ABSTRACT
Global projections on the climate change and the dynamic environmental perturbations indicate severe impacts on food security in general, and crop yield, vigor and the quality of produce in particular. Sessile plants respond to environmental challenges such as salt, drought, temperature, heavy metals at transcriptional and/or post-transcriptional levels through the stress-regulated network of pathways including transcription factors, proteins and the small non-coding endogenous RNAs. Amongs these, the miRNAs have gained unprecedented attention in recent years as key regulators for modulating gene expression in plants under stress. Hence, tailoring of miRNAs and their target pathways presents a promising strategy for developing multiple stress-tolerant crops. Plant stress tolerance has been successfully achieved through the over expression of microRNAs such as Os-miR408, Hv-miR82 for drought tolerance; OsmiR535A and artificial DST miRNA for salinity tolerance; and OsmiR535 and miR156 for combined drought and salt stress. Examples of miR408 overexpression also showed improved efficiency of irradiation utilization and carbon dioxide fixation in crop plants. Through this review, we present the current understanding about plant miRNAs, their roles in plant growth and stress-responses, the modern toolbox for identification, characterization and validation of miRNAs and their target genes including in silico tools, machine learning and artificial intelligence. Various approaches for up-regulation or knock-out of miRNAs have been discussed. The main emphasis has been given to the exploration of miRNAs for development of bioengineered climate-smart crops that can withstand changing climates and stressful environments, including combination of stresses, with very less or no yield penalties.
1. Introduction
Climate change is one of the greatest challenges that have emerged in this century, with impacts ranging from fluctuating weather patterns, amplifying temperature, acidifying oceans with declining corals, and imperiling ecosystems, thus impacting the vital food security. According to the Intergovernmental Panel on Climate Change (IPCC) climate change refers to ‘a change in the state of the climate that can be identified (e.g. using statistical tests) by changes in the mean and/or the variability of its properties, and that persists for an extended period, typically decades or longer’ [Citation1]. Agricultural practices and agro-ecosystems are susceptible to climate change as variations in temperature or precipitation pattern affect the growth and maintenance of crops in a particular region thereby severely influencing crop yield and quality. Furthermore, climate change aggravates land degradation through increase in rainfall intensity, flooding, drought severity, heat stress, dry spells, wind, sea-level rise, and permafrost thaw that have been modulated by land management [Citation2]. For instance, studies published between 1980 to 2015 have shown a decline of 21%, 40% and 10% in the yield of wheat, maize and cereal crops, respectively, at global scale due to climate change associated drought conditions. Further, with every °C rise in global temperature, yields of wheat, maize, rice and soybean decrease by 6 ± 2.9%, 7.4 ± 4.5%, 3.2 ± 3.7% and 3.1%, respectively [Citation3–6]. In order to maintain yield throughout the course of time, farmers may be forced to change cultivation practice, the time of cultivation or even type of crops grown [Citation7], and in this context, bioengineered crops may hold the key . The crop varieties that show resilience to challenging environments and complex climate change conditions with zero or less yield penalties are desired to sustain food security for the growing population [Citation8].
Conventional breeding techniques have been explored to improve crop durability against specific biotic and abiotic stresses using techniques such as mutation breeding, hybridization and selection from landraces resulting in significant improvement and release of new cultivars. However, these approaches are limited by challenging environmental hurdles, long generation time making the process time consuming and inefficacious. Genetic engineering has a potential to bypass these hurdles through utilizing the modern biotechnological tools such as genomics, proteomics and transcriptomics for developing stress-tolerant crop plants with less impacts on yield. Many studies have evaluated the influence of individual or combined abiotic stress factors as reviewed by Raza et al. 2019 [Citation8]. Though most of these studies were conducted under either single stress or under controlled laboratory environment, these cannot narrate and represent true adaptations for potential impacts of climate change under field ecosystems [Citation9]. Furthermore, the single-gene approach is largely becoming outdated as pleiotropic effect makes the agronomic traits genetically complex, in addition to this many agronomic traits are controlled by multi-gene products or multi-gene pathways making selection of single gene difficult [Citation10]. Therefore, keeping an eye on the projected futuristic climate changes/fluctuations and their impacts on crops, there is an urgent need to identify and explore new and effective biotechnological tools for developing climate-resilient or climate-smart crops [Citation11].
With the discovery of microRNAs (miRNAs) and their roles in almost all facets of plant life, increasing attention has been laid on elucidating complex mechanisms of miRNA mediated transcriptional or post-transcriptional regulation of genes involved in plant responses and adaptations to stress conditions [Citation12]. MiRNAs are a class of small, conserved, endogenous non-coding RNA (ncRNAs) species that regulate many biological processes by down and up-regulation of gene expression [Citation13]. In plants, miRNAs pair with their respective targets imperfectly in their coding site of mRNAs/transcripts to destabilize/degrade them or inhibit protein translation [Citation14–16]. Furthermore, miRNAs mediated gene regulation via miRNA-directed mRNA cleavage, translational repression, chromatin remodeling and epigenetic modification has also been recently explored for potential use in crop improvement [Citation17].
MiRNAs have gained considerable attention in recent years with an increasing number of reports confirming their key role in various plant molecular processes. To appraise the international scenario on filed inventions related to the miRNA, we have mined the Google Patent search tool, with a set of queries using the keywords (‘miRNA’ ‘abiotic stress’ ‘plants’ ‘transgenics’) on 4th Aug’2021. The retrieved data showed that, a total 13,651 patent applications and grants were filed so far, with China accounting for the highest numbers followed by USA, WO, EP and other countries (https://patents.google.com/). The data includes patents filed for diverse areas like miRNA and its role in drought tolerance/salt resistance/heat tolerance (CN-106434654-A, CN-103114091-A, WO-2013048255-A2), biotic stress (US-2012278929-A1, WO-2013048255-A2), plant growth and development (WO-2006034368-A2), artificial miRNAs (US-2013111634-A1), genome editing (WO-2020178099-A1), methods of developing transgenic plants for enhancing agronomic traits (US-2011197316-A1, US-2015135372-A1), heavy metal stress (CN-112662701-A, CN-106754913-A) and others. It is noteworthy here that a substantial gap was observed between the patented applications and available literature data. This survey demands integrating the miRNA biotechnological tools and scientific knowledge to develop strategies for improving plant growth and productivity under stressed environment. The increasing application of high throughput sequencing and techniques such as degradome sequencing, bisulfite sequencing, massive parallel signature sequencing (MPSS) in addition to computational tools have further assisted in identification of novel miRNAs involved in defining agronomic traits. Along with this, emerging tools such as artificial miRNAs (amiRNAs) and clustered regularly interspaced short palindromic repeats (CRISPR) based genome editing for improvement of traits and better resilience to climate change have been made possible to avoid non-targeted effects [Citation18–22]. While surveying for the global agenda 2030 ‘Sustainable Development’, authors have also considered the key aspects of an important mission; The UN 17 Sustainability development goals (SDGs), established in the year 2016 stating ‘A blueprint to achieve a better and more sustainable future for all people and the world by 2030’ [Citation23]. This mission targets 17 SDGs involving policy makers, academicians and agro-industries people. This review article is also an attempt to integrate multiple research views allied to the five SDGs namely; Goal 2 (zero hunger), Goal 3 (good health and well-being), Goal 9 (industry, innovation and infrastructure), Goal 13 (climate action) and ultimately the cumulative Goal 17 (partnerships for the goals).
This review aims to present the current trends in crop engineering via the miRNAs and their target genes, biogenesis and regulatory mechanisms in adaptations and response under climate conditions. Modern tools and techniques such as amiRNAs, target mimicry and emerging machine learning techniques in identifying and characterizing miRNAs in plants have been described. Strategies for up-regulation or knock-out of miRNAs of interest, successful events, challenges and future prospects are also discussed.
2. Brief overview of plant miRNAs
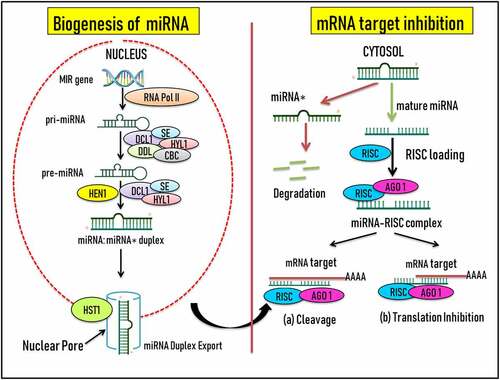
MicroRNAs (miRNAs) belonging to the class of single-stranded non-coding RNA molecules are considered to be effectual regulators of gene expression of the plant kingdom [Citation12]. In the past several years, there has been significant research outcome on the role of small ncRNAs in the life cycle of plants growing under stressed environment [Citation13]. Amongs these small RNA family members, panorama of miRNAs has been identified as a tiny promising pervasive class of key post-transcriptional/post-translational growth regulators often responsive to single/multiple/combinatorial stress stimuli [Citation24,Citation25]. First reported in Caenorhabditis elegans, a transparent nematode [Citation26,Citation27], miRNAs are single stranded 19–25nt highly conserved ncRNA molecules [Citation12,Citation28]. Plant-specific miRNAs were first reported by Reinhart et al. [Citation29], adding further to the presence of evolutionary conservation across eukaryotes and prokaryotes [Citation29]. During stressful conditions, miRNAs trigger up- or down-regulation of their target genes via four different pathways namely; mRNA cleavage, chromatin remodeling, translational repression and/or methylation of DNA [Citation17,Citation30]. They are synthesized by the plants during physiological, molecular, and developmental and plant growth hormone regulation processes [Citation28]. Looking at the origin, biosynthesis and mode of action, both plant and animal miRNAs have been reported for sharing in part the similarity and differences across them. This can be further supported by the fact that several plants have more than 100 types of nuclear encoded miRNA (MIR) genes [Citation31]. Growing evidence shows that the role of miRNAs in regulating the expression of their target genes under the given set of environmental signaling components is a tightly modulated process in both plants and animals. The regulation is intricate in the sense that single or multiple external signals can activate individual or multiple miRNAs causing differential expression of single or multiple target genes. Henceforth, miRNAs have been characterized as master regulators in response to multitude signals. Biogenesis and miRNA mode of action is a fine-tuned mechanism involving atlases of cellular and molecular switches in order to affect the dual regulation of target genes. On the basis of their genomic location, miRNAs are classified into two main types namely ‘intronic’ or ‘intergenic’. The transcription of both types of miRNAs varies; intronic types are transcribed from the introns region present in their respective host transcript whereas intergenic types are transcribed by an independent RNA Polymerase II enzyme. MIR genes are transcribed by either RNA Pol II or Pol III into primary miRNA transcripts, pri-miRNAs (). Thereafter, pri-miRNA undergoes an imperfectly folding mechanism resulting in a double-stranded stem loop hairpin precursor RNAs, pre-miRNA. Numerous cellular proteins are known to support this folding mechanism of pri-miRNA namely, Dicer-like RNase III endonucleases (DCL-1), RNA-binding protein DAWDLE (DDL), Zinc Finger protein SERRATE (SE), dsRNA binding protein HYPONASTIC LEAVES1 (HYL1), and nuclear cap binding complex (CBC) [Citation32]. The plant stem loop structure varies in size from 60–500 nt whilst in the animal system it is ∼70 nucleotides [Citation33]. Also, in case of plants , the processing of pri miRNAs can happen from either the loop-proximal to loop-distant site or contrariwise [Citation34]. Moving ahead, DCL1, HYL1, and SE assisted processing results in the formation of miRNA: miRNA*; duplex (19–25 nt) having 3ʹ overhangs at their ends [Citation35,Citation36]. Further, methylation occurs at only 2ʹ – OH position supported by another class of protein, small RNA methyltransferase HUA Enhancer 1 (HEN1). From here the duplex is exported from the nucleus into the cytosol with the help of an exportin protein HASTY (HST). Once into the cytoplasm, the duplex gets separated wherein, the next protein, ARGONAUTE (AGO), is ready to be assembled with the guide strand, miRNA. The other strand of the duplex, the passenger strand, miRNA*, gets degraded. RNA-induced silencing complex (RISC) mediated negative and/or positive regulation of the expression of target mRNA/mRNAs is highly relied on the complementary base pairing between the sequences present on the mRNA and miRNA respectively [Citation12,Citation24,Citation37,Citation38]. Presuming the given set of conditions, degree of base pairing is one of the pivotal factors responsible for either expression or repression of the target mRNA through four different pathways. Of note is the base pairing, which is sterner in plant systems then their animal counterparts.
Figure 1. Schematic representation of the biogenesis of miRNA and modulation of target mRNAs via cleavage/translation inhibition. Abbreviations- DCL-1: Dicer-like RNase III endonucleases, DDL: RNA-binding protein DAWDLE, HYL1: ds-RNA binding protein HYPONASTIC LEAVES1, SE: Zinc Finger protein SERRATE, CBC: Nuclear cap binding complex, HST: Exportin protein HASTY, HEN1: HUA Enhancer 1, AGO1: ARGONAUTE, RISC: RNA-induced silencing complex
Till date, there are several reports of specific plant miRNAs in response to different types of environmental stresses and changing climate conditions across diverse plant species [Citation38–40]. Plants under stress respond concomitantly with the complex yet controlled modulation of MIR genes and it is one of the fundamental steps in miRNA biogenesis that needs to be explored at a larger perspective for better understanding of the complex stress regulatory pathways. Eventually several reports have been published showing utilization of miRNA in various crop improvement programs without impacting agronomic traits by using an array of biotechnology and bioinformatics techniques. Owing to their ubiquity and complexity, miRNAs-based climate smart crops will be the possible strategy to balance the food and nutritional health of the rising global population rate [Citation13,Citation41].
3. Identification and characterization of plant miRNAs and their targets
The hereditary information which determines the function, structure, organization and cellular responses of an organism are stored in the genome. Hence understanding the mechanistic insights of plant responses and adaptation to challenging environmental conditions via decoding the genetic sequences and their transcripts has been in the focus historically. The genome, transcriptome, proteome and metabolomics play a vital role in understanding the hidden codes [Citation42]. The miRNAs present in an organism play regulatory roles as discussed earlier and hence their sequencing, identification and characterization are of utmost importance. The derived information can be used for imparting the necessary tolerance to plants and crops based on futuristic climatic conditions [Citation13]. The miRNA modification would keep the base genome untouched and would rather focus on enhancing or retarding the regulatory mRNAs . A brief description of modern tools and techniques currently in use for miRNA related work is given below.
3.1 Deep sequencing
Next generation sequencing (NGS) or deep sequencing technologies have revolutionized the discovery [Citation43] and identification of various novel and conserved miRNAs helping to fill the missing gaps in our knowledge. The small size of miRNAs along with their repetitive nature in the genome conferred significant challenges for sequencing. Sanger’s dideoxy synthesis method [Citation44,Citation45] and Maxam–Gilbert’s chemical cleavage method [Citation46] are the founding techniques based on which the NGS came into existence. Illumina (Solexa) sequencing is one of popular sequencing platforms with its removable fluorescent labels. It is sub-classified into platforms like HiSeq, MiSeq, MiniSeq, NextSeq and NovaSeq. At 600 Gb of sequence yield per run and much greater resolution, it is significantly better in comparison with other techniques like Roche 454 pyrosequencing, Sequencing by Oligonucleotide Ligation and Detection (SOLiD) and Ion Torrent. When used for transcriptome sequencing specializing on small RNA, the Illumina NGS is a preferred choice for identifying novel miRNAs under varying environmental conditions. Third generation, an advanced version of NGS, is cheaper, faster, accurate and reliable [Citation47]. Single-molecule real-time (SMRT) sequencing (Pacific biosciences), Helicos sequencing (genetic analysis system) and Nanopore are some examples of Third generation sequencing.
The degradome sequencing technique provides a plant profile containing miRNA along with their target mRNAs, allowing one to identify novel miRNAs and target mRNAs. It is most commonly utilized technique for degraded RNA under which sequencing and identification of ncRNAs, microRNA (miRNA), short-interfering RNA (siRNA), turnover RNA fragments, and miRNA-/siRNA-cleaved target RNA fragments can be performed. It is a result of combining modified 5ʹ rapid amplification of cDNA ends (5ʹ-RACE) and ‘NGS’ [Citation48]. The 3ʹ RNA fragment is usually cleaved under miRNA action, and degraded by XRN4. When compared to 5ʹ-cleaved RNA fragment XRN4 exhibits a lower degradation rate. Hence quantification of 3ʹ-cleaved RNA fragments provides reliable data to demonstrate the miRNA-target relationship. Using NGS and various computational approaches the 3ʹ-cleaved RNA fragments in the total RNAs can be converted to cDNAs by 5ʹ-RACE [Citation48].
Recently Selvi et al identified 1224 conserved miRNAs and 435 novel miRNAs from Saccharum under drought stress using high-throughput miRNA deep sequencing method [Citation49]. Xu et al [Citation50]used PacBio RSII and the Illumina sequencing platform to identify heat responsive LncRNA associated with heat shock protein in Populus qiongdaoensis seedlings. Pacbio sequencing technology was used to discover full length transcripts of Pinus elliottii for a phylogeny study [Citation51]. 24 putative novel miRNAs were identified from a leaf of Persicaria minor, a non-model plant, transcriptome using 2500 HiSeq Illumina [Citation52]. Similarly, transcriptome sequencing was done in Jatropha curcas by high-throughput sequencer called Illumina NextSeq500 resulting in identification of 266 conserved miRNA belonging to 78 miRNA families [Citation53]. In Populus, degradome sequencing has been applied to identify cold responsive miRNA and their targets using Illumina HiSeq 2000 [Citation54]. Identification of drought stress responsive miRNA present in Macleaya cordata was done by Illumina Hiseq xten Sequencer which showed presence of 355 miRNAs belonging to 68 families, miR166 being the most prominent [Citation55]. Deep sequencing using Illumina Hiseq 2500 platform by Novogene (Beijing, China) revealed 88 conserved and 13 novel miRNAs in Reaumuria songorica under salt stress during seed germination [Citation56]. With the use of high throughput deep sequencing technology, 67 conserved and 66 novel miRNAs were identified in Gossypium hirsutum L. in response to root-knot nematode Meloidogyne incognita infection [Citation57]. In another study, the high-throughput sequencing platform was utilized to perform a transcriptome-wide analysis for the identification of miRNAs produced in response to DNA damage in rice. The study identified 513 known and 72 novel miRNAs in the roots of bleomycin-treated and control rice specimens [Citation58]. Wu et al utilized the Illumina NovaSeq6000 by Gene Denovo Biotechnology Co. (Guangzhou, China) to identify miR9736, miR5059, miR167, miR5665, miR866, miR10186, miR8165, miR857, miR399, miR399, miR163,and miR393 that plays a key role in regulating seed germination-genes [Citation59].
3.2 In silico tools and databases
Recent advancement in improving crops that can withstand changing climatic conditions is mostly based on understanding the multidimensional regulatory network that occurs in plant cells at a molecular level. The candidate genes along with their targets are often identified from a wide pool of datasets. These are then characterized structurally, functionally and physiologically by comparing them with a homologous base. High throughput techniques have been established that are faster, more reliable and efficient resulting in the need for an omics approach in plant stress biology [Citation60]. The high usage of miRNAs for conferring tolerance to plants has flooded the databases with a big set of information on this subject. Hence miRNAomics have been developing at a faster rate.
Plant miRNA database (PMRD) (http://bioinformatics.cau.edu.cn/PMRD/) [Citation61], Tarbase (http://carolina.imis.athenainnovation.gr/diana_tools/web/index.php) [Citation62], MiRDB (http://www.mirdb.org/) [Citation63] and miRbase (https://www.mirbase.org/) [Citation64] are most commonly used databases that contain all published miRNAs based on which one can compare and identify novel miRNAs in plant species. miRNEST 2.0 represents a wide-ranging database of animal, plant, and virus miRNAs (http://mirnest.amu.edu.pl) [Citation65]. Plant miRNA Target Expression Database (PMTED) (http://pmted.agrinome.org/) [Citation66], Target prediction for plant miRNAs (TAPIR) (http://bioinformatics.psb.ugent.be/webtools/tapir/) [Citation67] are some other databases that provide expressional details along with target prediction. Other than these databases, PlantCircNet: a database for plant circRNA–miRNA–mRNA regulatory networks [Citation68] (http://bis.zju.edu.cn/plantcircnet/index.php) and GreenCircRNA: a database for plant circRNAs that act as miRNA decoys (http://greencirc.cn) [Citation69] are currently used for identification of miRNAs that are interactive with cirRNA. PlanTE-MIR DB [Citation70](http://bioinfo-tool.cp.utfpr.edu.br/plantemirdb/), is a database that harbors transposable element-related microRNAs in plant genomes that are publicly available.
The miRBase (http://www.mirbase.org/) is a most common searchable online repository that compiles all published miRNA sequences and annotations [Citation64]. A predicted portion of hairpin structure of putative miRNA transcript is made available along with precise location and sequence information of mature miRNA. The data available on miRBase is easily downloadable and accepted for publication. As of October 2018, under release 22.1, a total of 38,589 miRNA entries have been deposited in this database. The miRNA data from 271 organisms belonging to 6 major clades (Alveolata, Chromalveolata, Metazoa, Mycetozoa, Viridiplantae and Viruses) have been made available.
Plant miRNA Encyclopedia (PmiREN, http://www.pmiren.com/) [Citation71], is a knowledge-based database which includes processed small RNA libraries using miRDeep-P2 and a manual curation. As of now PmiREN harbors 16,422 high confidence novel miRNA loci in 88 plant species and 3,966 retrieved from miRBase. Information on precursor sequence, precursor secondary structure, expression pattern, clusters and synteny in the genome, potential targets supported by Parallel Analysis of RNA Ends (PARE) sequencing, and references is attached.
MepmiRDB (http://mepmirdb.cn/mepmirdb/index.html) [Citation72] is a freely available medicinal plant microRNA database. The miRNA information on sequences, expression patterns and regulatory networks has been included along with organ/growth condition-specific expression information of the mature miRNAs. The ‘Interaction’ module offers the information of the degradome-validated miRNA – target pairs of eight plant species.
In recent times, due to an increase in the Small RNA sequencing procedures, a huge amount of data has been created. Hence various databases are utilized to store, characterize and keep record of all published, annotated miRNAs. A summarized list of such databases has been given in .
Table 1. Computational tools and databases useful for plant miRNAs and their characterization
3.3 Identification of miRNAs and their targets
Identification of miRNAs can be done via experimental and computational approaches. The initial attempts started in the early 2000s when Llave et al. [Citation101] and Reinhart et al.[Citation29] identified miRNA using direct-cloning approaches. The advancement in computational approaches jumpstarted their identification along with the target genes these miRNAs degrade. Several computational tools and software have been made available in recent times that predict the targets of miRNAs using well established databases as mentioned earlier [Citation38].Most of the programs use different steps including to acquire data and predict/finding the targets, the base-pairing pattern of miRNA and 3ʹUTR of target mRNA sequence, the miRNA-mRNA duplex thermodynamic stability and third, the evolutionary conservation of miRNA-target sites in different species. RNA22 [Citation102], PITA (https://www.mybiosoftware.com/pita-6-microrna-prediction-tool.html), psRNATarget (http://plantgrn.noble.org/psRNATarget/home) [Citation103], and microInspector (http://mirna.imbb.forth.gr/microinspector/) [Citation104] are some of the commonly used tools for finding targets of identified miRNAs.
The miRNA-truncation and tailing analysis (miTRATA) uses truncation and tailing analysis of miRNA that is used to analyze 3 ̀ modifications [Citation105]. PAREameters (http://srna-workbench.cmp.uea.ac.uk/pareameters/) is an offline downloadable tool for identifying computational inference of plant miRNA–mRNA targeting rules using small RNA and degradome sequencing data. It is used to identify and quantify the differences between subsets of miRNA–mRNA interactions in model and non-model organisms [Citation106]. Iwa-miRNA is a Galaxy-based framework that can be utilized to annotate miRNAs by computational analysis and manual curation. It generates a comprehensive list of candidate miRNAs from known databases and predicts novels sets from sRNA-Seq datasets [Citation107].A summarized list of tools along with their descriptions has been given in .
4. Approaches for developing miRNA-mediated bioengineered crops
4.1 Transgenesis, cisgenesis and intragenesis
With the advancement of transformation techniques, a new arena has opened up where the MIR genes could be transferred within and between the various crossable and non-crossable plant species. Transgenesis (transfer of one or more MIR gene between non-crossable plant species), cisgenesis (transfer of MIR gene between crossable species) and intragenesis (transfer of MIR gene between same species) utilize the T-DNA of Agrobacterium and engineer it to carry the gene of interest for further (and desired) expression [Citation108,Citation109]. Cisgenesis, as described by Schouten et alis the modification done in the genetic background of a recipient plant that is cross compatible with the donor plant. It is a faster method when compared with traditional breeding wherein desired genes are introduced rapidly without any changes in other characteristics of the plant [Citation110]. Rommens et al. [Citation111] introduced intragenesis transformation allowing customized cassettes designing to transform plants that are cross compatible. Intragenesis transformation utilizes additional promoter and termination sequences that overcome the challenges faced under overexpression of MIR genes [Citation109].
4.2 Topical delivery of miRNA (pre- and mature-miRNA)
Topical delivery can also be called a transgene-free approach which was recently optimized from the topical delivery of nanostructured dsRNA molecules in model and/or crop plants. Although the technology is still in its developing phase for transfer of pre-miRNA and miRNA, the higher stability and internalization capacity of pre-miRNA creates possibilities of using this technology for transgenic free manipulation of transcriptional profile of plant. This system uses an organic (ribonucleoprotein, cross linkers) or nanoparticle (biopolymers of chitosan, silicon, carbon and clay nanosheets) carrier to deliver the internalized RNA in plant cell [Citation112–114].These pre-miRNAs are structured like viroids flanked by pH-dependent ribozymes, engineered such that they are not processed by the plant RNAi machinery but interact with insect digestive tract cells [Citation109]. A topical delivery of miRNA of 22 nt was done in two species namely Nicotiana benthamiana and Amaranthus cruentus to induce biogenesis of secondary transitive miRNA that cleaved the targeted mRNAs. The delivered miRNA was able to silence green fluorescent protein (GFP) transgene and 3 endogenous genes: magnesium chelatase subunit I (CHL-I), magnesium chelatase subunit H (CHL-H), and GUN4 in treated leaves of Nicotiana benthamiana [Citation115].
4.3 Artificial miRNA and target mimicry
To overcome the problem of nonspecific gene silencing, a side effect of siRNA-based gene silencing, artificial miRNAs (amiRNAs) were developed [Citation116]. These amiRNAs are highly efficient, specific and are proven to have more potential to silence undesirable gene expression. The desired gene is designed and integrated into a stem-loop backbone forming a precursor (which helps in in vivo processing of amiRNA) resulting in decreased accumulation of mature amiRNAs in plant cells [Citation117]. Thus the original sequence structured as miRNA-5p:miRNA-3p is replaced with a bioengineered sequence that is specific for inhibiting target mRNA. A significant feature, processing of pre-amiRNAs to form mature amiRNAs, makes them very specific in nature and avoids off-target interactions [Citation118]. These mature amiRNAs are created for known targets, have restricted systemic movement, stable in nature and limited production of secondary siRNAs from pre-amiRNA strands [Citation109].
Target mimicry is often utilized to regulate the biological processes in plants that are induced due to different plant behaviors. This process can be divided into two types based on their origin, endogenous target mimicry (eTMs) and artificial. The eTMS include actions of long noncoding RNA (lncRNA) and circular noncoding RNA (cirRNA) that’s been transcribed from the plant genome under varying stress conditions. Similar in function, artificial short tandem target mimic (STTM) was developed that modulated the accumulation of miRNAs. The STTMs are expressed transiently or constitutively based on the promoter (induced or tissue specific) they are built with. Some are engineered with high nucleotide sequences on an lncRNA basal template that identify and target mRNA [Citation119]. Another class of artificial target mimicry includes miRNA SPONGES that are produced from transgenes containing multiple miRNA binding sites located in tandem repeats. These SPONGES are utilized to target and inhibit whole families of related miRNAs [Citation120,Citation121]. An artificial Vitis vinifera miRNA319e (amiR319e) was introduced in transgenic Nicotiana benthamiana using overlapping long primers and was able to silence pre-inserted green fluorescent protein (GFP) gene [Citation122]. Von et al. [Citation123], in their comparative study conducted on Arabidopsis thaliana, showed that the efficiency of amiRNA declined based on their environmental conditions and developmental stages of plant as compared to miRNAs. The research showed amiRNAs were unable to silence the targeted gene with age during its vegetative growth stage in a temperature dependent manner.
4.4 MIR gene editing
A sequence specific editing is required for specific and efficient modulation of a particular gene. This genome editing tool involves modulation of a gene that is inherently present inside the plant and no integration of foreign DNA into the host plant is conducted [Citation124]. There are four types of editing tools used for MIR gene modification: (1) zinc finger nucleases (ZFN), (2) meganucleases, (3) transcription activator-like effector nucleases (TALENs), and (4) CRISPR systems [Citation125]. By using basic cellular repair mechanism like non-homologous end-joining (NHEJ) and homology-directed recombination (HDR) pathways for repairing the double stranded breaks caused by these endonucleases, allows the insertion and deletion (INDELs) of desired gene [Citation126].
The CRISPR-associated protein-9 nuclease (CRISPR/Cas9), is the most popular, modern, simplest Nobel Prize winning (https://www.nobelprize.org/prizes/chemistry/2020/press-release/) method of genome editing. CRISPR/Cas9 works by retarding the miRNA biogenesis through introduction of INDELs at miRNA processing sites of MIR genes [Citation127,Citation128]. Apart from biogenesis, the INDELs can also hamper miRNA-mRNA target pairing, full deletion and knock-in of the MIR genes and/or their promoters [Citation20]. CRISPR-derived systems, such as dCas9 nickase79, fCas980, Cpf181, Cpf1 and Csm1 are being searched and studied for more flexible, efficient and applicable usage [Citation129].Despite being most used, CRISPR/Cas9 faces some complexities and challenges while editing MIR genes. MIR genes have highly complex regulatory networks which makes knock-down or knock-in experiments harder to conduct. Along with this, the smaller length of MIR genes requires shorter guide RNA (gRNA) reducing the number of possible gRNA/Cas9 targets thus reducing possible hits on mature miRNA. Furthermore, CRISPR/Cas13a which uses novel nuclease type (class II type VI-A endoribonuclease) is under study but has not yet been established for pre-, pri-miRNA or mature miRNA editing in plants [Citation109]. Recombinant codon-optimized Cas9 (rCas9) was prepared for rice with the use of single-gRNA that were specifically targeting mature miRNA sequences or sites required for the biogenesis of mature miRNA. 409 transgenic plants of rice were formed in a CRISPR/Cas9 mediated mutation on 13 independent miRNAs involved in drought tolerance responses [Citation130].
5. Exploring machine learning and/or artificial intelligence for miRNA research
Machine learning (ML) or artificial intelligence (AI) is a discipline that uses computational and statistical approaches to predict from unseen data based from experiences learned from previous experiments. These ML algorithms learn from a set of known labeled data to make future predictions, thus reducing the false positives and increasing the specificity of the ML models [Citation131]. Feature selection is one of the fundamental problems faced by ML during ranking of miRNAs. When it comes to prediction of miRNAs, ML has been used to determine stress responsive miRNAs wherein it learns the complex non-linear patterns between the miRNA expression (input) and plant stress (output). Supervised learning methods like decision tree (DT), support vector machines (SVMs), least-square support vector machines (LSSVMs), and naïve Bayes (NB) can be used to find the patterns in a database [Citation132]. The ML prediction of novel miRNAs has a high-class imbalance because fewer well-known miRNAs are being compared with the whole genome of the candidate plant. When the known dataset is prepared in a manner where the model, experiments and performance are not properly addressed, the pre-miRNA prediction works in a biased and nonspecific manner. Hence the labeling of positive as well as negative datasets are of utmost importance [Citation133].
Bugnon et al. [Citation134] used ML to conduct a genome wide identification of pre-miRNAs. The research study included comparison of genome of five model organisms (Arabidopsis thaliana, Caenorhabditis elegans, Anopheles gambiae, Drosophila melanogaster, Homo sapiens) with known pre-miRNAs as their labeled dataset. The study concluded that for smaller genomes and imbalances, all methods perform in a similar way but for larger datasets such as the Homo sapiens genome, deep learning approaches using raw information from the sequences reached the best scores. PlantMiRNAPred (compares all miRNA plants in miRBase as positive and Pseudo hairpins from the protein coding sequences of Arabidopsis thaliana and Glycine max as negative) [Citation135], miRPara (dataset made from animals, plants and virus in miRBase and sequences with pri-miRNAs as positive and negative selection respectively) [Citation136] and SMIRP (Species-specific positive sets from miRBase and ncRNA as negative set) [Citation137] are some of the ML approaches available for novel miRNA prediction in plants [Citation133]. Thus, though the use of ML and/or AI for plant miRNA work is still in infancy and far from full potential exploration, these initial attempts are very encouraging and are expected to attract more studies that will help their exploration further.
6. MiRNA-based biotechnology for developing bioengineered climate resilient crops
Challenging or unfavorable environments are known to confer negative impacts on plant growth, development and yield. The expression levels of key genes are often altered in such stress conditions due to differential regulators, transcription factors and miRNAs expression as part of the plant’s responsive mechanism. Numerous miRNAs have been identified, characterized and used for transgenic plant development in response to confer tolerance to various abiotic stresses [Citation13,Citation28].Along with singular stresses, the plants often face multiple stress factors in nature, or more specifically in agro-ecosystems. The environment mimicry is thus attempted in contemporary times via studying the impact of combinational or multi-factorial stresses as well as sequential stresses which are seen to cause more than 50% of crop loss worldwide [Citation138,Citation139]. Recent reports confirm that the sound platform miRNAs can be used for engineering crops that can withstand the singular as well as multiple environmental stresses, or climate changes [Citation13]. depicts recently identified miRNAs from crops involved in plant responses to multiple environmental challenges.
Figure 2. Schematic representation of the miRNAs responsive to multiple abiotic stress factors and their respective targets. Abbreviations- NFY: Nuclear Factor Y, SPL: SQUAMOSA Promoter-binding Protein-Like, TF: Transcription Factor, AGO1: Argonaute, MYB: Myeloblastosis, ARF: Auxin Response Transcription Factor, GRL: Growth Factor-Like, AFB: Auxin-binding F-Box AP2: APETALA2. Reproduced with permission from Xu et al. 2019, Copyright 2019, Elsevier [Citation13]
![Figure 2. Schematic representation of the miRNAs responsive to multiple abiotic stress factors and their respective targets. Abbreviations- NFY: Nuclear Factor Y, SPL: SQUAMOSA Promoter-binding Protein-Like, TF: Transcription Factor, AGO1: Argonaute, MYB: Myeloblastosis, ARF: Auxin Response Transcription Factor, GRL: Growth Factor-Like, AFB: Auxin-binding F-Box AP2: APETALA2. Reproduced with permission from Xu et al. 2019, Copyright 2019, Elsevier [Citation13]](/cms/asset/f5d2ac3c-7110-4f9e-b98c-c13042f9e88e/kbie_a_1997244_f0002_oc.jpg)
6.1 Individual stresses
6.1.1 Drought stress
Water availability is an important factor in plant growth and development processes and thus drought stress in considered a major challenge for optimal crop production and yield. Changing precipitation rate due to increase in global warming has changed the seasonal pattern of rainfall. In the near future there is a possible shift in the EI Nino-Southern Oscillation (ENSO) and Indian monsoon rainfall due to global warming [Citation140]. Similar observations were done regarding the East Asian summer monsoon (EASM) rainfall under global warming based on the historical and representative concentration pathway (RCP) [Citation141]. Hence there is an intermittent drought, and flash floods that damper the crop growth, along with hampering the soil physiology.
Hajyzadeh et al. [Citation142] created transgenic chickpea line overexpressing miR408 to examine its role in drought conditions. The study reported overexpression of miR408 caused reduced plastocyanin transcript expression which resulted in dehydration-responsive element-binding protein (DREB) regulation under drought conditions conferring enhanced tolerance as compared to the wild-type line. Similarly, Hang et al. [Citation143]worked on miR408 of Oryza sativa (Os-miR408) to develop transgenic Lolium perenne (perennial ryegrass) line to confer tolerance against drought. The transgenic perennial ryegrass showing heterologous expression of Os-miR408 exhibited higher leaf relative water content (RWC), lower electrolyte leakage (EL) and less lipid peroxidation resulting in improved drought tolerance as compared to wild-type plants, under drought stress. The results suggested that the improved drought tolerance in Os-miR408 transgenic plants could be due to leaf morphological changes favoring the maintenance of water status and increased antioxidative capacity protecting against the reactive oxygen species damages that occurs under such conditions [Citation143]. Similarly, transgenic Hordeum vulgare (barley) overexpressing Hv-miR82 under drought conditions conferred drought tolerance to plant when the transgene was regulated by a drought induced promoter [Citation144]. Visenti et al. [Citation145] compared wild-type and transgenic tomato overexpressing miR156-oe under drought conditions with strigolactone-depleted and strigolactone-treated plants. The study showed miR156 overexpression and strigolactone treatment led to lower stomatal conductance and higher ABA sensitivity conferring faster drought recovery. MiR156 has also been reported to play a key role in conferring drought tolerance by its interaction with SPLs (MsSPL6, MsSPL12, and MsSPL13) in Medicago sativa [Citation146].
In contrast, Zhou et al. [Citation147] reported that gma-miR398c negatively regulated the peroxisome-related genes (GmCSD1a/b, GmCSD2a/b/c and GmCCS) under drought conditions. When gma-miR398c was overexpressed in transgenic Arabidopsis thaliana, it resulted in reduced survivability under water deficient condition, sensitivity to drought during seed growth and germination leading to decline in percentage germination rate along with higher rate of water loss in leaf. Similarly, when gma-miR398c was overexpressed in transgenic Glycine max, the expression levels of GmCSD1a/b, GmCSD2a/b/c and GmCCS were decreased phenomenally as compared to the knocked out miR398c and wild-type soybean under drought conditions [Citation147].
6.1.2 Salinity stress
Hyper soil salinity is a secondary effect of irrational irrigation practices, improper drainage, overuse of fertilizers as well as the climatic changes. The predicted increase in the temperature and decline in precipitation leads to drying of soil, inducing an increase in the ionic concentrations [Citation148]. Besides, the global warming is projected to rise the sea water levels causing increased infusion of salinity within the inland soil [Citation139]. The salt stress dramatically affects the crop growth and yield resulting in low harvest, poor quality of seed and less availability of food from the agricultural sector [Citation149].Ma et al. [Citation150], analyzed miR156/SPL from Malus domestica (apple) which is seen to regulate embryogenesis, morphogenesis, life cycle stage transformation, flower formation and other processes. The overexpression of MIR156a induced salt sensitivity in apples, whereas,MdSPL13overexpression stabilized the tolerance levels.MiRNA408 is an abiotic stress responsive candidate and is reported to be involved in conferring salinity tolerance in Nicotiana benthamiana [Citation151]. Overexpression of Sm-MIR408 (the miR408 precursor sequence) and its promoter sequence in tobacco promoted seed germination, reduced the accumulation of reactive oxygen species by increasing the expression of NbSOD, NbPOD and NbCAT under salt stress [Citation151]. A transgenic BRRIdhan 28 (BR28) (rice cultivar) was developed with DST_artificial miRNA (Drought and Saline tolerance; DST_amiRNA). Transformed plants showed vigorous growth with longer panicle length and higher primary branching resulting in higher yield under saline stress [Citation152].
The miRNAs also known as negative regulators of stress responses including salinity stress, and their overexpression can make the plant stress sensitive. For instance, using the CRISPR/Cas9 knockout system, inhibiting OsmiR535 in transgenic rice enhanced the tolerance to salt along with other stresses. The overexpression of OsmiR535 reduced seedling setting under stress conditions [Citation153]. Wang et al. [Citation146] studied the interaction of miR414c with iron superoxide dismutase gene GhFSD1 in Gossypium under saline stress. In cotton the ghr-miR414c targets the coding sequence region of GhFSD1, thus inhibiting its expression. The patterns were proven when transgenic Arabidopsis thaliana conferred salinity stress tolerance under ectopic expression of GhFSD1 and salinity sensitivity when ghr-miR414c was constitutively expressed. The overexpression of ghr-miR414c in cotton decreased the expression of GhFSD1 similar to GhFSD1-silenced cotton [Citation146].
6.1.3 UV light
Light is the first element sensed by a plant when it emerges from the soil. Being the inducer of various metabolic activities, its mechanism of induction is utmost necessary. That being said, its action and interaction with miRNAs is a lesser known area of research [Citation154]. The ultraviolet (UV) radiations are usually deflected by the earth’s stratospheric ozone layer. A slight change in its thickness would create a strong biological effect due to the radiation emitted by the rays. With changing climate and increasing global warming, although unpredictable, there would be a slight change in the ozone layer thickness in the near future [Citation155]. The role of BrmiR828 in the light-induced synthesis of anthocyanin in Brassica rapawas explored by Zhou et al [Citation156]. The study observed that light-induced down-regulation of BrmiR828 can target BrTAS4, BrPAP1 (Bra039763), MYB82 (Bra022602) by repressing their expression levels leading to the accumulation of MYB transcription factors that confer positive regulation of anthocyanin biosynthesis in light-exposed seedlings of Brassica rapa [Citation156]. Li et al. [Citation157] showed action of LONG HYPOCOTYL 5 (HY5) on light regulated transcription of miR163 in Arabidopsis thaliana seedlings. Overexpression of miR163 in transgenic Arabidopsis thaliana showed inhibited primary root elongation due to inhibition of its target PXMT1 under light conditions [Citation157]. Thus, though there are only a handful of studies, the results are encouraging that miRNA-biotechnology can play a key role in developing bioengineered crops that can survive under harsh UV-radiations.
6.1.4 Heavy metals
Heavy metal availability in soil has been predicted to increase with changing climatic conditions, leading to higher rates of contamination and environmental concern [Citation158,Citation159]. It is particularly concerning considering the high environmental health risk related with the elements, such as Lead (Pb), Arsenic (As), Mercury (Hg), Cadmium (Cd) etc. The source of contamination is not only limited to the presence of such metals in soil, but their high concentration in the air through automobile exhausts [Citation160–162].Ding et al. [Citation163], analyzed the role of miR166 during Cd accumulation in rice. The study observed, overexpression of miR166 improved tolerance against Cd in transgenic rice by reducing the translocation of Cd from roots to shoots. The class-III homeodomain-Leu zipper (HD-Zip) family proteins along with HOMEODOMAIN CONTAINING PROTEIN4 (OsHB4) gene (Os03g43930) are usually up regulated during Cd stress, but were observed to be down regulated under overexpressing miR166 [Citation163]. Similarly, Shen et al. [Citation164] worked on overexpressing IamiR-4-3p from Ipomoea aquatic in transgenic Arabidopsis thaliana against Cd stress. It was observed that expression level of GST3 was reduced by 20%, shoot MDA and H2O2 were higher in concentration along with shorter root and shoot length in transgenic Arabidopsis thaliana [Citation164].Another study showed grafting of Solanum lycopersicum onto Solanum melongena reduces Cd accumulation in tomato by increasing the miR395b expression levels that target Phavoluta (PHV), Revolute (REV), Class III (HD-Zip), Sulfate transporter 2;1 (SULTR2;1), ATP sulfurylase (APS) (APS1/3), and High-affinity sulfate transporter (HAST) genes, which are directly or indirectly responsible for sulfate transport [Citation165]. The study reaffirms the correlation between Cd and sulfur accumulation in tomato scions. Two Cd responsive miRNAs, miR172b-3p and miR398-3p targeting copper chaperone for superoxide dismutase (ATCCS) and Fasciclin-Like Arabinogalactan-protein 9 (FLA9) respectively, were down-regulated in leaves of Brassica juncea under Cd stress [Citation166]. The study observed bra-miR172b-3p as a negative regulator that enhances inhibition of resistant responses against cd stress.
6.2 Combined stresses
Plants repetitively face environmental stresses whether in individual/multiple/combined form which is drastically reducing the fertile land and thus heightening the issues of loss in agricultural productivity . Looking at the preponderance of the evidence from the recent scientific literature, the niche of miRNAs is widespread in modulating the differential expression of stress responsive target genes, in particular, to the individual or multiple stresses across biota [Citation167–169]. However, till date the role of miRNA in plant growth and development during the combined stresses are not well understood and requires more studies. Identification and understanding the up/down-regulation of miRNAs to the combination of different stresses can be a challenging task but it is a new window for proper utilization of the biotechnological tools [Citation38,Citation109,Citation170,Citation171] in developing miRNA-mediated climate-resilient futuristic crops [Citation172]. In view of the above scenario, there are few reports utilizing various advent transgenic approaches to untangle the complex and multitude expression patterns of miRNAs and mRNAs in response to combined abiotic stresses in plants. Transgenic tobacco plant was generated expressing Zea mayszm-miR156for enhanced drought and salt stress tolerance using either cauliflower mosaic virus or stress inducible promoter [Citation173]. In another study, overexpression of Glycine maxmiR172c was attempted by engineering Arabidopsis thaliana plants in response to water deficit and salt tolerance. The resulting data identified gma-miR172c as a positive regulator in response to the combined stresses [Citation174]. In another genetic engineering approach using CRISPR/Cas9 knockout system technique, expression of Oryza sativa miR535 was studied in response to salt, drought, polyethylene glycol (PEG) and abscisic acid (ABA) stresses. It was shown that either knockout/inhibition results in tolerance of rice plants to the combined stresses and thus making it as a negative regulator of target gene expression in stressed rice plants [Citation153].
6.3 Climate change
Changing climate has emerged by inflating CO2 concentration causing greenhouse effect and subsequently increasing global ambient temperature. Atmospheric CO2 concentration is predicted to rise 550–700 ppm by 2050 and to 650–1,200 ppm by 2100, which will subsequently result in a global climatic warming of 1.1 to 6.4°C by the end of this century [Citation175]. Based on predicted climate change there will be a shift in the temperature and precipitation zones from the tropical area toward the polar caps, which will lead to an increase in crop yield in North-Western Europe and decrease the crop yield in the Mediterranean area [Citation8]. Therefore, for creating future climate proof crops it is essential to develop crops that sustain at elevated CO2 and variable temperature. Chilling and heat stress have generally negative impacts on yield and growth on crops. Heat stress causes an excessive increase in membrane fluidity, a disruption of protein function and turnover, and metabolic imbalances. Plant cells sense and respond to this change in ambient temperature using various molecular mechanisms and signaling pathways [Citation176]. miRNA induced cellular regulation is one such prominent mechanism currently widely manipulated for developing temperature resilient crops [Citation13]. In one such study, comparing miR156 overexpressing alfalfa plants with empty vector control under heat stress of 40°C revealed altered protein expression patterns under heat stress. Furthermore,miR156OE plants showed stress tolerance characteristics with unique protein expression patterns related to metabolism, photosynthesis, stress-response and plant defenses [Citation177]. Chilling and heating stress studies demonstrated the role of miR319 and its target GAMYB-like1 in tomato, potato and Solanum habrochaites [Citation178–180]. Overexpression of sha-miR319d by Shi et al [Citation181]conferred chilling and heat stress tolerance in tomato. Under stress conditions, sha-miR319dOE lines showed enhanced stress tolerance, including lower relative electrolyte leakage, malondialdehyde concentration, O2− generation as well as H2O2 concentration while higher chlorophyll contents and Fv/Fm values than wild-type plants. Furthermore, sha-miR319dOE lines showed curly leaf phenotypes and suppressed plant height suggesting potential role of miR319d in plant growth and development.
Along with overexpression strategies, synonymous mutation, amiRNA and knockdown strategies for miRNA mediated thermotolerance have also been used for crop improvement. For instance, Zhao et al. [Citation182] carried out a synonymous mutation in Growth Regulating Factor (GRF) 15 at the target site of miR396a in poplar that showed enhanced heat tolerance as well as increased photosynthesis. Transgenic PagGRF15 overexpression plants were created using primer mutation of six nucleotides at the sites targeted by miR396 in such a way that it won’t hamper the encoded amino acids sequence. Transgenic plants showed significantly higher transcript as well as protein levels of GRF15 than in the wild-type plants with significantly greater tolerance to heat stress and higher values of instantaneous WUE (photosynthesis/transpiration) throughout the course of the HS treatment. In parallel, under heat stress expression of miR160 and its precursors is also considerably increased with decrease in miR160 targets, ARF10, ARF16, and ARF17 [Citation183]. To investigate role of miR160 and its targets in Arabidopsis thaliana under heat stress, Lin et al. [Citation184] created transgenic lines of artificial miRNA (MIM160) that mimics an inhibitor of miR160 miRNA, T-DNA insertion mutants of miR160 targets and overexpressing miR160 precursor a (160OE). It was demonstrated that miR160OE lines showed improved seed germination and seedling survival under heat stress while those of MIM160 had reduced adaptation to heat.
In plant systems, ambient CO2 (Ca) uptake by stomata depends on the difference in concentration of ambient and internal CO2 (Ci) level and on stomatal conductance [Citation185]. Therefore, elevated CO2 levels due to climate change might raise photosynthetic rate and plant primary net production or biomass. However, in open chamber experiment with elevated CO2 and temperature, it was seen that grain yield increased but at the cost of decrease in concentration of grain NKP and protein content, although it has also been seen that C4 plants are less responsive to elevated CO2 than that of C3 plants [Citation186]. Hence it is imperative to develop crops that are better in fixing ambient CO2 along with increasing yield and quality to tackle yield and nutrient loss due to climate change. In this context, miRNA miR408 was overexpressed to improve photosynthesis in Arabidopsis thaliana, rice and tobacco [Citation187]. Overexpression resulted in increased copper content in the chloroplast, elevated abundance of plastocyanin, and an induction of photosynthetic genes. Along with this, miR408OE line also showed improved efficiency of irradiation utilization and the capacity for carbon dioxide fixation. Similar studies of overexpressing and/or down regulating miRNA for improving crop tolerance to climate change are cited in , while gives a workflow of developing miRNA-mediated bioengineered climate-resilient crops.
Table 2. Plant miRNAs and their exploration for engineering crops for conferring single and combined environmental stress tolerance
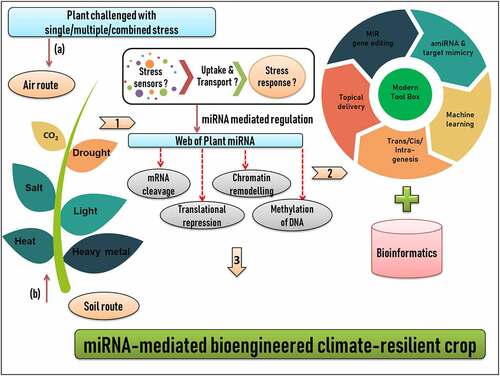
Figure 3. Workflow of developing miRNA-mediated bioengineered climate-resilient crops. (1) Plant growing in diverse stress environment conditions. In general, there are two common routes by which stress stimulus can reach the plant system; (a) Air route and (b) Soil route. Stress stimulus is sensed, perceived, uptake and transport by a complex set of cellular and molecular soldiers, which still demands for a critical mining (2) Identification and characterization of plant miRNAs using a technical blend of modern tool box and bioinformatics (3) Candidate miRNAs can be engineered for developing climate smart crops
7. Conclusion and future prospects
Climate change and environmental challenges severely hamper the crop production and thus global food security. As a common approach, the crops require an immediate yet effective genetic (or epigenetic) upgrade to thrive well and give better yields and vigor in the predicted challenging environmental conditions. Toward this, emphasis has been laid heavily on understanding and exploring the role of effective regulators of plant responses to stress factors, and the miRNAs are emerging as key signaling regulators for their differential gene expression in response to single/multiple/combined stress conditions. Thus, the target is to exploit the miRNAs/mRNAs and their potential interference in diverse cellular and molecular signaling cascades by utilizing genomics and other biotechnological tools. Recent developments in bioinformatics have enabled the identification, characterization and validation of data regarding miRNAs and their role in stress responses. Several microRNAs including miR408, miR156, miR393 are involved in various abiotic stresses. Additionally, considering the fact that plants suffer combined stresses in their native habitat, three miRNAs namely; miR156, miR172c and miR535 are shown to be involved conferring tolerance against multiple stress (Salt + drought, Water deficit + salt and Salt + drought + PEG + ABA) respectively. There is a renewed interest on finding the cross-talk associated microRNAs for combinational stresses, for example miRNAs responsive to CO2 and light stress. This opens up a new arena for research that needs to be explored to establish the data on miRNAs responsive to combination of stress factors. The algorithms established by bioinformatics researchers have paved the way for machine learning that has made studies on miRNAs faster, accurate and much more efficient. The advances in genome editing like CRISPR/cas9 have been highlighted for producing transgenic crops that are bioengineered to sustain harsh conditions. Techniques like artificial miRNA, topical delivery and target mimicry have been discussed as a means for crop improvement.
To mimic the changing climate conditions, researchers need to change their outlook in developing a single stress tolerant/resistant crop to the multiple stress tolerant crops. The signaling routes involved in modulating the miRNA directed multiple target genes should also be identified. A smart research shift is required wherein current artificial intelligence and machine learning based programs can be developed and explored for empowering the looming crop improvement. Broader applicability of the miRNA-based technology, public awareness about the dynamic climate change and linked practices, funding from the government bodies and landmark discoveries by the scientific community are the suggested steps to create miRNA based futuristic smart crops and a safe agro-ecosystem for all. The thrust of 3S- ‘SAFETY, SECURITY and SUSTAINABILITY’ activities happening globally are the obvious indications that mankind has accepted the interconnected impending climate changes and SDGs and is thus making themselves future ready by optimal utilization of tools leading to the development of miRNA-bioengineered climate resilient crops.
Acknowledgements
VK acknowledge the use of facilities created under DST-FIST (SR/ FST/COLLEGE-/19/568), DBT Star Status (BT/HRD/11/030/ 2012) and DBT-BUILDER Schemes implemented at Modern College of Arts, Science and Commerce, Ganeshkhind, Pune, India.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cubasch U, Wuebbles D, Chen D, et al. AR5 climate change 2013: the physical science basis — IPCC. United Kingdom and New York: Cambridge University Press, Cambridge; 2021. p. 1585.
- Shukla PR, Skea J, Buendia EC, et al. IPCC, 2019: climate change and land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems, [2019, accessed 2021 Aug 26]. https://www.ipcc.ch/site/assets/uploads/2019/11/SRCCL-Full-Report-Compiled-191128.pdf
- Daryanto S, Wang L, Jacinthe P-A. Global synthesis of drought effects on maize and wheat production. PLoS One. 2016;11:e0156362.
- Asseng S, Ewert F, Martre P, et al. Rising temperatures reduce global wheat production. Nat Clim Chang. 2015;5:143–147.
- Kim W, Iizumi T, Nishimori M. Global patterns of crop production losses associated with droughts from 1983 to 2009. J Appl Meteorol Climatol. 2019;58:1233–1244.
- Zhao C, Liu B, Piao S, et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci. 2017;114:9326–9331.
- Tito R, Vasconcelos HL, Feeley KJ. Global climate change increases risk of crop yield losses and food insecurity in the tropical Andes. Glob Chang Biol. 2018;24:e592–e602. Epub ahead of print 15 February 2018.
- Raza A, Razzaq A, Mehmood S, et al. Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants. 2019;8:34.
- Lobell DB. Climate change adaptation in crop production: beware of illusions. Glob Food Sec. 2014;3:72–76.
- Chaudhary S, Grover A, Sharma PC. MicroRNAs: potential targets for developing stress-tolerant crops. Life. 2021;11:289.
- Zhang B, Wang Q. MicroRNA, a new target for engineering new crop cultivars. Bioengineered. 2016;7:7–10.
- Wani SH, Kumar V, Khare T, et al. miRNA applications for engineering abiotic stress tolerance in plants. Biologia. 2020;75:1063–1081.
- Xu J, Hou QM, Khare T, et al. Exploring miRNAs for developing climate-resilient crops: a perspective review. Sci Total Environ. 2019;653:91–104.
- Chen X. Small RNAs and their roles in plant development. Epub ahead of print 2009. DOI: 10.1146/annurev.cellbio.042308.113417.
- Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203.
- Reis RS, Eamens AL, Waterhouse PM. Missing pieces in the puzzle of plant MicroRNAs. Trends Plant Sci. 2015;20:721–728.
- Kumar V, Khare T, Shriram V, et al. Plant small RNAs: the essential epigenetic regulators of gene expression for salt-stress responses and tolerance. Plant Cell Rep. 2018;37:61–75.
- Cedillo-Jiménez CA, Hernández-Salazar M, Escobar-Feregrino T, et al. MicroRNAs sequencing for understanding the genetic regulation of plant genomes. In: Plant genomics. InTech; 2016; p. 137–144.
- Sablok G, Pérez-Quintero ÁL, Hassan M, et al. Artificial microRNAs (amiRNAs) engineering – on how microRNA-based silencing methods have affected current plant silencing research. Biochem Biophys Res Commun. 2011;406:315–319.
- Zhou J, Deng K, Cheng Y, et al. CRISPR-Cas9 based genome editing reveals new insights into MicroRNA function and regulation in rice. Front Plant Sci. 2017:1598.DOI:10.3389/fpls.2017.01598.
- Kis A, Tholt G, Ivanics M, et al. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol Plant Pathol. 2016;17:427–437.
- Zhou J, Yuan M, Zhao Y, et al. Efficient deletion of multiple circle RNA loci by CRISPR‐Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol J. 2021;19:1240.
- United Nations. THE 17 GOALS, 2016. https://sdgs.un.org/goals.
- Noman A, Aqeel M. miRNA-based heavy metal homeostasis and plant growth. Environ Sci Pollut Res. 2017;24:10068–10082.
- Srivastava S, Suprasanna P. MicroRNAs: tiny, powerful players of metal stress responses in plants. Plant Physiol Biochem. 2021;166:928–938.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854.
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862.
- Shriram V, Kumar V, Devarumath RM, et al. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci. 2016;7. Epub ahead of print 14 June 2016. DOI:10.3389/fpls.2016.00817.
- Reinhart BJ. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626.
- Chinnusamy V, Zhu J, Zhou T, et al. Small Rnas: big role in abiotic stress tolerance of plants. In: Jenks M.A., Hasegawa P.M., Jain S.M. (eds) Advances in molecular breeding toward drought and salt tolerant crops. Dordrecht: Springer Netherlands; 2007; p. 223–260.
- Budak H, Akpinar BA. Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics. 2015;15:523–531.
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell. 2013;25:2383–2399.
- Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in arabidopsis. Annu Rev Plant Biol. 2014;65:473–503.
- Werner S, Wollmann H, Schneeberger K, et al. Structure determinants for accurate processing of miR172a in arabidopsis thaliana. Curr Biol. 2010;20:42–48.
- Chaves SS, Fernandes-Brum CN, Silva GFF, et al. New insights on coffea miRNAs: features and evolutionary conservation. Appl Biochem Biotechnol. 2015;177:879–908.
- Stepien A, Knop K, Dolata J, et al. Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip Rev RNA. 2017;8:e1403.
- Ding Y, Ding L, Xia Y, et al. Emerging roles of microRNAs in plant heavy metal tolerance and homeostasis. J Agric Food Chem. 2020;68:1958–1965.
- Zhou X, Khare T, Kumar V. Recent trends and advances in identification and functional characterization of plant miRNAs. Acta Physiol Plant. 2020;42:25.
- D’Ario M, Griffiths-Jones S, Kim M. Small RNAs: big impact on plant development. Trends Plant Sci. 2017;22:1056–1068.
- Patel P, Yadav K, Ganapathi TR, et al. Plant miRNAome: cross talk in abiotic stressful times. pp. 25–52.
- Khare T, Shriram V, Kumar V. RNAi technology: the role in development of abiotic stress-tolerant crops. In: Biochemical, physiological and molecular avenues for combating abiotic stress tolerance in plants. Elsevier, Academic Press; 2018; p. 117–133.
- Kumar V, Datir S, Khare T, Shriram V. Advances in biotechnological tools: improving abiotic stress tolerance in rice. In: Hasanuzzaman M., Fujita M., Nahar K., Biswas J. (eds) Advances in rice research for abiotic stress tolerance 2019; pp. 839-867. DOI:10.1016/B978-0-12-814332-2.00030-7. Duxford, United Kingdom: Woodhead Publishing.
- Si X, Wang Q, Zhang L, et al. Survey of gene splicing algorithms based on reads. Bioengineered. 2017;8:750–758.
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467.
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. pp. 499–560.
- Kulski JK. Next-generation sequencing — an overview of the history, tools, and “Omic” applications. In: Kulski J. (eds) Next generation sequencing - advances, applications and challenges. Epub ahead of print 14 January 2016; pp. 3-60. Intechopen. DOI:10.5772/61964.
- Lin SS, Chen Y, Lu MYJ. Degradome sequencing in plants. Methods Mol Biol. 2019;1932:197–213.
- Selvi A, Devi K, Manimekalai R, et al. High-throughput miRNA deep sequencing in response to drought stress in sugarcane. 3 Biotech. 2021;11:312.
- Xu J, Zheng Y, Pu S, et al. Third-generation sequencing found LncRNA associated with heat shock protein response to heat stress in Populus qiongdaoensis seedlings. BMC Genomics. 2020;21:1–14.
- Diao S, Ding X, Luan Q, et al. A complete transcriptional landscape analysis of Pinus elliottii Engelm. Using third-generation sequencing and comparative analysis in the Pinus phylogeny. Forests. 10. Epub ahead of print 2019. DOI:10.3390/f10110942.
- Samad AFA, Nazaruddin N, Murad AMA, et al. Deep sequencing and in silico analysis of small RNA library reveals novel miRNA from leaf Persicaria minor transcriptome. 3 Biotech. 2018;8. Epub ahead of print 2018. DOI:10.1007/s13205-018-1164-8.
- Prakash VN, Jadeja VJ. Identification and characterization of miRNA transcriptome in Jatropha curcas by high-throughput sequencing. Plant Genet. 2018;15:44–50.
- Bao H, Chen M, Chen H, et al. Transcriptome-wide identification of miRNA targets and a TAS3-homologous gene in Populus by degradome sequencing. Genes Genomics. 2019;41:849–861.
- Yu L, Zhou L, Liu W, et al. Identification of drought resistant miRNA in Macleaya cordata by high-throughput sequencing. Arch Biochem Biophys. 2020;684:108300.
- Zhang H, Liu X, Yang X, et al. miRNA–mRNA integrated analysis reveals roles for miRNAs in a typical halophyte, reaumuria soongorica, during seed germination under salt stress. Plants. 2020;9:1–16.
- Cai C, Li C, Sun R, et al. Small RNA and degradome deep sequencing reveals important roles of microRNAs in cotton (Gossypium hirsutum L.) response to root-knot nematode Meloidogyne incognita infection. Genomics. 2021;113:1146–1156.
- Zhang J, Xu C, Liu K, et al. Deep sequencing discovery and profiling of known and novel miRNAs produced in response to DNA damage in rice. Int J Mol Sci. 2021;22:9958.
- Wu Y, Zheng L, Bing J, et al. Deep sequencing of small RNA reveals the molecular regulatory network of AtENO2 regulating seed germination. Int J Mol Sci. 2021;22:5088.
- Jamla M, Khare T, Joshi S, et al. Omics approaches for understanding heavy metal responses and tolerance in plants. Curr Plant Biol. 2021;27:100213.
- Zhang Z, Yu J, Li D, et al. PMRD: plant microRNA database. Nucleic Acids Res. 2010;38:D806–D813.
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018;46:D239–D245.
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131.
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162.
- Szcześniak MW, Makałowska I. miRNEST 2.0: a database of plant and animal microRNAs. Nucleic Acids Res. 2014;42:D74–D77.
- Sun X, Dong B, Yin L, et al. PMTED: a plant microRNA target expression database. BMC Bioinformatics. 2013;14:174.
- Bonnet E, He Y, Billiau K, et al. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26:1566–1568.
- Zhang P, Meng X, Chen H, et al. PlantCircNet: a database for plant circRNA-miRNA-mRNA regulatory networks. Database (Oxford). 2017;2017:1–8.
- Zhang J, Hao Z, Yin S, et al. GreenCircRNA: a database for plant circRNAs that act as miRNA decoys. Database. 2020;2020:1–9.
- Lorenzetti APR, de Antonio GYA, Paschoal AR, et al. PlanTE-MIR DB: a database for transposable element-related microRNAs in plant genomes. Funct Integr Genomics. 2016;16:235–242.
- Guo Z, Kuang Z, Wang Y, et al. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020;48:D1114–D1121.
- Yu D, Lu J, Shao W, et al. MepmiRDB: a medicinal plant microRNA database. Database. 2019;2019:1–6.
- Liu J, Liu X, Zhang S, et al. TarDB: an online database for plant miRNA targets and miRNA-triggered phased siRNAs. BMC Genomics. 2021;22:348.
- Chen C, Li J, Feng J, et al. sRNAanno—a database repository of uniformly annotated small RNAs in plants. Hortic Res. 8. Epub ahead of print 2021. DOI:10.1038/s41438-021-00480-8.
- Vasconcelos AM, Carmo MB, Ferreira B, et al. IsomiR_window: a system for analyzing small-RNA-seq data in an integrative and user-friendly manner. BMC Bioinformatics. 2021;22:37.
- Huang H-Y, Lin Y-C-D, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. Epub ahead of print 24 October 2019. DOI:10.1093/nar/gkz896.
- Fei Y, Wang R, Li H, et al. DPMIND: degradome-based plant miRNA-target interaction and network database. Bioinformatics. 2018;34:1618–1620.
- Remita MA, Lord E, Agharbaoui Z, et al. A novel comprehensive wheat miRNA database, including related bioinformatics software. Curr Plant Biol. 2016;7–8:31–33.
- Zhang Y, Zang Q, Xu B, et al. IsomiR bank: a research resource for tracking IsomiRs. Bioinformatics. 2016;32:2069–2071.
- Li J-H, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97.
- Yi X, Zhang Z, Ling Y, et al. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2015;43:D982–D989.
- Zhang S, Yue Y, Sheng L, et al. PASmiR: a literature-curated database for miRNA molecular regulation in plant response to abiotic stress. BMC Plant Biol. 2013;13:33.
- Meng Y, Gou L, Chen D, et al. PmiRKB: a plant microRNA knowledge base. Nucleic Acids Res. 2011;39:D181–D187.
- Liu H, Jin T, Liao R, et al. miRFANs: an integrated database for Arabidopsis thaliana microRNA function annotations. BMC Plant Biol. 2012;12:68.
- Cagirici HB, Sen TZ, Budak H. mirMachine: a one-stop shop for plant miRNA annotation. J Vis Exp. 2021; Epub ahead of print 1 May 2021. doi:10.3791/62430
- Russell PH, Vestal B, Shi W, et al. miR-MaGiC improves quantification accuracy for small RNA-seq. BMC Res Notes. 2018;11:296.
- Kuang Z, Wang Y, Li L, et al. miRDeep-P2: accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics. 2019;35:2521–2522.
- Ma X, Liu C, Gu L, et al. TarHunter, a tool for predicting conserved microRNA targets and target mimics in plants. Bioinformatics. 2018;34:1574–1576.
- Vitsios DM, Kentepozidou E, Quintais L, et al. Mirnovo: genome-free prediction of microRNAs from small RNA sequencing data and single-cells using decision forests. Nucleic Acids Res. 2017;45:e177–e177.
- Yang K, Sablok G, Qiao G, et al. isomiR2Function: an integrated workflow for identifying MicroRNA variants in plants. Front Plant Sci. 8. Epub ahead of print 21 March 2017. DOI:10.3389/fpls.2017.00322.
- Tav C, Tempel S, Poligny L, et al. miRNAFold: a web server for fast miRNA precursor prediction in genomes. Nucleic Acids Res. 2016;44:W181–W184.
- Urgese G, Paciello G, Acquaviva A, et al. isomiR-SEA: an RNA-Seq analysis tool for miRNAs/isomiRs expression level profiling and miRNA-mRNA interaction sites evaluation. BMC Bioinformatics. 2016;17:148.
- Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466.
- Bandyopadhyay S, Ghosh D, Mitra R, et al. MBSTAR: multiple instance learning for predicting specific functional binding sites in microRNA targets. Sci Rep. 2015;5:8004.
- Zhang Z, Jiang L, Wang J, et al. MTide: an integrated tool for the identification of miRNA-target interaction in plants. Bioinformatics. 2015;31:290–291.
- An J, Lai J, Sajjanhar A, et al. miRPlant: an integrated tool for identification of plant miRNA from RNA sequencing data. BMC Bioinformatics. 2014;15:275.
- Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;39:W132–W138.
- Wang W-C, Lin F-M, Chang W-C, et al. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328.
- Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284.
- Wang X, Zhang J, Li F, et al. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614.
- Llave C, Kasschau KD, Rector MA, et al. Endogenous and silencing-associated small RNAs in plants[W]. Plant Cell. 2002;14:1605–1619.
- Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–3323.
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46:W49–W54.
- Rusinov V, Baev V, Minkov IN, et al. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:696–700.
- Patel P, Ramachandruni SD, Kakrana A, et al. miTRATA: a web-based tool for mi croRNA Tr uncation a nd T ailing A nalysis. Bioinformatics. 2016;32:450–452.
- Thody J, Moulton V, Mohorianu I. PAREameters: a tool for computational inference of plant miRNA-mRNA targeting rules using small RNA and degradome sequencing data. Nucleic Acids Res. 2020;48:2258–2270.
- Zhang T, Zhai J, Zhang X, et al. Interactive web-based annotation of plant MicroRNAs with iwa-miRNA. Genomics Proteomics Bioinformatics. Epub ahead of print July 2021. DOI:10.1016/j.gpb.2021.02.010.
- Wani SH, Sah SK, Hossain MA, Kumar V, Balachandran SM. Transgenic approaches for abiotic stress tolerance in crop plants. In: Al-Khayri J., Jain S., Johnson D. (eds) Advances in plant breeding strategies: agronomic, abiotic and biotic stress traits. Cham: Springer International Publishing. 2016: p. 345–396.
- Basso MF, Ferreira PCG, Kobayashi AK, et al. MicroRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol J. 2019;17:1482–1500.
- Mujjassim NE, Mallik M, Krishna Kumar Rathod N, et al. Cisgenesis and intragenesis a new tool for conventional plant breeding: a review. J Pharmacogn Phytochem. 2019;8:2485–2489.
- Rommens CM, Haring MA, Swords K, et al. The intragenic approach as a new extension to traditional plant breeding. Trends Plant Sci. 2007;12:397–403.
- Cunningham FJ, Goh NS, Demirer GS, et al. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 2018;36:882–897.
- Joga MR, Zotti MJ, Smagghe G, et al. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol. 2016;7. Epub ahead of print 17 November 2016. DOI:10.3389/fphys.2016.00553.
- McLoughlin AG, Wytinck N, Walker PL, et al. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci Rep. 2018;8:7320.
- Hendrix B, Zheng W, Bauer MJ, et al. Topically delivered 22 nt siRNAs enhance RNAi silencing of endogenous genes in two species. bioRxiv. Epub ahead of print 2020. DOI:10.1101/2020.04.20.052076.
- Zhang N, Zhang D, Chen SL, et al. Engineering artificial MicroRNAs for multiplex gene silencing and simplified transgenic screen. Plant Physiol. 2018;178:989–1001.
- Van Vu T, Nang Do V. Customization of artificial microRNA design. Methods Mol Biol. 2017;1509:235–243.
- Gualtieri C, Leonetti P, Macovei A. Plant miRNA cross-kingdom transfer targeting parasitic and mutualistic organisms as a tool to advance modern agriculture. Front Plant Sci. 2020;11:1–7.
- Reichel M, Millar AA. Specificity of plant microRNA target MIMICs: cross-targeting of miR159 and miR319. J Plant Physiol. 2015;180:45–48.
- Reichel M, Li Y, Li J, et al. Inhibiting plant microRNA activity: molecular SPONGE s, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J. 2015;13:915–926.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283.
- Castro Á, Quiroz D, Sánchez E, et al. Synthesis of an artificial Vitis vinifera miRNA 319e using overlapping long primers and its application for gene silencing. J Biotechnol. 2016;233:200–210.
- Von Born P, Bernardo-Faura M, Rubio-Somoza I. An artificial miRNA system reveals that relative contribution of translational inhibition to miRNA-mediated regulation depends on environmental and developmental factors in Arabidopsis thaliana. PLoS One. 2018;13:1–14.
- Shanmugavadivel PS, Prakash C, Amitha Mithra SV. Molecular approaches for dissecting and improving drought and heat tolerance in rice. In: Hasanuzzaman M., Fujita M., Nahar K., Biswas J. (eds) Advances in rice research for abiotic stress tolerance. Duxford, United Kingdom:Woodhead Publishing. 2019; p. 839–867.
- Jain M. Function genomics of abiotic stress tolerance in plants: a CRISPR approach. Front Plant Sci. 6. Epub ahead of print 27 May 2015. DOI:10.3389/fpls.2015.00375.
- Jaganathan D, Ramasamy K, Sellamuthu G, et al. CRISPR for crop improvement: an update review. Front Plant Sci. 2018;9. Epub ahead of print 17 July 2018. DOI:10.3389/fpls.2018.00985.
- Tiwari P, Khare T, Shriram V, et al. Plant synthetic biology for producing potent phyto-antimicrobials to combat antimicrobial resistance. Biotechnol Adv. 2021;48:107729.
- Perez Rojo F, Nyman RKM, Johnson AAT, et al. CRISPR-Cas systems: ushering in the new genome editing era. Bioengineered. 2018;9:214–221.
- Liu X, Wu S, Xu J, et al. Application of CRISPR/Cas9 in plant biology. Acta Pharm Sin B. 2017;7:292–302.
- Chung PJ, Chung H, Oh N, et al. Efficiency of recombinant CRISPR/rCas9-mediated miRNA gene editing in rice. Int J Mol Sci. 2020;21:1–14.
- Jayasundara S, Lokuge S, Ihalagedara P, et al. Machine learning for plant microRNA prediction: a systematic review, 2021. http://arxiv.org/abs/2106.15159.
- Asefpour Vakilian K. Machine learning improves our knowledge about miRNA functions towards plant abiotic stresses. Sci Rep. 2020;10:1–10.
- Stegmayer G, Di Persia LE, Rubiolo M, et al. Predicting novel microRNA: a comprehensive comparison of machine learning approaches. Brief Bioinform. 2019;20:1607–1620.
- Bugnon LA, Yones C, Milone DH, et al. Genome-wide discovery of pre-miRNAs: comparison of recent approaches based on machine learning. Brief Bioinform. 2021;22:1–8.
- Xuan P, Guo M, Liu X, et al. PlantMiRNAPred: efficient classification of real and pseudo plant pre-miRNAs. Bioinformatics. 2011;27:1368–1376.
- Wu Y, Wei B, Liu H, et al. MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinformatics. 2011;12:107.
- Bradley J-C, Samuel B. SMIRP-A systems approach to laboratory automation. JALA J Assoc Lab Autom. 2000;5:48–53.
- Rodziewicz P, Swarcewicz B, Chmielewska K, et al. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiol Plant. 2014;36:1–19.
- Kaur N, Kaur G, Pati PK. Deciphering strategies for salt stress tolerance in rice in the context of climate change. In: Hasanuzzaman M., Fujita M., Nahar K., Biswas J. (eds) Advances in rice research for abiotic stress tolerance. Duxford, United Kingdom: Woodhead Publishing. 2019; p. 113–132.
- Azad S, Rajeevan M. Possible shift in the ENSO-Indian monsoon rainfall relationship under future global warming. Sci Rep. 2016;6:3–8.
- Zhou S, Huang G, Huang P. Changes in the East Asian summer monsoon rainfall under global warming moisture budget decompositions and the sources of uncertainty. Clim Dyn. 2018;51:1363–1373.
- Hajyzadeh M, Turktas M, Khawar KM, et al. miR408 overexpression causes increased drought tolerance in chickpea. Gene. 2015;555:186–193.
- Hang N, Shi T, Liu Y, et al. Overexpression of Os-microRNA408 enhances drought tolerance in perennial ryegrass. Physiol Plant. 2021;172:733–747.
- Ferdous J, Whitford R, Nguyen M, et al. Drought-inducible expression of Hv-miR827 enhances drought tolerance in transgenic barley. Funct Integr Genomics. 2017;17:279–292.
- Visentin I, Pagliarani C, Deva E, et al. A novel strigolactone‐miR156 module controls stomatal behaviour during drought recovery. Plant Cell Environ. 2020;43:1613–1624.
- Wang K, Liu Y, Teng F, et al. Heterogeneous expression of Osa-MIR156bc increases abiotic stress resistance and forage quality of alfalfa. Crop J. 2021;9:1135–1144.
- Zhou Y, Liu W, Li X, et al. Integration of sRNA, degradome, transcriptome analysis and functional investigation reveals gma-miR398c negatively regulates drought tolerance via GmCSDs and GmCCS in transgenic Arabidopsis and soybean. BMC Plant Biol. 2020;20:190.
- Aamir M, Rai KK, Dubey MK, et al. Impact of climate change on soil carbon exchange, ecosystem dynamics, and plant-microbe interactions. Duxford, United Kingdom: Woodhead Publishing . 2019. DOI:10.1016/B978-0-12-816483-9.00020-7.
- Wani SH, Kumar V, Khare T, et al. Engineering salinity tolerance in plants: progress and prospects. Planta. 2020;251:76.
- Ma Y, Xue H, Zhang F, et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol J. 2021;19:311–323.
- Guo X, Niu J, Cao X. Heterologous expression of Salvia miltiorrhiza MicroRNA408 enhances tolerance to salt stress in Nicotiana benthamiana. Int J Mol Sci. 2018;19:3985.
- Faisal A-RM, Biswas S, Zerin T, et al. Downregulation of the DST transcription factor using artificial microRNA to increase yield, salt and drought tolerance in rice. Am J Plant Sci. 2017;8:2219–2237.
- Yue E, Cao H, Liu B. OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa. Plants. 2020;9:1337.
- Yang B, Tang J, Yu Z, et al. Light stress responses and prospects for engineering light stress tolerance in crop plants. J Plant Growth Regul. 2019;38:1489–1506.
- Kosobryukhov A, Khudyakova A, Kreslavski V. Impact of UV radiation on photosynthetic apparatus: adaptive and damaging mechanisms. In: Hasanuzzaman M. (eds) Plant ecophysiology and adaptation under climate change: mechanisms and perspectives I. Singapore: Springer. p. 555–576.
- Zhou B, Leng J, Ma Y, et al. BrmiR828 targets BrPAP1, BrMYB82, and BrTAS4 Involved in the light induced anthocyanin biosynthetic pathway in Brassica rapa. Int J Mol Sci. 2020;21:4326.
- Li T, Lian H, Li H, et al. HY5 regulates light‐responsive transcription of microRNA163 to promote primary root elongation in Arabidopsis seedlings. J Integr Plant Biol. 2021;63:1437–1450.
- Jalmi SK, Bhagat PK, Verma D, et al. Traversing the links between heavy metal stress and plant signaling. Front Plant Sci. 2018;9:1–21.
- Nechita C, Iordache AM, Lemr K, et al. Evidence of declining trees resilience under long term heavy metal stress combined with climate change heating. J Clean Prod. 2021;317:128428.
- Ozaki H, Watanabe I, Kuno K. Investigation of the heavy metal sources in relation to automobiles. Water Air Soil Pollut. 2004;157:209–223.
- Adamiec E, Jarosz-Krzemińska E, Wieszała R. Heavy metals from non-exhaust vehicle emissions in urban and motorway road dusts. Environ Monit Assess. 2016;188:369.
- Xu Z, Lei Y, Patel J. Bioremediation of soluble heavy metals with recombinant Caulobacter crescentus. Bioeng Bugs. 2010;1:207–212.
- Ding Y, Gong S, Wang Y, et al. MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 2018;177:1691–1703.
- Shen C. Overexpression of a novel microRNA IamiR-4-3p from water spinach (Ipomoea aquatica Forsk.) increased Cd toxicity and accumulation in Arabidopsis thaliana.
- He L, Wang H, Zhao Q, et al. Tomato grafting onto Torubamu (Solanum melongena): miR166a and miR395b reduce scion Cd accumulation by regulating sulfur transport. Plant Soil. 2020;452:267–279.
- Liu L, Yin H, Liu Y, et al. Analysis of cadmium-stress-induced microRNAs and their targets reveals bra-miR172b-3p as a potential Cd2+-specific resistance factor in Brassica juncea. Processes. 2021;9:1099.
- Manavella PA, Yang SW, Palatnik J. Keep calm and carry on: mi RNA biogenesis under stress. Plant J. 2019. tpj.14369. DOI:10.1111/tpj.14369.
- Demirel U, Morris WL, Ducreux LJM, et al. Physiological, biochemical, and transcriptional responses to single and combined abiotic stress in stress-tolerant and stress-sensitive potato genotypes. Front Plant Sci. 2020;11. Epub ahead of print 27 February 2020. DOI:10.3389/fpls.2020.00169.
- Raza A, Su W, Gao A, et al. Catalase (CAT) gene family in rapeseed (Brassica napus L.): genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int J Mol Sci. 2021;22:4281.
- Sanan-Mishra N, Tripathi A, Goswami K, et al. ARMOUR – a rice miRNA: mRNA interaction resource. Front Plant Sci. 2018;9. Epub ahead of print 8 May 2018. DOI:10.3389/fpls.2018.00602.
- Chen L, Heikkinen L, Wang C, et al. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019;20:1836–1852.
- Kamthan A, Chaudhuri A, Kamthan M, et al. Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci. 2015;6. Epub ahead of print 2 April 2015. DOI:10.3389/fpls.2015.00208.
- Kang T, Yu C-Y, Liu Y, et al. Subtly manipulated expression of ZmmiR156 in tobacco improves drought and salt tolerance without changing the architecture of transgenic plants. Front Plant Sci. 2019;10. Epub ahead of print 10 January 2020. DOI:10.3389/fpls.2019.01664.
- Li W, Wang T, Zhang Y, et al. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J Exp Bot. 2016;67:175–194.
- Hoegh-Guldberg O, Jacob D, Taylor M, et al. Impacts of 1.5°C Global Warming on Natural and Human Systems. In: global Warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty. 2019.
- Li B, Gao Z, Liu X, et al. Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock–induced gene expression in arabidopsis. Plant Cell. 2019;31:2353–2369.
- Arshad M, Puri A, Simkovich AJ, et al. Label-free quantitative proteomic analysis of alfalfa in response to microRNA156 under high temperature. BMC Genomics. 21. Epub ahead of print 1 December 2020. DOI:10.1186/s12864-020-07161-1.
- Cao X, Wu Z, Jiang F, et al. Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genomics. 2014;15:1130.
- Zhou R, Wang Q, Jiang F, et al. Identification of miRNAs and their targets in wild tomato at moderately and acutely elevated temperatures by high-throughput sequencing and degradome analysis. Sci Rep. 2016;6:1–13.
- Valiollahi E, Farsi M, Kakhki AM. Sly-miR166 and Sly-miR319 are components of the cold stress response in Solanum lycopersicum. Plant Biotechnol Rep. 2014;8:349–356.
- Shi X, Jiang F, Wen J, et al. Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum). BMC Plant Biol. 2019;19:1–17.
- Zhao Y, Xie J, Wang S, et al. Synonymous mutation in growth regulating factor 15 of miR396a target sites enhances photosynthetic efficiency and heat tolerance in poplar. J Exp Bot. 2021;72:4502–4519.
- Şanlı BA, Öztürk Gökçe ZN. Investigating effect of miR160 through overexpression in potato cultivars under single or combination of heat and drought stresses. Plant Biotechnol Rep. 2021;15:335–348.
- Lin J-S, Kuo -C-C, Yang I-C, et al. MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front Plant Sci. 9. Epub ahead of print 1 February 2018. DOI:10.3389/fpls.2018.00068.
- Saminathan T, Alvarado A, Lopez C, et al. Elevated carbon dioxide and drought modulate physiology and storage-root development in sweet potato by regulating microRNAs. Funct Integr Genomics. 2019;19:171–190.
- Abebe A, Pathak H, Singh SD, et al. Growth, yield and quality of maize with elevated atmospheric carbon dioxide and temperature in north-west India. Agric Ecosyst Environ. 2016;218:66–72.
- Pan J, Huang D, Guo Z, et al. Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J Integr Plant Biol. 2018;60:323–340.
- Zhang J-P, Yu Y, Feng Y-Z, et al. MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 2017;175:1175–1185.
- Wang W, Liu D, Chen D, et al. MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 2019;16:362–375.
- Yu Y, Ni Z, Wang Y, et al. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 2019;285:68–78.
- Sun M, Jing Y, Wang X, et al. Gma-miR1508a confers dwarfing, cold tolerance, and drought sensitivity in soybean. Mol Breed. 40. Epub ahead of print 2020. DOI:10.1007/s11032-020-01116-w.
- Chen L, Meng J, Luan Y. miR1916 plays a role as a negative regulator in drought stress resistance in tomato and tobacco. Biochem Biophys Res Commun. 2019;508:597–602.
- Zhao J, Yuan S, Zhou M, et al. Transgenic creeping bentgrass overexpressing Osa‐miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol J. 2019;17:233–251.
- Sun Z, Shu L, Zhang W, et al. Cca-miR398 increases copper sulfate stress sensitivity via the regulation of CSD mRNA transcription levels in transgenic Arabidopsis thaliana. PeerJ. 2020;8:e9105. Epub ahead of print 2020.
- Cui N, Sun X, Sun M, et al. Overexpression of OsmiR156k leads to reduced tolerance to cold stress in rice (Oryza Sativa). Mol Breed. 2015;35:214.
- Li Y, Li X, Yang J, et al. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat Commun. 2020;11:5351.
