ABSTRACT
As a currently identified small non-coding RNAs (ncRNAs) category, the PIWI-interacting RNAs (piRNAs) are crucial mediators of cell biology. The human genome comprises over 30.000 piRNA genes. Although considered a new field in cancer research, the piRNA pathway is shown by the existing evidence as an active pathway in a variety of different types of cancers with critical impacts on main aspects of cancer progression. Among the regulatory molecules that contribute to maintaining the dynamics of cancer cells, the P-element Induced WImpy testis (PIWI) proteins and piRNAs, as new players, have not been broadly studied so far. Therefore, the identification of cancer-related piRNAs and the assessment of target genes of piRNAs may lead to better cancer prevention and therapy strategies. This review articleaimed to highlight the role and function of piRNAs based on existing data. Understanding the role of piRNA in cancer may provide perspectives on their applications as particular biomarker signature in diagnosis in early stage, prognosis and therapeutic strategies.
KEYWORDS:
1. Introduction
Despite the transcription of over 90% of the human genomes, only 1–2% of the genome is composed of genes that code proteins, while the remaining portion of the transcriptome consists of non-coding RNAs (ncRNAs) [Citation1, D.-Y. Citation2]. ncRNAs play a pivotal role in the development and homeostasis [Citation3]. In particular, frequent deregulation of ncRNAs in cancer can contribute to the initiation, progression, and metastasis of tumors [Citation4, Citation5, Z. Citation6]. There are two major ncRNA categories, including regulatory and housekeeping/structural ncRNAs (tRNAs, rRNAs, and snoRNAs). Regulatory ncRNAs are grouped into large (exceeding 200 nucleotides) and small (below 200 nucleotides) ncRNAs [Citation7]. Large ncRNAs are primarily comprised of long non-coding RNAs (lncRNAs) as well as circular RNAs (circRNAs) [Citation8, Citation9. Citation10]. Moreover, small ncRNAs are heterogeneous, including P-element-Induced WImpy testis (PIWI)-interacting RNAs (piRNAs), microRNAs (miRNAs), transfer RNAs (tRNAs), small interfering RNAs (siRNAs), ribosomal RNAs (rRNAs), small cytoplasmic RNAs (scRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) [Citation11–13]. In , we have described some of the main features of three major types of small ncRNAs, siRNAs, miRNAs, and piRNAs [Citation14–18 J. C. Citation19, Citation20, L. Citation21]. Concerning small non-coding RNAs, it is noteworthy that they have a critical role in the cell growth, proliferation, apoptosis, and differentiation [Citation22]. Moreover, their regulatory functions are performed through the Argonaute (AGO) protein family as a major component of RNA silencing complexes, which is highly maintained in all organisms [Citation23].
Table 1. Characteristics of major noncoding RNAs (ncRNAs)
The third objective of Sustainable Development Goals (SDGs) (https://www.un.org/sustainabledevelopment/sustainable-development-goals/), that were adopted by United Nations member states in 2015 to be achieved by 2030, is to ensure healthy lives and promote well-being for all at all ages. Cancers are among the main noncommunicable diseases that are regarded as important targets for SDGs. Cancers have a significant health burden, and their incidence rates have increased worldwide since last decades [Citation24]. Efforts for finding new effective cancer biomarkers have become an integral part of cancer research because they have multiple clinical implications in different stages of disease [Citation25–27, S. Citation28–31].
Cancer detection at early stage and treatment of metastatic and chemoresistant tumors need specific and effective biomarkers [Citation25,Citation27,Citation32,Citation33]. Recently, the role of piRNAs as potential biomarkers and therapeutic tools in cancers had been attracting much attention. In this review, biological and clinical aspects of piRNAs in cancer have been reviewed.
1.1. An overview of piRNAs
In 2001, Aravin et al. identified the presence of piRNAs as repeat-associated small interfering RNAs (rasiRNAs), derived from repeated genomic components in testis and initial embryos of D. melanogaster [A. A. Citation34, A. A. Citation35, Citation36]. However, at that time, it was unknown that those small ncRNAs were actually piRNAs. They were ultimately named piRNAs in 2006 [A. Citation37]. Upon this discovery in Drosophila, it was later identified that there are plenty of piRNAs in gonads of different vertebrate and nonvertebrate species such as humans and mice [Citation38]. Immunoprecipitation and northern blot analyses demonstrated the existence of piRNAs in ovaries and testes of fetal and adult human [Z. Citation39].
As essential mediators in the biology of cells, piRNAs are considered as the latest, largest, and most various series of ncRNAs in somatic and germ cells [Citation40,Citation41]. A piRNA is a small noncoding single-stranded RNA, made up of 23–35 nucleotides, with a monophosphate group at 5′ end, a strong uracil base, and 2′-O-methylation at 3′ end, which is indicative of highly conserved functions in different species [A. Citation36, Citation37, Citation42, Citation43]. The human genome contains over 30,000 piRNA genes [Citation14] that are mainly derived from the intergenic regions [Citation15]. In particular, contrary to other ncRNAs, piRNAs are composed of few numbers of RNA precursors, which are long and single-stranded. Moreover, their transcription from transposons, known as ‘piRNA clusters’, is performed through a Dicer-independent pathway. These clusters are mainly situated in the peri-centromeric and sub-telomeric parts of the chromosomes [Citation44–46].
In accordance with multiple origins, piRNAs are divided into three main types: mRNA-derived, lncRNA-derived, and transposon-derived piRNA [Citation47,Citation48]. Given the presence of some piRNA genes in snoRNAs and tRNAs, these can be regarded as sources of piRNAs as well [Citation49–51]. It is noteworthy that piRNAs have a silencing impact on genes and protein regulation, PIWI-dependent transposon silencing, as well as on epigenetic regulation, germ stem cell maintenance, genome rearrangement, and reproduction and fertility regulation through attaching to PIWI proteins and forming a silencing ribonucleoprotein complex [A. A. Citation52–55, W. Citation56, Citation57, S. Citation58].
1.2. PIWI proteins
PIWI proteins belong to the Argonaute protein family. These proteins attach to small RNAs and are expressed in a wide range of organisms [Citation59,Citation60]. There are four major types of them in humans: PIWI-like protein 1 (PIWIL1 or human PIWI homolog (HIWI)), PIWIL2 (HILI (Hiwi-like)), PIWIL4 (HIWI2), and PIWIL3 (HIWI3)) [Citation61]. These proteins are expressed in different cell types, like follicle cells and germ cells in drosophila ovaries [C. Citation59, Citation62], along with stem cells [Citation63–66], cancer cells [J. H. Citation67, X. Citation68, Citation69], or even adult somatic cells [Citation70, C. Citation71, Citation72]. Nevertheless, it is worth mentioning that PIWI expression in out-of-germline tissues is specific to certain species [Citation73]. Additionally, PIWI proteins’ expression can be observed in a broad spectrum of body organs, such as heart, pancreas, lung, liver, kidney and brain [Citation74,Citation75]. The PIWI proteins comprise three major functional domains that are as follows: PIWI-Argonaute-Zwille (PAZ) domain that acts as binding site of the 3′-end of guide small RNA; the middle domain (MID) that identifies 5′-uridine (1 U-bias) of piRNAs and prescribes transposon orientation in terms of strand bound to piRNA; the PIWI-domain comprises an endonuclease region (RNase-H) which contributes to mRNA cleavage [Citation76,Citation77]. Traditionally, coimmunoprecipitation and subsequent sequencing or RT-PCR of RNAs revealed piRNAs binding to PIWI proteins [Citation78]. Even though these kinds of proteins were initially found to show a crucial role in self-renewal and preservation of germ-line stem-cells (GSC) [Citation55,Citation79], they have been shown to be vital to piRNAs biogenesis and their chief function via transcriptional and post-transcriptional suppression of Transposable Elements (TEs) or transposons in the cytoplasm and nucleus [Citation80].
2. Biogenesis of piRNAs
The recently emerging piRNAs need to be post-transcriptionally processed before they become entirely matured. The biogenesis of mature piRNAs comprises two main steps: the primary pathway and the ‘ping-pong’ amplification mechanism () [Citation41]. The first one occurs in Drosophila’s somatic and germline cells; however, the ‘ping-pong’ cycle was only described in germline cells [Citation7].
Figure 1. The two pathways of piRNA biogenesis: primary and secondary (Ping-Pong cycle) amplification Aub, Aubergine; Ago3, Argonaute-3; PIWI, P-element-induced wimpy testis; TE, Transposable Element
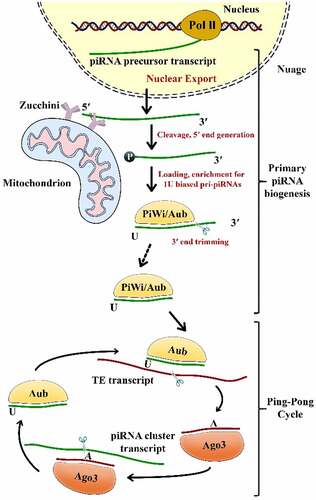
2.1. Primary amplification
In the nucleus, Riboendonuclease Zucchini (Zuc) cleaves the primary transcripts of piRNAs, to form a 5ʹ-phosphate residue*** [Citation81]. Subsequently, PIWI is combined with a 3ʹ fragment of the transcripts and snipped to the ultimate length by a 3ʹ to 5ʹ exonuclease. It is noteworthy that the 2ʹ-hydroxy group is methylated at the 3ʹ end through a small RNA 2ʹ-O-methyltransferase enzyme, known as Hen1**** [Citation82]. It has been shown that there is a substantial bias toward U residues on the part of the PIWI-incorporated 5ʹ-end of the piRNAs. The piRNAs attach PIWI proteins sequentially to form a piRNA/PIWI complex after they are transferred into the cytoplasmic production centers and processed into their final length [Citation83]. Later, the complex is translocated to the nucleus where it inhibits the target gene transcription via a complementary base pairing of piRNAs with DNA. As transcriptional regulators, piRNAs recruit histone methyltransferases, thereby functioning on TE sequences via establishing heterochromatin structures [B. W. Citation44, Citation84].
2.2. Secondary amplification
Upon the formation of a primary piRNA, piRNA accumulation needs to be augmented by the cytoplasmic ‘ping–pong’ mechanism [Citation54]. On the contrary to the primary pathway that correlates with PIWI proteins, piRNAs connect with AUB (Aubergine) or AGO3 proteins in order to form complexes with these proteins with complementary sequences [Citation72]. Given its slicer activity, a primary piRNA-associated AUB present in fly germ cells identifies and cleaves active transposon transcripts. These cleavages form the 5ʹ ends of new piRNAs with a sense of orientation to transposons. Next, AGO3–piRNA complexes identify and cut cluster transcripts so that they could create antisense piRNAs subsequent to load into AGO3 and become mature through trimming or Zuc [B. W. Citation85, Citation86]. The piRNAs are amplified and gathered in the cytoplasm through a ping pong cycle that is dependent on sequence-complementary [Citation41]. Although ping-pong characteristics have been observed in Drosophila melanogaster, zebrafish, and sponges, they were not identified in mice, suggesting that this pathway is present in the early evolution [Citation87].
3. Biological functions of PIWI-piRNAs pathway
On account of the fact that various species have distinct piRNA sequences and functioning, piRNAs biological functions have been recognized to a limited extent. Nevertheless, according to ample evidence, piRNAs play crucial roles in genes and protein regulation, epigenetic, TEs silencing, genome rearrangement, as well as in the maintenance of homeostasis, fertilization, somatic and germline stem cell self-renewal, and embryogenesis [Y. Citation7, Citation88].
3.1. Retrotransposons silencing
TEs comprise a number of genetic units that are capable of moving and propagating within the genome [Citation89]. In spite of TEs’ vital role in improving genetic diversity, they lead to genetic instability via mutations, epigenetic/genetic deregulations, and chromosomes rearrangements [Citation90]. As an innate immune-like system, piRNAs can oppose the negative impacts of TEs on the genome to conserve the genome integrity. DNA methylation and chromatin alterations of CpG islands in germline cells to preserve normal gametogenesis are the mechanisms employed by piRNAs to silence TEs [Citation15]. Additionally, antisense piRNAs can attach to repeat sequences of TEs and lead to their silencing through degradation of mRNAs, which comprise these repeats, and also via translational inhibition [Citation91].
3.2. Genes and proteins regulation
Since piRNAs have a broad array of applications, this group of ncRNAs is recognized for its master gene expression regulatory function both in germinal and somatic cells. piRNA-mediated gene regulation is performed through a number of processes. Epigenetic mechanisms are the first one as they are involved in histone modifications and DNA methylation [Citation92], and the other one is gene regulation mediated by piRNA at the post-transcriptional level, which is conducted through controlling mRNA stability or alternative splicing, plus interaction with RNAs or RNA endonucleolytic splitting [Citation93]. The last point, which is of equal significance, is that interfering RNAs/piRNA-like (iRNAs/piRNA-L) are possibly involved in translational or post-translational level of gene expression via the direct connection with the protein-coding genes in physiological and pathophysiological contexts [Citation94].
Furthermore, the first studies demonstrated that PIWI proteins participate in the regulation of translation. It has been found that at different steps of mouse spermatogenesis, mouse PIWI homolog (MIWI) and Miwi-like (MILI) are associated with the mRNA cap-binding complex, which is essential for the activation of translation [Citation95,Citation96]. In addition, AUB was shown to play a crucial role in translational regulation in germline stem cells of Drosophila ovaries [Citation97]. Also, in Drosophila cultured cells, a link was found between Aub and eIF4 translation initiation factors [X. Citation98].
3.3. Differentiation
During early embryogenesis, PIWI proteins assume great significance in cell differentiation. As the main PIWI protein involved in cell differentiation by its inhibitory effect on the Transforming Growth Factor-β (TGF-β) signaling pathway, PIWIL2 has a direct interaction with Heat Shock Protein 90 (HSP90) and Mothers against decapentaplegic homolog 4 (Smad4) and impedes the formation of HSP90 – TGF-β Receptor (TβR) complex; thereby the TGF-β signaling pathway is inhibited. Moreover, PIWIL2 contributes to TβR and Smad degradation through its involvement in the upregulation of TβR ubiquitination as well as degradation by the ubiquitin E3 ligase Smurf2. Likewise, PIWI proteins assist in the differentiation of germline stem cells through post-transcriptional-level suppression of c-Fos proto-oncogene. Hence, these proteins improve piRNAs’ synthesis from 3′ UTR region of c-Fos mRNA, leading to instability of c-Fos mRNA and translation repression [Citation99, Citation100, J. C. Citation19, Citation101].
3.4. Cell survival
Activation of numerous pro-survival molecules is enhanced by the PIWI-piRNA axis. Also, through upregulation of the anti-apoptotic molecule Fibroblast Growth Factor 8 (FGF8) expression and downregulation of pro-apoptotic p21 and Bax expression,PIWIL1 induces cell survival. Additionally, PIWIL2 mainly regulates p53 by directly interacting with c-Src and STAT3 as well as improving a PIWIL2/STAT3/c-Src complex. It would repress p53 phosphorylation and inhibit Fas-mediated apoptosis. The STAT3 activation, induced by PIWIL2, was also reported to lead to the up-regulation of the anti-apoptotic Bcl-XL [X. ZhaCitation102].
3.5. Fertilization and development
In Drosophila and murine GSCs, the PIWI-piRNA pathway is largely engaged in germline cell biology, which includes differentiation, maintenance, and function of these cells. Given their transgenerational epigenetic effects, piRNAs result in epigenetic activation of gene expression through euchromatin induction by activating H3K4me3 and inhibiting H3K27me3 in sub-telomeric heterochromatin [Citation79,Citation99–101,Citation103,Citation104].
The embryonic development, such as nuclear division, cell cycle development, chromatin organization, control of mRNA translation, chromosome integrity in pro-survival molecules during mitosis, and embryonic sex determination are the processes in which the PIWI-piRNA axis is implicated [Citation105–107, H. Citation108]. During mammalian spermatogenesis and oogenesis, along with early embryogenesis, organogenesis, and post-natal, the spatial-temporal regulation and activation of piRNAs and PIWI proteins assume utmost importance [S. Citation58]. The regulation of PIWIL2 is performed in germline cells and contributes to genome stability maintenance by preventing gene dysregulation, chromosome rearrangements, oncogenic mutations, and TEs propagation [D.-T. Citation109].
3.6. Mitochondrial functions
piRNAs also have mitochondrial expression, wherein they originate from a sequence of 12S rRNA, tRNA, 16S rRNA, and protein-coding genes of mitochondria, including NADH-ubiquinone oxidoreductase chain 4 L (ND4L), CycloOxygenase2 (COX2), and NADH-ubiquinone oxidoreductase chain 5 (ND5) [Citation110]. It shows that nuclear and mitochondrial piRNA transcripts have a potential cross-talk, and mitochondrial (mt)-piRNAs are involved in cell responses to oxidative stress and bioenergetics. Regarding the fact that mitochondria is an organelle involved in different cellular key processes like apoptosis and bioenergetics, functional studies are of the utmost significance to better comprehend the piRNAs role in mitochondrial epigenetics and how it affects the development of health and diseases [Citation111].
3.7. Somatic functions
At the somatic level, it was reported that piRNAs are expressed in body fluids and somatic cells [Citation112–115]. Nevertheless, their functions in the soma are still unknown because there is not enough knowledge on functional assays of piRNAs. A variety of studies have indicated the likelihood of piRNAs involvement in genes’ epigenetic regulation associated with neuronal activity, neurogenesis, plasticity [E. J. Citation116–118], and that pseudogenes-derived piRNAs may contribute to the regulation of parental genes [Citation119]. Antisense piRNAs were also found to lead to regulation of immune response as well as self-tolerance genes [Citation89]. In addition, it has been reported that sno-piRNAs exist in primary CD4 + T-lymphocytes and regulate interleukin-4 (IL-4) levels, thereby hindering their differentiation into T-helper 2 (Th2) T-cells [Citation51].
3.8. Autophagic regulation
The regulatory role of piRNAs in cell cycle, apoptosis, and proliferation is becoming increasingly evident. Phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian Target Of Rapamycin (mTOR) is a classical pathway that serves as a signal transduction pathway and plays a vital biological role in different cellular processes, including autophagy [Citation120]. However, the exact role of piRNAs in autophagy needs further investigations since very limited related publications are available.
4. The roles of PIWI-piRNAs pathway in cancer
4.1. piRNAs and cancer
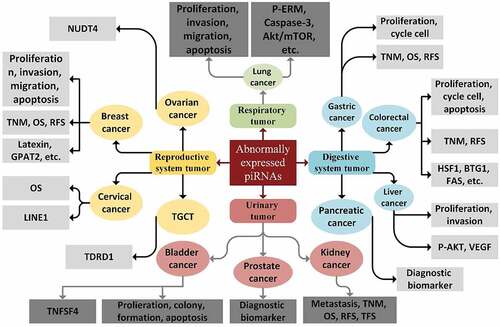
Given our insufficient knowledge of how piRNAs function, they have long been considered as ‘dark matter’ of ncRNAs in the genome. Nevertheless, the current document has altered our perception of piRNAs as vital clinical and biological agents in different diseases [Citation41]. Likewise, based on recent studies, aberrant piRNAs expression might be introduced as a particular signature of cancer and could have a correlation with clinical characteristics of tumor tissues, suggesting a significant role of these molecules in a wide variety of cancers, such as colorectal, prostate, gastric, bladder, breast, lung, and hepatic cancers, as well as malignant melanoma () [Citation14,Citation121,Citation122]. Dysregulated expression of piRNAs suggests that they could be considered as tumor suppressors or oncogenes in tumorigenesis (depending on the function of their targeted gene/mRNA/protein), and as such can act as crucial regulators of the major characteristics of cancer, including cancer cell differentiation, proliferation, progression, and metastasis [Citation93,Citation123].
Table 2. Cancer-associated piRNAs
The functions of piRNA pathways are shown to be associated with several processes that occur in cancer, such as genomic instability, aneuploidy, DNA methylation, cell cycle progression, repetitious expression, and cell metabolism [Citation17, Citation121, Y. Citation139]. piRNA pathway genes have been reported to be linked to the regulation of genes that are implicated in the establishment of stem cell features [L. Citation140, Y. Citation139]. depicts a trace of the piRNAs in a variety of cancer types [Citation93].
RT-qPCR analyses, next-generation sequencing and microarray screening have revealed the association between piRNAs and carcinogenesis [J. Citation141, Citation142]. As one of the most prominent discoveries, piRNAs can influence two of the most classical pathways with involvement in cancer progression, PI3K/PTEN/Akt/mTOR and Ras/Raf/MEK/ERK pathways [Citation143]. These pathways are crucial in the regulation of a wide variety of genes, and their stimulation can enhance metabolism, growth, and survival of cancer cells [Citation144]. Therefore, the identification of piRNAs, which are related to cancer and functional analysis of their target genes, could be capitalized on in preventive and therapeutic strategies in cancer [J. Citation141].
4.2. piRNAs as potential biomarkers and therapeutic tools
Cancers often become symptomatic when they have already spread, and this makes the treatments and interventions less effective. An effective screening biomarker must be able to distinguish the cancer at an initial stage to decrease the morbidity rate and increase the survival chances. As shows, and in accordance with previous findings, it has been identified that aberrant piRNAs expression is associated with different characteristics of cancer patients. piRNAs are early products of different regulatory and signaling pathways. So, at least theoretically, they might be very important for early detection and treatment of cancers [Citation41,Citation121,Citation145]. With a tissue-specific expression profile, piRNAs are considered as promising tissue-based diagnostic and prognostic biomarkers for cancer [Citation146]. In addition, piRNAs are also noninvasive biomarkers that are present in exosomes or as free-circulating in serum, plasma, saliva, and stool [L. Citation147–151, Z. Citation152, X. Citation153]. piRNAs can be detected using microarray, sequencing, and qRT-PCR [Citation78,Citation146]. Alterations in circulating piRNA expression levels might be good cancer biomarkers, with higher specificity and sensitivity compared to circulating miRNAs and lncRNA-based biomarkers [L. Citation147]. It should be noted that no enzyme is needed for piRNA processing in the synthesis pathway. Therefore, it is beneficial to utilize synthetic piRNAs with better specificity of targets [Citation121]. Fortunately, these small RNAs are capable of passing through the cell membrane, without degradation in the circulation and remain stable in human serum and plasma samples even after frequent freeze-thaw cycles or long-term incubation at room temperature [Citation115,Citation154]. This requires identifying novel piRNA biomarkers for cancer diagnosis at an early stage and their validation in independent studies with larger sample sizes and long-term clinical trials. Therefore, the combination of circulating miRNAs and piRNAs could increase its specificity and sensitivity cancer diagnosis and provide new approaches to develop a particular panel for early diagnosis and prognosis [Citation146].
Table 3. Clinical applications of piRNAs in cancer
Table 4. Cancer-related PIWI Proteins
Despite the fact that we know little about the molecular mechanisms of piRNAs functioning in tumorigenesis, some of their characteristics would make them ideal targets for therapeutic interventions. As an example, the piRNAs’ role in DNA methylation, which results in gene silencing at the transcriptional level, is applicable in inhibiting the expression of certain oncogenes, or their functions in post-transcriptionally RNA degradation could have therapeutic advantages. Moreover, gene silencing that is mediated by RNA interference (RNAi) utilized for cancer treatment could contribute to selective silencing of oncogenic piRNAs [Citation123]. Nevertheless, the efficiency of piRNAs as therapeutic targets is dependent on effective in-vivo delivery strategies, and prior to any application of piRNAs for therapeutic purposes, it is required that in-depth studies be conducted [Citation155].
A number of different studies have been documented the clinical significance of piRNAs as potential biomarkers and therapeutic tools in the most common cancers. Recent studies revealed that overexpression of piR-823 in gastric cancer cells led to suppress growth of tumor, suggesting it may be considered as a promising therapeutic agent [J. Citation156]. In addition, serum level of piR-823 and piR-651 were significantly lower in gastric cancer patients compared to normal, highlighting its clinical value as a diagnostic biomarker [H. Citation157]. In lung cancer, as the most common cause of cancer death, abnormal expression of piRNAs was associated with the growth of lung cancer cells [Citation158]. In breast cancer, the most common cancer among females, it was revealed that tumor tissues express eight piRNAs that are believed to be independent prognostic biomarkers (piR-34,736, piR-31,106, piR-36,249, piR-36,026, piR-36,318, piR-34,377, piR-35,407, and piR-36,743) [Citation41]. Prognostic biomarkers of breast cancer are of significant importance, since local breast cancer has a 5-year survival rate of at least 90%; however, this rate declines to 60% in metastatic patients if it is regional and 30% if distant [Citation159]. Interestingly, a cohort study documented the correlation of these breast cancer prognostic biomarkers with the survival rate. However, comparing sensitivity, specificity, and practicality of piRNAs with traditional tumor markers is a neglected area in this field.
The clinical relevance of piRNAs with cancer is not limited to the above-mentioned examples. As it is shown in , several studies reported other applications of piRNAs in the common cancers such as colorectal cancer, multiple myeloma, clear cell renal cancer, etc. Despite that, clinical researches about applications of piRNAs in targeted therapy are limited. Also, the mechanisms by which piRNA expression is altered in several cancers have not been studied yet.
4.3. PIWI proteins and cancer
Investigations about piRNAs have mainly been focused at the transcriptional and post-transcriptional levels, and only few researches have studied piRNA function at the post-translational level. It is noteworthy that in addition to clinicopathologic analysis, both in-vivo and in-vitro functional research has recognized all of the four human PIWI proteins as novel molecular agents involved in carcinogenesis [Citation174]. The expression of PIWI in cancer was, for the first time, reported in seminoma [Citation175]. Other reports on different cancers also revealed that PIWI protein expression profiles had been potentially and functionally associated with a broad range of human cancers, with both somatic and germline origin. Moreover, they have correlations with poor clinical outcomes and aggressive cancers () [Citation41,Citation91,Citation123]. Hence, these proteins were shown to be involved in the proliferation of cancer cells, metastasis, invasion, apoptosis, migration, division, and survival. As found by immunohistochemical and western blot analysis of PIWI protein expression in tissue specimens from cancer patients, PIWI proteins could be proposed as promising biomarkers for cancer prognosis and diagnosis [Y. Citation45, Citation88, Citation176, Y. Citation177].
Since the expression of PIWI proteins is not associated with the presence of large amounts of piRNAs, the mechanisms of their actions in cancers are still debated [Citation178]. In addition, it has been reported that PIWI proteins have independent functions of piRNAs in cancer progression and metastasis. Previous studies revealed the independent function of PIWIL1 in pancreatic [F. Citation179] and gastric cancer [S. Citation180] metastasis. In mouse spermatogenesis, it has also been described that MIWI exerts an independent role of piRNAs in protein regulation [Citation181]. Whether PIWI proteins and piRNAs affect the cancer cells independently, or they act on cancer cells together, remains a challenging issue yet to be addressed.
5. Database for piRNAs and functional predictions
The rapid increase in studies conducted on piRNAs has allowed for the generation of several databases, such as piRNA Bank (http://pirnabank.ibab. ac.in/) and piRBase (http://www.regulatoryrna.org/database/piRNA/) for the analysis of piRNA function, homologous piRNAs, piRNA clusters, and anticipation of the targeted RNAs [Y. Citation203].
Considered as the first piRNA database and a web resource about the grouped and classified piRNAs, the piRNA Bank was established by Lakshmi et al. group [Citation204]. The database presents extensive information about 20 million identified sequences and other relevant data on reported piRNAs in human, mouse, rat, and Drosophila [Citation41]. This above-mentioned database supports extensive search features of the organism and chromosome such as sequence homology-based search, accession numbers, name or symbol of the gene, localization on chromosomes, clusters, and corresponding repeat elements and genes. It also represents each piRNA or piRNA cluster as a graphical map of genomes (http://pirnabank.ibab.ac.in/) [Y.-N. Citation205]. However, it should be noted that the above-mentioned database contains only restricted amounts of piRNAs obtained from some species, and the data on the piRNA functions are rarely documented [Citation206].
piRBase is a recently produced and special piRNA database included in RNAcentral, [K. P. Citation207] and is the primary database with a systematic integration of different piRNA-related data to sustain functional analysis of piRNA. In the latest release of piRBase, the unique piRNA sequences exceeded 173 million, which include 21 species*** [Citation208]. Also, the intended mRNA records of piRNA were extended and the piRNA target lncRNAs were included*** [Citation209]. The data concerning eight piRNAs associated cancer types such as breast, pancreas, colorectal, gastric, bladder, myeloma, kidney, and liver cancer were also appended to the recent version. It should be noted that the released piRBase v2.0 (http://www.regulatoryrna.org/database/piRNA/) introduces new web tools and enhances user interface [Citation206].
More recently, Junyi Xin et al. constructed a user-friendly database, piRNA – expression quantitative trait locus (eQTL) (http://njmu-edu.cn:3838/piRNAeQTL/, which is also available at http://222.190.246. 206:3838/piRNA-eQTL/), using R package Shiny. As the first online database that provides cis-piRNA eQTL results via mixing genotype and piRNA expression data across 33 cancer types, it can act as a central source to find the roles of piRNAs and genetic variants in human cancer development [Citation210].
6. Conclusion and future extension
piRNAs have currently been revealed to display abnormal expression in a cancer-specific manner in diverse types of cancers. PIWI proteins and piRNAs could be valid prognostic or diagnostic biomarkers in targeted therapies. As a biomarker candidate, piRNA needs to be validated by multiple centers in multiple independent, preferably prospective cohorts with large sample size, and demonstrate benefit over any existing markers. In the future, more studies and clinical trials are required to thoroughly comprehend the underlying biological mechanisms of piRNAs and their interruption. Given the continuous attempts of scientists as well as the advent of modern technologies, effective perspectives research should evaluate the association of piRNAs and cancers, which leads to an elevated piRNAs knowledge to hopefully improve cancer prevention or treatment possibilities for patients.
In spite of the importance of revealing their precise roles in cancer, there are indeed a number of unanswered questions, like whether the piRNAs’ abnormal expression explains these types of cancers, or it is a byproduct of other molecular activities. Is there a suitable threshold to differentiate healthy people from patients with specific cancer? Another question is whether metastasis, apoptosis, invasion, and proliferation of cancer cells are independently affected by piRNAs and PIWI proteins, or whether PIWI proteins and piRNAs together impact cancer cells. Likewise, as the abnormal expression of piRNA pathway could provoke stemness, analyzing the association of piRNAs and PIWI proteins with cancer stem cells is likely to establish a new course in future research about the origin of cancer. These ideas mount challenges that should be addressed before using piRNA-based treatments.
Furthermore, since piRNAs involved in cancers were clarified vastly in recent years, an unbelievable amount of data will be produced in the near future. Therefore, piRNA–cancer relationship is able to provide promising insights into piRNAs functional relationship in wide range of cancers. Novel and unique database resource will lead toward further research ideas in the field of cancer. Moreover, it is beneficial to utilize synthetic piRNAs with better specificity of targets via some piRNA target web-based prediction database resource or software based on some innovative algorithms to provide detailed information about piRNAs’ role in various cancers.
Abbreviations
piRNAs: PIWI-interacting RNAs; PIWI: P-element Induced Wimpy testis; lncRNAs: long non-coding RNAs; ncRNAs: non-coding RNAs; circRNAs: circular RNAs; miRNAs: microRNAs; mRNAs: messenger RNAs; tRNAs: transfer RNAs; siRNAs: small interfering RNAs; rRNAs: ribosomal RNAs; scRNAs: small cytoplasmic RNAs; snRNAs: small nuclear RNAs; snoRNAs: small nucleolar RNAs; AGO: Argonaute; rasiRNAs: repeat-associated small interfering RNAs; UTRs: UnTranslated Regions; GSC: Germ-line Stem-Cells; PIWIL1: PIWI-Like protein 1; PIWIL2: PIWI-Like protein 2; PIWIL3: PIWI-Like protein 3; PIWIL4: PIWI-Like protein 4; HIWI: Human PIWI homolog; HILI: Hiwi-Like; PAZ: PIWI-Argonaute-Zwille; MID: Middle Domain; TEs: Transposable Elements; Zuc: Zucchini; AUB: Aubergine; MIWI: Mouse PIWI homolog; MILI: MIwi-Like; iRNAs/piRNA-L: interfering RNAs/piRNA-Like; eIF4: eukaryotic Initiation Factor 4; TGF-β: Transforming Growth Factor-β; HSP90: Heat Shock Protein90; Smad4: Mothers against decapentaplegic homolog; TβR: TGF-β Receptor; FGF8: Fibroblast Growth Factor 8; STAT3: Signal Transducer and Activator; ND4L: NADH-ubiquinone oxidoreductase chain 4L; COX2: CycloOXygenase 2; ND5: NADH-ubiquinone oxidoreductase chain 5; mt-piRNAs: mitochondrial piRNAs; IL-4: Interleukin-4; Th2: T helper 2; PI3K: Phosphatidyl-Inositol 3-Kinase; mTOR: mammalian Target Of Rapamycin; PTEN: Phosphatase and TENsin homolog; MEK: Mitogen-activated protein kinase; ERK: Extracellular signal-Regulated Kinase; RNAi: RNA interference; cfRNAs: circulating free RNAs; eQTL: expression Quantitative Trait Locu
Highlights
As early products of different regulatory and signaling pathways, piRNAs might be very important for early detection and treatment of cancers.
Alterations in circulating piRNA expression levels might be good cancer biomarkers, with higher specificity and sensitivity compared to circulating miRNAs and lncRNA-based biomarkers.
All of the four human PIWI proteins have been recognized as novel molecular agents involved in carcinogenesis.
Authors’ contributions
All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
Data sharing is not applicable to this article as no new data are created or analyzed in this study.
Additional information
Funding
References
- Amaral PP, Dinger ME, Mercer TR, et al. The eukaryotic genome as an RNA machine. science. 2008;319(5871):1787–1789.
- Cui D-Y, Wang -J-J, He S-M. The research advance of the piRNA function. Sheng li ke xue jin zhan [Progress in physiology]. 2016;47(3):182–186.
- Zhu X, Jiang S, Wu Z, et al. Long non-coding RNA TTN antisense RNA 1 facilitates hepatocellular carcinoma progression via regulating miR-139-5p/SPOCK1 axis. Bioengineered. 2021;12(1):578–588.
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell. 2005;120(1):15–20.
- Shi Z, Shen C, Yu C, et al. Long non-coding RNA LINC00997 silencing inhibits the progression and metastasis of colorectal cancer by sponging miR-512-3p. Bioengineered. 2021;12(1):627–639.
- Meseure D, Alsibai KD. Part 1: The PIWI-piRNA pathway is an immune-like surveillance process that controls genome integrity by silencing transposable elements. In: Colin Logie and Tobias Aurelius Knoch (eds) Chromatin and Epigenetics. IntechOpen; 2018:233-251. London, UK: IntechOpen.
- Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214.
- Zhang Y-P, Cheng Y-B, Li S, et al. An epithelial-mesenchymal transition-related long non-coding RNA signature to predict overall survival and immune microenvironment in kidney renal clear cell carcinoma. Bioengineered. 2021;12(1):555–564.
- Zhao C, Tolkach Y, Schmidt D, et al. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma. World J Urol. 2019;37(8):1639–1647.
- Farazi TA, Juranek SA, Tuschl T (2008a). The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members.
- Hamilton AJ, Baulcombe Dc. A species of small antisense RNA in posttranscriptional gene silencing in plants. science. 1999;286(5441):950–952.
- Leung YY, Kuksa PP, Amlie-Wolf A, et al. DASHR: database of small human noncoding RNAs. Nucleic Acids Res. 2016;44(D1)(D1):D216–D222.
- Chalbatani GM, Dana H, Memari F, et al. Biological function and molecular mechanism of piRNA in cancer. Pract Lab Med. 2019;13:e00113.
- Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem. 2015;84(1):405–433.
- Kang J, Huang X, Dong W, et al. MicroRNA-1269b inhibits gastric cancer development through regulating methyltransferase-like 3 (METTL3). Bioengineered. 2021;12(1):1150–1160.
- Ng KW, Anderson C, Marshall EA, et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol Cancer. 2016;15(1):1–13.
- Nozawa M, Kinjo S (2016). Noncoding RNAs, origin and evolution of.
- Peng JC, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr Opin Cell Biol. 2013;25(2):190–194.
- Rayford KJ, Cooley A, Rumph JT, et al. piRNAs as modulators of disease pathogenesis. Int J Mol Sci. 2021;22(5):2373.
- Wang L, Ge S, Zhou F. MicroRNA-487a-3p inhibits the growth and invasiveness of oral squamous cell carcinoma by targeting PPM1A. Bioengineered. 2021;12(1):937–947.
- Xu J, Yang X, Zhou Q, et al. Biological significance of piRNA in liver cancer: a review. Biomarkers. 2020;25(6):436–440.
- Höck J, Weinmann L, Ender C, et al. Proteomic and functional analysis of Argonaute‐containing mRNA–protein complexes in human cells. EMBO Rep. 2007;8(11):1052–1060.
- Fitzmaurice C, Dicker D, Pain A, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527.
- Ashktorab H, Brim H. Blood-based liquid biopsies: a noninvasive and cost-effective tool for improved risk assessment and identification of lymph node metastasis in patients with submucosal T1 colorectal cancer. Gastroenterology. 2021;161(1):29–31.
- Aziz N, Hong YH, Jo M, et al. Molecular signatures of JMJD10/MINA53 in gastric cancer. Cancers (Basel). 2020;12(5):5.
- Brim H, Ashktorab H. Integrating microbiomics in cancer management. Nat Rev Cancer. 2021;21(11):684–685.
- Deng S, Zhang X, Qin Y, et al. miRNA‐192 and‐215 activate Wnt/β‐catenin signaling pathway in gastric cancer via APC. J Cell Physiol. 2020;235(9):6218–6229.
- Erfani M, Hosseini SV, Mokhtari M, et al. Altered ARID1A expression in colorectal cancer. BMC Cancer. 2020;20(1):1–13.
- Igder S, Mohammadiasl J, Azadpour S, et al. KRAS mutation and abnormal expression of Cripto‐1 as two potential candidate biomarkers for detection of colorectal cancer development. J Cell Biochem. 2020;121(4):2901–2908.
- Zhang X, Peng Y, Yuan Y, et al. Histone methyltransferase SET8 is regulated by miR-192/215 and induces oncogene-induced senescence via p53-dependent DNA damage in human gastric carcinoma cells. Cell Death Dis. 2020;11(10):937.
- Igder S, Mohammadiasl J, Mokarram P. Altered miR-21, miRNA-148a expression in relation to KRAS mutation status as indicator of adenoma-carcinoma transitional pattern in colorectal adenoma and carcinoma lesions. Biochem Genet. 2019;57(6):767–780.
- Poursheikhani A, Bahmanpour Z, Razmara E, et al. Non-coding RNAs underlying chemoresistance in gastric cancer. Cell Oncol. 2020;43:961–988.
- Aravin AA, Lagos-Quintana M, Yalcin A, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5(2):337–350.
- Aravin AA, Naumova NM, Tulin AV, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11(13):1017–1027.
- Vagin VV, Sigova A, Li C, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. science. 2006;313(5785):320–324.
- Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207.
- Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20(15):1993–1997.
- Williams Z, Morozov P, Mihailovic A, et al. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 2015;13(4):854–863.
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008b;135(7):1201–1214.
- Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: emerging biological concepts and potential clinical implications. Biochim Biophys Acta (BBA)- Rev Cancer. 2019;1871(1):160–169.
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14(4):347–348.
- Saito K, Sakaguchi Y, Suzuki T, et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21(13):1603–1608.
- Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14(8):523–534.
- Suzuki R, Honda S, Kirino Y. PIWI expression and function in cancer. Front Genet. 2012;3:204.
- Yamanaka S, Siomi MC, Siomi H. piRNA clusters and open chromatin structure. Mob DNA. 2014;5(1):1–12.
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annual Review of Cell and Developmental. 2009;25(1):355–376.
- Weick E-M, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141(18):3458–3471.
- Keam SP, Young PE, McCorkindale AL, et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014;42(14):8984–8995.
- Krishnan P, Ghosh S, Wang B, et al. Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci Rep. 2016;6(1):32843.
- Zhong F, Zhou N, Wu K, et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43(21):10474–10491.
- Aravin AA, Sachidanandam R, Girard A, et al. Developmentally regulated piRNA clusters implicate MILI in transposon control. science. 2007;316(5825):744–747.
- Batista PJ, Ruby JG, Claycomb JM, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31(1):67–78.
- Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. cell. 2007;128(6):1089–1103.
- Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–514.
- Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2(6):819–830.
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129(4):1119–1136.
- Zhao S, Gou L-T, Zhang M, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24(1):13–25.
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. cell. 2009;136(4):656–668.
- Siomi MC, Sato K, Pezic D, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258.
- Sasaki T, Shiohama A, Minoshima S, et al. Identification of eight members of the Argonaute family in the human genome☆. Genomics. 2003;82(3):323–330.
- Li C, Vagin VV, Lee S, et al. Without Argonaute3, Aubergine-bound piRNAs collapse but Piwi-bound piRNAs persist. cell. 2009b;137(3):509.
- Alié A, Leclère L, Jager M, et al. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev Biol. 2011;350(1):183–197.
- Rinkevich Y, Rosner A, Rabinowitz C, et al. Piwi positive cells that line the vasculature epithelium, underlie whole body regeneration in a basal chordate. Dev Biol. 2010;345(1):94–104.
- Rinkevich Y, Voskoboynik A, Rosner A, et al. Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev Cell. 2013;24(1):76–88.
- Sharma AK, Nelson MC, Brandt JE, et al. Human CD34+ stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood J Am Soc Hematol. 2001;97(2):426–434.
- Lee JH, Schütte D, Wulf G, et al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15(2):201–211.
- Liu X, Sun Y, Guo J, et al. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer. 2006;118(8):1922–1929.
- Miller F, Pozniak C, Walsh G. Neuronal life and death: an essential role for the p53 family. Cell Death Differ. 2000;7(10):880–888.
- Brower-Toland B, Findley SD, Jiang L, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21(18):2300–2311.
- Li C, Vagin VV, Lee S, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. cell. 2009a;137(3):509–521.
- Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–359.
- Lenart P, Novak J, Bienertova-Vasku J. PIWI-piRNA pathway: setting the pace of aging by reducing DNA damage. Mech Ageing Dev. 2018;173:29–38.
- Henaoui IS, Jacovetti C, Mollet IG, et al. PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia. 2017;60(10):1977–1986.
- Nandi S, Chandramohan D, Fioriti L, et al. (2016). Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proceedings of the National Academy of Sciences,USA, 113( 45), 12697–12702.
- Bamezai S, Rawat VP, Buske C. Concise review: the Piwi‐piRNA axis: pivotal beyond transposon silencing. Stem Cells. 2012;30(12):2603–2611.
- Cora E, Pandey RR, Xiol J, et al. The MID-PIWI module of Piwi proteins specifies nucleotide-and strand-biases of piRNAs. Rna. 2014;20(6):773–781.
- Krishnan P, Damaraju S. The challenges and opportunities in the clinical application of noncoding RNAs: the road map for miRNAs and piRNAs in cancer diagnostics and prognostics. Int J Genomics. 2018;2018:1–18.
- Cox DN, Chao A, Baker J, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727.
- Völler D, Linck L, Bruckmann A, et al. Argonaute family protein expression in normal tissue and cancer entities. PLoS One. 2016;11(8):e0161165.
- Malone CD, Brennecke J, Dus M, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. cell. 2009;137(3):522–535.
- Malone CD, Brennecke J, Dus M, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. cell. 2009;137(3):522–535.
- Bernstein BE, Meer A, Lander ES. The mammalian epigenome. cell. 2007;128(4):669–681.
- Han BW, Zamore PD. PiRNAs. Curr Biol. 2014;24(16):R730–R733.
- Han BW, Wang W, Li C, et al. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. science. 2015;348(6236):817–821.
- Tsaytler P, Harding HP, Ron D, et al. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. science. 2011;332(6025):91–94.
- Grimson A, Srivastava M, Fahey B, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197.
- Cheng Y, Wang Q, Jiang W, et al. Emerging roles of piRNAs in cancer: challenges and prospects. Aging (Albany NY). 2019;11(21):9932.
- Cichocki F, Lenvik T, Sharma N, et al. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185(4):2010–2012.
- Kazazian HH. Mobile elements: drivers of genome evolution. science. 2004;303(5664):1626–1632.
- Moyano M, Stefani G. piRNA involvement in genome stability and human cancer. J Hematol Oncol. 2015;8(1):38.
- Wei JW, Huang K, Yang C, et al. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9.
- Yu Y, Xiao J, Hann SS. <p>The emerging roles of PIWI-interacting RNA in human cancers. <![cdata[cancer Management and Research]]>. 2019;11:5895.
- Mei Y, Wang Y, Kumari P, et al. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat Commun. 2015;6(1):7316.
- Grivna ST, Pyhtila B, Lin H (2006). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences, 103( 36), 13415–13420.
- Unhavaithaya Y, Hao Y, Beyret E, et al. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284(10):6507–6519.
- Rojas‐Ríos P, Chartier A, Pierson S, et al. Aubergine and pi RNA s promote germline stem cell self‐renewal by repressing the proto‐oncogene Cbl. EMBO J. 2017;36(21):3194–3211.
- Ma X, Zhu X, Han Y, et al. Aubergine controls germline stem cell self-renewal and progeny differentiation via distinct mechanisms. Dev Cell. 2017;41(2):157–169. e155.
- Ashe A, Sapetschnig A, Weick E-M, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. cell. 2012;150(1):88–99.
- Pek JW, Kai T. Non-coding RNAs enter mitosis: functions, conservation and implications. Cell Div. 2011;6(1):1–6.
- Szakmary A, Cox DN, Wang Z, et al. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15(2):171–178.
- Zhang X, He X, Liu C, et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. 2016;196(4):1591–1603.
- Shirayama M, Seth M, Lee H-C, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. cell. 2012;150(1):65–77.
- Stuwe E, Toth KF, Aravin AA. Small but sturdy: small RNAs in cellular memory and epigenetics. Genes Dev. 2014;28(5):423–431.
- Kawaoka S, Izumi N, Katsuma S, et al. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43(6):1015–1022.
- Khurana JS, Xu J, Weng Z, et al. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6(12):e1001246.
- Schwager EE, Meng Y, Extavour CG. vasa and piwi are required for mitotic integrity in early embryogenesis in the spider Parasteatoda tepidariorum. Dev Biol. 2015;402(2):276–290.
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–308.
- Yin D-T, Wang Q, Chen L, et al. Germline stem cell gene PIWIL2 mediates DNA repair through relaxation of chromatin. PLoS One. 2011;6(11):e27154.
- Kwon C, Tak H, Rho M, et al. Detection of PIWI and piRNAs in the mitochondria of mammalian cancer cells. Biochem Biophys Res Commun. 2014;446(1):218–223.
- Fonseca Cabral G, Azevedo Dos Santos Pinheiro J, Vidal AF, et al. piRNAs in gastric cancer: a new approach towards translational research. Int J Mol Sci. 2020;21(6):2126.
- Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–230.
- Roy J, Sarkar A, Parida S, et al. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol Biosyst. 2017;13(3):565–576.
- Wang S, Wang Z, Tao R, et al. The potential use of Piwi-interacting RNA biomarkers in forensic body fluid identification: a proof-of-principle study. Forensic Sci Int Genet. 2019;39:129–135.
- Yang X, Cheng Y, Lu Q, et al. Detection of stably expressed piRNAs in human blood. Int J Clin Exp Med. 2015;8(8):13353.
- Lee EJ, Banerjee S, Zhou H, et al. Identification of piRNAs in the central nervous system. Rna. 2011;17(6):1090–1099.
- Rajasethupathy P, Antonov I, Sheridan R, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. cell. 2012;149(3):693–707.
- Roque CG, Hengst U. Wimpy nerves: piRNA pathway curbs axon regrowth after injury. Neuron. 2018;97(3):477–478.
- Pantano L, Jodar M, Bak M, et al. The small RNA content of human sperm reveals pseudogene-derived piRNAs complementary to protein-coding genes. Rna. 2015;21(6):1085–1095.
- Zeng Q, Wan H, Zhao S, et al. Role of PIWI‐interacting RNAs on cell survival: proliferation, apoptosis, and cycle. IUBMB Life. 2020;72(9):1870–1878.
- Meseure D, Alsibai KD. Part 2: Deregulated expressions of PIWI proteins and piRNAs as new candidate biomarkers and potential therapeutic tools in cancer. In: Colin Logie and Tobias Aurelius Knoch (eds)., Chromatin and epigenetics. London, United Kingdom: IntechOpen; 2020:263-274.
- Siddiqi S, Matushansky I. Piwis and piwi‐interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113(2):373–380.
- Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336(1):46–52.
- Lim SL, Ricciardelli C, Oehler MK, et al. Overexpression of piRNA pathway genes in epithelial ovarian cancer. PLoS One. 2014;9(6):e99687.
- Yan H, Wu Q, Sun C, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29(1):196–206.
- Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5(1):1–17.
- Tan L, Mai D, Zhang B, et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18(1):1–15.
- Ding X, Li Y, Lü J, et al. piRNA-823 is involved in cancer stem cell regulation through altering DNA methylation in association with luminal breast cancer. Front Cell Dev Biol. 2021;9. DOI:10.3389/fcell.2021.641052
- Weng W, Liu N, Toiyama Y, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018;17(1):16.
- Jacobs DI, Qin Q, Lerro MC, et al. PIWI-interacting RNAs in gliomagenesis: evidence from post-GWAS and functional analyses. Cancer Epidemiol Prev Biomarkers. 2016;25(7):1073–1080.
- Li Y, Wu X, Gao H, et al. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med. 2015;21(1):381–388.
- Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356(2):561–567.
- Xiao Z, Shen J, Zhang L, et al. Therapeutic targeting of noncoding RNAs in hepatocellular carcinoma: recent progress and future prospects. Oncol Lett. 2018;15(3):3395–3402.
- Tang X, Xie X, Wang X, et al. The combination of piR-823 and eukaryotic initiation factor 3 B (EIF3B) activates hepatic stellate cells via upregulating TGF-β1 in liver fibrogenesis. Med Sci Monit. 2018;24:9151.
- Zhang L, Meng X, Li D, et al. piR-001773 and piR-017184 promote prostate cancer progression by interacting with PCDH9. Cell Signal. 2020;76:109780.
- Qi T, Cao H, Sun H, et al. piR-19166 inhibits migration and metastasis through CTTN/MMPs pathway in prostate carcinoma. Aging (Albany NY). 2020;12(18):18209.
- Ma H, Wang H, Tian F, et al. PIWI-interacting RNA-004800 is regulated by S1P receptor signaling pathway to keep myeloma cell survival. Front Oncol. 2020;10:438.
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734.
- Tan Y, Liu L, Liao M, et al. Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin (Shanghai). 2015;47(5):315–324.
- Liu L, Dai Y, Chen J, et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial‐mesenchymal transition by way of Akt/GSK‐3β/Snail signaling. Hepatology. 2014;59(2):531–543.
- Cheng J, Guo J-M, Xiao B-X, et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412(17–18):1621–1625.
- Huang G-L, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15(7):563–568.
- Law PT-Y, Qin H, Ching A-K-K, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58(6):1165–1173.
- Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: Structural and pharmacological perspectives. Eur J Med Chem. 2016;109:314–341.
- Wu X, Pan Y, Fang Y, et al. The biogenesis and functions of piRNAs in human diseases. In Molecular Therapy-Nucleic Acids. 2020a;21:108-120. USA: Elsevier.
- Jeong H, Park K-H, Lee Y, et al. The regulation and role of piRNAs and PIWI proteins in cancer. Processes. 2021;9(7):1208.
- Cui L, Lou Y, Zhang X, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44(13):1050–1057.
- Gu X, Wang C, Deng H, et al. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim Biophys Sin (Shanghai). 2020;52(5):475–484.
- Iliev R, Fedorko M, Machackova T, et al. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res. 2016;36(12):6419–6423.
- Ogawa Y, Taketomi Y, Murakami M, et al. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36(1):66–75.
- Vychytilova-Faltejskova P, Stitkovcova K, Radova L, et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiology Biomarkers & Prevention. 2018;27(9):1019–1028.
- Wang Z, Yang H, Ma D, et al. Serum PIWI-interacting RNAs piR-020619 and piR-020450 are promising novel biomarkers for early detection of colorectal cancer. Cancer Epidemiol Prev Biomarkers. 2020;29(5):990–998.
- Zhou X, Liu J, Meng A, et al. Gastric juice piR‐1245: a promising prognostic biomarker for gastric cancer. J Clin Lab Anal. 2020;34(4):e23131.
- Houwing S, Kamminga LM, Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. cell. 2007;129(1):69–82.
- Sarkar A, Ghosh Z. Rejuvenation of piRNAs in emergence of cancer and other diseases. In: Bibekanand Mallick AGO-driven non-coding RNAs. USA: Elsevier; 2019. p. 319–333.
- Cheng J, Deng H, Xiao B, et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315(1):12–17.
- Zhou H, Guo J-M, Lou Y-R, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med. 2010;88(7):709–717.
- Wu X, Pan Y, Fang Y, et al. The biogenesis and functions of piRNAs in human diseases. Mol Ther Nucleic Acids. 2020b;21:108–120.
- Ren Z, Li Y, Hameed O, et al. Prognostic factors in patients with metastatic breast cancer at the time of diagnosis. Pathol Res Pract. 2014;210(5):301–306.
- Zhang H, Ren Y, Xu H, et al. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22(4):217–223.
- Krishnan P, Ghosh S, Graham K, et al. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget. 2016;7(25):37944.
- Li D, Luo Y, Gao Y, et al. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38(3):927–936.
- Yao J, Wang YW, Fang BB, et al. piR-651 and its function in 95-D lung cancer cells. Biomed Rep. 2016;4(5):546–550.
- Peng L, Song L, Liu C, et al. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumor Biol. 2016;37(2):2749–2756.
- Chu H, Xia L, Qiu X, et al. Genetic variants in noncoding PIWI‐interacting RNA and colorectal cancer risk. Cancer. 2015;121(12):2044–2052.
- Mai D, Zheng Y, Guo H, et al. Serum piRNA-54265 is a new biomarker for early detection and clinical surveillance of human colorectal cancer. Theranostics. 2020;10(19):8468.
- Yin J, Jiang XY, Qi W, et al. piR‐823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF 1. Cancer Sci. 2017;108(9):1746–1756.
- Yin J, Qi W, Ji CG, et al. Small RNA sequencing revealed aberrant piRNA expression profiles in colorectal cancer. Oncol Rep. 2019;42(1):263–272.
- Öner Ç, Turgut Coşan D, Çolak E. Estrogen and androgen hormone levels modulate the expression of PIWI interacting RNA in prostate and breast cancer. PLoS One. 2016;11(7):e0159044.
- Busch J, Ralla B, Jung M, et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015b;34(1):61.
- Busch J, Ralla B, Jung M, et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015a;34(1):1–11.
- Chang Z, Ji G, Huang R, et al. PIWI-interacting RNAs piR-13643 and piR-21238 are promising diagnostic biomarkers of papillary thyroid carcinoma. Aging (Albany NY). 2020;12(10):9292.
- Han H, Fan G, Song S, et al. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood. 2021;137(12):1603–1614.
- Litwin M, Szczepańska‑Buda A, Piotrowska A, et al. The meaning of PIWI proteins in cancer development. Oncol Lett. 2017;13(5):3354–3362.
- Qiao D, Zeeman A-M, Deng W, et al. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21(25):3988–3999.
- Gao C-L, Sun R, Li D-H, et al. PIWI-like protein 1 upregulation promotes gastric cancer invasion and metastasis. Onco Targets Ther. 2018;11:8783.
- Wang Y, Liu Y, Shen X, et al. The PIWI protein acts as a predictive marker for human gastric cancer. Int J Clin Exp Pathol. 2012;5(4):315.
- Genzor P, Cordts SC, Bokil NV, et al. (2019). Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proceedings of the National Academy of Sciences,USA, 116( 23), 11111–11112.
- Li F, Yuan P, Rao M, et al. piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive pancreatic cancer metastasis. Nat Cell Biol. 2020;22(4):425–438.
- Shi S, Yang -Z-Z, Liu S, et al. (2020). PIWIL1 promotes gastric cancer via a piRNA-independent mechanism. Proceedings of the National Academy of Sciences, USA, 117( 36), 22390–22401.
- Gou L-T, Kang J-Y, Dai P, et al. Ubiquitination-deficient mutations in human Piwi cause male infertility by impairing histone-to-protamine exchange during spermiogenesis. cell. 2017;169(6):1090–1104. e1013.
- Su C, Ren Z-J, Wang F, et al. PIWIL4 regulates cervical cancer cell line growth and is involved in down-regulating the expression of p14ARF and p53. FEBS Lett. 2012;586(9):1356–1362.
- Litwin M, Dubis J, Arczyńska K, et al. Correlation of HIWI and HILI expression with cancer stem cell markers in colorectal cancer. Anticancer Res. 2015;35(6):3317–3324.
- Martinez VD, Enfield KS, Rowbotham DA, et al. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric Cancer. 2016;19(2):660–665.
- Sugimoto K, Kage H, Aki N, et al. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem Biophys Res Commun. 2007;359(3):497–502.
- Liang D, Yang Y, Liu Y. The role Hiwi gene in the maintenance of lung cancer stem cell populations. Neoplasma. 2013. DOI:10.4149/neo_2014_022
- Singh G, Roy J, Rout P, et al. Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers. PLoS One. 2018;13(1):e0190485.
- Lu Y, Zhang K, Li C, et al. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One. 2012;7(1):e30999.
- Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061.
- Taubert H, Greither T, Kaushal D, et al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene. 2007;26(7):1098–1100.
- Sun G, Wang Y, Sun L, et al. Clinical significance of Hiwi gene expression in gliomas. Brain Res. 2011;1373:183–188.
- Ye Y, Yin D-T, Chen L, et al. Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One. 2010;5(10):e13406.
- Oh S-J, Kim S-M, Kim Y-O, et al. Clinicopathologic implications of PIWIL2 expression in colorectal cancer. Korean J Pathol. 2012;46(4):318.
- Yan Z, Qu L-K, Lin M, et al. HIWI expression profile in cancer cells and its prognostic value for patients with colorectal cancer. Chin Med J (Engl). 2011;124(14):2144–2149.
- Xie Y, Yang Y, Ji D, et al. Hiwi downregulation, mediated by shRNA, reduces the proliferation and migration of human hepatocellular carcinoma cells. Mol Med Rep. 2015;11(2):1455–1461.
- Lee JH, Jung C, Javadian-Elyaderani P, et al. Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Cancer Res. 2010;70(11):4569–4579.
- Klein JD, Qu C, Yang X, et al. c-Fos repression by piwi regulates Drosophila ovarian germline formation and tissue morphogenesis. PLoS Genet. 2016;12(9):e1006281.
- He G, Chen L, Ye Y, et al. Piwil2 expressed in various stages of cervical neoplasia is a potential complementary marker for p16INK4a. Am J Transl Res. 2010;2(2):156.
- Jiang L, Wang W-J, Li Z-W, et al. Downregulation of Piwil3 suppresses cell proliferation, migration and invasion in gastric cancer. Cancer Biomarkers. 2017;20(4):499–509.
- Wang D-W, Wang Z-H, Wang -L-L, et al. Overexpression of hiwi promotes growth of human breast cancer cells. Asian Pac J Cancer Prev. 2014;15(18):7553–7558.
- Qu X, Liu J, Zhong X, et al. PIWIL2 promotes progression of non-small cell lung cancer by inducing CDK2 and Cyclin A expression. J Transl Med. 2015;13(1):301.
- Li L, Yu C, Gao H, et al. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer. 2010;10(1):38.
- Liu Y, Dou M, Song X, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18(1):123.
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36(suppl_1):D173–D177.
- Han Y-N, Li Y, Xia S-Q, et al. PIWI proteins and PIWI-interacting RNA: emerging roles in cancer. Cell Physiol Biochem. 2017;44(1):1–20.
- Wang J, Zhang P, Lu Y, et al. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019;47(D1):D175–D180.
- Williams KP, Lau BY. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2016;45:SAND-2017-0752J.
- Wang X, Tong X, Gao H, et al. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45(6):2385–2392.
- Wang Z, Ma K, Pitts S, et al. Novel circular RNA circNF1 acts as a molecular sponge, promoting gastric cancer by absorbing miR-16. Endocr Relat Cancer. 2019;26(3):265–277.
- Xin J, Du M, Jiang X, et al. Systematic evaluation of the effects of genetic variants on PIWI-interacting RNA expression across 33 cancer types. Nucleic Acids Res. 2021;49(1):90–97.