopore fabrication. Chem. Soc. Rev. 2021;50(8):4974–4992.
- Kim MJ, McNally B, Murata K, et al. Characteristics of solid-state nanometre pores fabricated using a transmission electron microscope. Nanotechnology. 2007;18(20):205302.
- Prakash S, Pinti M, Bellman K. Variable cross-section nanopores fabricated in silicon nitride membranes using a transmission electron microscope. J Micromech Microeng. 2012;22(6):67002.
- Spinney PS, Howitt DG, Smith RL, et al. Nanopore formation by low-energy focused electron beam machining. Nanotechnology. 2010;21(37):375301.
- Lanyon YH, De Marzi G, Watson YE, et al. Fabrication of nanopore array electrodes by focused ion beam milling. Anal. Chem. 2007;79(8):3048–3055.
- Briggs K, Charron M, Kwok H, et al. Kinetics of nanopore fabrication during controlled breakdown of dielectric membranes in solution. Nanotechnology. 2015;26(8):84004.
- Kwok H, Briggs K, Tabard-Cossa V. Nanopore fabrication by controlled dielectric breakdown. PLoS One. 2014;9(3):e92880.
- Marshall MM, Yang J, Hall AR. Direct and transmission milling of suspended silicon nitride membranes with a focused helium ion beam. Scanning. 2012 Mar;34(2):101–106.
- Gilbert SM, Dunn G, Azizi A, et al. Fabrication of subnanometer-precision nanopores in hexagonal boron nitride. Sci. Rep. 2017;7(1):1–7.
- Mara A, Siwy Z, Trautmann C, et al. An asymmetric polymer nanopore for single molecule detection. Nano Lett. 2004;4(3):497–501.
- Deng T, Chen J, Si W, et al. Fabrication of silicon nanopore arrays using a combination of dry and wet etching. J Vac Sci Technol B, Nanotechnol Microelectron Mater Process Meas Phenom. 2012;30(6):61804.
- Park SR, Peng H, Ling XS. Fabrication of nanopores in silicon chips using feedback chemical etching. Small. 2007;3(1):116–119.
- Bian F, Tian YC, Wang R, et al. Ultrasmall silver nanopores fabricated by femtosecond laser pulses. Nano Letters. 2011 Aug;11(8):3251–3257.
- Chung NX, Gatty HK, Lu X, et al. Optimized electrochemical breakdown etching using temporal voltage variation for formation of nanopores in a silicon membrane. Sens Actuators B Chem. 2021;331(October 2020):1–9.
- Prakash S, Pinti M, Bhushan B. Review article: theory, fabrication and applications of microfluidic and nanofluidic biosensors. Philos Trans R Soc A Math Phys Eng Sci. 2012;370(1967):2269–2303.
- Alahmad W, Varanusupakul P, Varanusupakul P. Recent developments and applications of microfluidic paper-based analytical devices for the detection of biological and chemical hazards in foods: a critical review. Crit Rev Anal Chem. 2021 Jul;1–20. DOI:10.1080/10408347.2021.1949695
- Fu J, Wu L, Qiao Y, et al. Microfluidic systems applied in solid-state nanopore sensors. Micromachines. 2020;11(3):332.
- Jain T, Guerrero RJS, Aguilar CA, et al. Integration of solid-state nanopores in microfluidic networks via transfer printing of suspended membranes. Anal Chem. 2013;85(8):3871–3878.
- Yanagi I, Akahori R, Aoki M, et al. Multichannel detection of ionic currents through two nanopores fabricated on integrated Si 3 N 4 membranes. Lab Chip. 2016;16(17):3340–3350.
- Tahvildari R, Beamish E, Tabard-Cossa V, et al. Integrating nanopore sensors within microfluidic channel arrays using controlled breakdown. Lab Chip. 2015;15(6):1407–1411.
- Tahvildari R, Beamish E, Briggs K, et al. Manipulating electrical and fluidic access in integrated nanopore microfluidic arrays using microvalves. small. 2017;13(10):1602601.
- Liu S, Zhao Y, Parks JW, et al. Correlated electrical and optical analysis of single nanoparticles and biomolecules on a nanopore-gated optofluidic chip. Nano Lett. 2014 Aug;14(8):4816–4820.
- Varongchayakul N, Hersey JS, Squires A, et al. A solid state hard microfluidic nanopore biosensor with multilayer fluidics and on chip bioassay/purification chamber. Adv Funct Mater. 2018;28(50):1804182.
- Wu H-C, Astier Y, Maglia G, et al. Protein nanopores with covalently attached molecular adapters. J Am Chem Soc. 2007 Dec;129(51):16142–16148.
- Lin Y, Ying Y-L, Gao R, et al. Single-molecule sensing with nanopore confinement: from chemical reactions to biological interactions. Chemistry - A European Journal. 2018 Sep;24(50):13064–13071.
- Eggenberger OM, Ying C, Mayer M. Surface coatings for solid-state nanopores. Nanoscale. 2019;11(42):19636–19657.
- Gu L-Q, Cheley S, Bayley H. Prolonged residence time of a noncovalent molecular adapter, -cyclodextrin, within the lumen of mutant -hemolysin pores. J Gen Physiol. 2001;118(5):481–494.
- Majd S, Yusko EC, Billeh YN, et al. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr Opin Biotechnol. 2010;21(4):439–476.
- Hlady V, Buijs J. Protein adsorption on solid surfaces. Curr Opin Biotechnol. 1996;7(1):72–77.
- Powell MR, Cleary L, Davenport M, et al. Electric-field-induced wetting and dewetting in single hydrophobic nanopores. Nat Nanotechnol. 2011;6(12):798–802.
- Sparreboom W, Van Den Berg A, Eijkel JCT. Principles and applications of nanofluidic transport. Nat Nanotechnol. 2009;4(11):713–720.
- Fologea D, Uplinger J, Thomas B, et al. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5(9):1734–1737.
- Yusko EC, Johnson JM, Majd S, et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011;6(4):253–260.
- Thangaraj V, Lepoitevin M, Smietana M, et al. Detection of short ssDNA and dsDNA by current-voltage measurements using conical nanopores coated with Al 2 O 3 by atomic layer deposition. Microchim Acta. 2016;183(3):1011–1017.
- Rollings R, Graef E, Walsh N, et al. The effects of geometry and stability of solid-state nanopores on detecting single DNA molecules. Nanotechnology. 2015;26(4):44001.
- Li X, Hu R, Li J, et al. Non-sticky translocation of bio-molecules through Tween 20-coated solid-state nanopores in a wide pH range. Appl. Phys. Lett. 2016;109(14):143105.
- Rosen MJ, Kunjappu JT. Surfactants and interfacial phenomena. Hoboken, NJ): John Wiley & Sons; 2012.
- Whitesides GM, Kriebel JK, Love JC. Molecular engineering of surfaces using self-assembled monolayers. Sci Prog. 2005;88(1):17–48.
- Ananth A, Genua M, Aissaoui N, et al. Reversible immobilization of proteins in sensors and solid state nanopores. Small. 2018;14(18):1703357.
- Martin CR, Nishizawa M, Jirage K, et al. Controlling ion-transport selectivity in gold nanotubule membranes. Adv Mater. 2001 Sep;13(18):1351–1362.
- Wei R, Gatterdam V, Wieneke R, et al. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat Nanotechnol. 2012;7(4):257–263.
- Yu S, Lee SB, Kang M, et al. Size-based protein separations in poly (ethylene glycol)-derivatized gold nanotubule membranes. Nano Lett. 2001;1(9):495–498.
- Emilsson G, Sakiyama Y, Malekian B, et al. Gating protein transport in solid state nanopores by single molecule recognition. ACS Cent. Sci. 2018;4(8):1007–1014.
- Zambrana-Puyalto X, Maccaferri N, Ponzellini P, et al. Site-selective functionalization of plasmonic nanopores for enhanced fluorescence emission rate and Forster resonance energy transfer. Nanoscale Adv. 2019;1(6):2454–2461.
- Wang C, Fu Q, Wang X, et al. Atomic layer deposition modified track-etched conical nanochannels for protein sensing. Anal. Chem. 2015;87(16):8227–8233.
- Hu R, Diao J, Li J, et al. Intrinsic and membrane-facilitated -synuclein oligomerization revealed by label-free detection through solid-state nanopores. Sci Rep. 2016;6(1):1–11.
- Tan S, Wang L, Liu H, et al. Single nanoparticle translocation through chemically modified solid nanopore. Nanoscale Res Lett. 2016;11(1):1–10.
- Lepoitevin M, Jamilloux B, Bechelany M, et al. Fast and reversible functionalization of a single nanopore based on layer-by-layer polyelectrolyte self-assembly for tuning current rectification and designing sensors. RSC Adv. 2016;6(38):32228–32233.
- Ali M, Yameen B, Cervera J, et al. Layer-by-layer assembly of polyelectrolytes into ionic current rectifying solid-state nanopores: insights from theory and experiment. J. Am. Chem. Soc. 2010;132(24):8338–8348.
- Alem H, Blondeau F, Glinel K, et al. Layer-by-layer assembly of polyelectrolytes in nanopores. Macromolecules. 2007;40(9):3366–3372.
- Djozan D, Ebrahimi B, Mahkam M, et al. Evaluation of a new method for chemical coating of aluminum wire with molecularly imprinted polymer layer. Application for the fabrication of triazines selective solid-phase microextraction fiber. Anal Chim Acta. 2010;674(1):40–48.
- Eggenberger OM, Leriche G, Koyanagi T, et al. Fluid surface coatings for solid-state nanopores: comparison of phospholipid bilayers and archaea-inspired lipid monolayers. Nanotechnology. 2019;30(32):325504.
- Yusko EC, Bruhn BR, Eggenberger OM, et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2017;12(4):360–367.
- Robertson JWF, Rodrigues CG, Stanford VM, et al. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc Natl Acad Sci. 2007;104(20):8207–8211.
- Trémeaux P, Caporossi A, Thélu M-A, et al. Hepatitis C virus whole genome sequencing: current methods/issues and future challenges. Crit. Rev. Clin. Lab. Sci. 2016 Sep;53(5):341–351.
- DeBlois RW, Bean CP, Wesley RKA. Electrokinetic measurements with submicron particles and pores by the resistive pulse technique. J Colloid Interface Sci. 1977;61(2):323–335.
- Feuer BI, Uzgiris EE, Deblois RW, et al. Length of glycoprotein spikes of vesicular stomatitis virus and sindbis virus, measured in Situ using quasi elastic light scattering and a resistive-pulse technique. Virology. 1978;90(1):156–161.
- Zhou K, Li L, Tan Z, et al. Characterization of hepatitis B virus capsids by resistive-pulse sensing. J Am Chem Soc. 2011;133(6):1618–1621.
- McMullen A, De Haan HW, Tang JX, et al. Buckling causes nonlinear dynamics of filamentous viruses driven through nanopores. Phys Rev Lett. 2018;120(7):78101.
- Miyagawa T, Hongo S, Nakamura N, et al. A novel diagnostic system for infectious diseases using solid-state nanopore devices. Proc Annu Int Conf IEEE Eng Med Biol Soc EMBS. 2018;2018-July:2833–2836.
- Taniguchi M, Minami S, Ono C, et al. Combining machine learning and nanopore construction creates an artificial intelligence nanopore for coronavirus detection. Nat. Commun. 2021;12(1):1–8.
- Cavanagh D. Innovation and discovery: the application of nucleic acid-based technology to avian virus detection and characterization. Avian Pathol. 2001 Dec;30(6):581–598.
- Nguyen BTT, Koh G, Lim HS, et al. Membrane-based electrochemical nanobiosensor for the detection of virus. Anal Chem. 2009;81(17):7226–7234.
- Nguyen BTT, Peh AEK, Chee CYL, et al., “Electrochemical impedance spectroscopy characterization of nanoporous alumina dengue virus biosensor,”. Bioelectrochemistry. 2012;88:15–21.
- Peh AEK, Li SFY. Dengue virus detection using impedance measured across nanoporous alumina membrane. Biosens Bioelectron. 2013;42(1):391–396.
- DeBlois RW, Wesley RK. Sizes and concentrations of several type C oncornaviruses and bacteriophage T2 by the resistive-pulse technique. J Virol. 1977;23(2):227–233.
- McMullen A, Liu X, Tang J, et al. Solid-state nanopores for detection of rod-like viruses and trapping of single DNA molecules. 2012 Lester Eastman Conf High Perform Devices, LEC. 2012;2012(c):8–9.
- Das N, RoyChaudhuri C. Reliability study of nanoporous silicon oxide impedance biosensor for virus detection: influence of surface roughness. IEEE Trans Device Mater Reliab. 2015;15(3):402–409.
- Wu H, Chen Y, Zhou Q, et al. Translocation of rigid rod-shaped virus through various solid-state nanopores. Anal. Chem. 2016;88(4):2502–2510.
- McMullen A, De Haan HW, Tang JX, et al. Stiff filamentous virus translocations through solid-state nanopores. Nat Commun. 2014;5(May). DOI:10.1038/ncomms5171
- Harms ZD, Mogensen KB, Nunes PS, et al. Nanofluidic devices with two pores in series for resistive-pulse sensing of single virus capsids. Anal Chem. 2011 Dec;83(24):9573–9578.
- Karawdeniya BI, Bandara YMNDY, Khan AI, et al. Adeno-associated virus characterization for cargo discrimination through nanopore responsiveness. Nanoscale. 2020;12(46):23721–23731.
- Arima A, Tsutsui M, Washio T, et al. Solid-State nanopore platform integrated with machine learning for digital diagnosis of virus infection. Anal Chem. 2021;93(1):215–227.
- Misiunas K, Ermann N, Keyser UF. QuipuNet: convolutional neural network for single-molecule nanopore sensing. Nano Lett. 2018;18(6):4040–4045.
- Fenton A, Pedersen AB. Community epidemiology framework for classifying disease threats. Emerg Infect Dis. 2005;11(12):1815.
- La Rosa G, Fontana S, Di Grazia A, et al. Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription-PCR assays. Appl Environ Microbiol. 2007;73(13):4152–4161.
- Mori A, Pomari E, Deiana M, et al. Molecular techniques for the genomic viral RNA detection of West Nile, Dengue, Zika and Chikungunya arboviruses: a narrative review. Expert Rev. Mol. Diagn. 2021 Jun;21(6):591–612.
- Beddow JA, Peterson IR, Heptinstall J, et al. Reconstitution of nicotinic acetylcholine receptors into gel-protected lipid membranes. Anal Chem. 2004;76(8):2261–2265.
- Shenoy DK, Barger WR, Singh A, et al. Functional reconstitution of protein ion channels into planar polymerizable phospholipid membranes. Nano Letters. 2005;5(6):1181–1185.
- Ding S, Gao C, Gu L-Q. Capturing single molecules of immunoglobulin and ricin with an aptamer-encoded glass nanopore. Anal Chem. 2009;81(16):6649–6655.
- Ervin EN, White RJ, White HS. Sensitivity and signal complexity as a function of the number of ion channels in a stochastic sensor. Anal Chem. 2009;81(2):533–537.
- Ryu S-W, Lee J-H, Kim J, et al. Comparison of two new generation influenza rapid diagnostic tests with instrument-based digital readout systems for influenza virus detection. Br. J. Biomed. Sci. 2016 Sep;73(3):115–120.
- Hauer P, Le Ru EC, Willmott GR. Co-ordinated detection of microparticles using tunable resistive pulse sensing and fluorescence spectroscopy. Biomicrofluidics. 2015;9(1):14110.
- Riaz N, Leung P, Barton K, et al. Adaptation of Oxford nanopore technology for hepatitis C whole genome sequencing and identification of within-host viral variants. BMC Genomics. 2021;22(1):1–12.
