ABSTRACT
Cerebral ischemia/reperfusion (CI/R) injury results in serious brain tissue damage, thereby leading to long-term disability and mortality. It has been reported that dexmedetomidine (DEX) exerted neuroprotective effects in CI/R injury. Herein, we intended to investigate whether and how circular RNA (circRNA) cerebellar degeneration-related protein 1 antisense RNA (circ-CDR1as) was involved in the DEX-mediated protection on hippocampal neurons. In our work, the mouse hippocampal neuronal cells (HT-22) were used to construct a hypoxia/reperfusion (H/R) model for CI/R injury. Cell proliferation and apoptosis were evaluated by CCK-8 and flow cytometry. Gene expressions were detected by RT-qPCR. Levels of pro-inflammatory cytokines (TNF-α, IL-6, and IL-1β) were measured by ELISA. The association between miR-28-3p and circ-CDR1as or TRAF3 was verified by dual-luciferase assay. The results indicated that DEX alleviated HT-22 cell dysfunction induced by H/R treatment. In addition, circ-CDR1as was downregulated after DEX treatment and reversed the effects of DEX on the proliferation, apoptosis, and inflammatory responses of H/R-treated HT-22 cells. Circ-CDR1as positively regulated TRAF3 expression via interaction with miR-28-3p in HT-22 cells. Circ-CDR1as aggravated H/R-treated HT-22 cell dysfunction through targeting miR-28-3p. Furthermore, TRAF3 inhibition partly abolished the effect of circ-CDR1as overexpression on cellular activities of H/R-treated HT-22 cells. To sum up, our findings, for the first time, demonstrated that DEX exerted neuroprotective effects on hippocampal neurons against H/R treatment via the circ-CDR1as/miR-28-3p/TRAF3 regulatory network, providing novel therapeutic targets for DEX administration in CI/R treatment.
Introduction
Stroke, an acute cerebrovascular disorder, is a leading cause of long-term disability and mortality in adults worldwide [Citation1]. Nearly 85% of stroke cases might be attributed to cerebral ischemia [Citation2,Citation3]. At present, the most effective method for ischemia stroke treatment in clinical practice is thrombolytic therapy through which blood supply to brain tissues can be timely restored, thereby ameliorating ischemic stroke-induced brain injury [Citation4]. However, sudden resumption of blood supply after cerebral ischemia may induce a succession of pathological reactions, such as aggravated apoptosis and inflammatory responses in neurons, and even cause secondary injury to local brain tissues, which is known as cerebral ischemia/reperfusion (I/R) injury [Citation5]. Although numerous drugs are of neuroprotective capability, many of them fail to therapeutic effects in cerebral I/R (CI/R) treatment [Citation6]. Hence, it is of great significance to find new therapeutic approaches for CI/R therapy.
Dexmedetomidine (DEX), an activator of Alpha2-adrenoceptor, is a widely applied anesthetic drug for sympathetic activity depression, analgesia, and sedation in clinical anesthesia, without causing respiratory depression [Citation7]. Besides, DEX also exerts essential pharmacological effects on reducing apoptosis, diminishing inflammation, and relieving neuropathic pain [Citation8]. For example, Kang et al. revealed that DEX eliminated diabetes-induced neuropathic pain in mice by regulating P2X4 and NLRP3 expressions [Citation9]. Li et al. found that DEX reduced renal I/R-induced apoptosis via the α2 Adrenoceptor/PI3K/Akt signaling [Citation10]. He et al. demonstrated that DEX alleviated doxorubicin-mediated apoptosis and inflammation of myocardial cells [Citation11]. Recently, there is a heated topic on DEX-mediated neuroprotection in CI/R [Citation12–14]. However, the pharmacological action of DEX in CI/R still needs further investigation. Hence, a deeper understanding of the underlying mechanisms of DEX in CI/R is imperative for the better improvement of DEX application in CI/R treatment.
Circular RNAs (circRNAs), a group of newly identified RNAs with covalent closed-loop structures, play vital roles in several diseases, including CI/R [Citation15–18]. To cite an instance, Zhang et al. disclosed circRNA CAMK4 aggravated CI/R injury by accelerating neuron cell death [Citation19]. Yang et al. revealed that circ_008018 exacerbated CI/R-induced neuronal cell apoptosis via regulating miR-99a [Citation20]. Liu et al. circ_002664 promoted CI/R-induced neuron cell apoptosis via regulating Herpud1 through interaction with miR-182-5p [Citation21]. As reported in a study by Quan et al., circRNA cerebellar degeneration-related protein 1 antisense (circ-CDR1as) was highly expressed in PD and cause cell damage in vitro [Citation22], indicating its promoting role in neurological disorder. Nevertheless, the functions of circ-CDR1as in CI/R remains poorly understood.
In this study, a hypoxia/reoxygenation (H/R)-induced neuronal cell model serves as an effective tool for the research on the cellular dysfunction caused by CI/R injury [Citation23]. It was hypothesized that DEX exerted protective effects on H/R-induced hippocampal neuron cells via regulating circ-CDR1as. Herein, we for the first time explored the specific role of circ-CDR1as/miR-28-3p/TRAF3 competing endogenous RNA (ceRNA) network in the DEX-mediated protection against H/R-induced hippocampal neuronal dysfunction, thereby providing novel molecular targets for CI/R treatment with DEX.
Materials and methods
Cell culture and DEX treatment
Mouse hippocampal neuronal cells (HT-22) purchased from BeNa Culture Collection (Beijing, China) were cultured in DMEM supplemented with 10% FBS in an incubator (5% CO2; 37°C) as per standard protocols. For DEX treatment, HT-22 cells were exposed to DEX (100 µM) for 24 h.
Establishment of hypoxia/reoxygenation (H/R) cell model
To construct an H/R cell model, HT-22 cells were cultivated in a low-oxygen atmosphere (94% N2 + 5% CO2 + 1% O2; 37°C) for 6 h. Thereafter, the HT-22 cells were transferred to fresh DMEM supplemented with 10% FBS and cultured in a regular incubator (95% O2 + 5% CO2; 37°C) for 6 h. Untreated HT-22 cells were used as the blank control group (Control group) [Citation24].
Cell transfection
Small interfering RNAs against circ-CDR1as (si-circ-CDR1as: 5′-UAAUGUGAGACGUCAUAGAAC-3′), TRAF3 (si-TRAF3: 5′-AUAGAGAAUAUAACCUGUCGA-3′), scramble control (si-NC: 5′-AUUGUAGAUAAUCAGCUUAAU-3′), pcDNA3.1 overexpression vector for circ-CDR1as (oe-circ-CDR1as), empty vector (Vector), miR-28-3p overexpression and inhibition plasmids (miR-28-3p mimics: 5′-UAGAUCACAGUCCUUUGUUAU-3′ and miR-28-3p inhibitor: 5′-AUCUAGUGUCAGGAAACAAAUA-3′), and corresponding negative controls (NC mimics: 5′-AUCUAGUCAGUCCUUUGUUUAU-3′ and NC inhibitor: 5′-AUCUAGUGUCAGGAAACAAAUA-3′) were provided by GenePharma (Shanghai, China) and transfected into HT-22 cells via Lipofectamine 2000 (Invitrogen, USA).
CCK-8 assay
CCK-8 assay was utilized for cell viability assessment. HT-22 cells were seeded into six-well plates (3 × 105 cells/well) and cultured for 24 h. Afterward, 10 μl CCK-8 reagent was added into each well. After incubation with CCK-8 reagent for 1 h, the absorbance (optical density) was measured with a microplate reader (Bio-Rad, USA) [Citation25].
Flow cytometry
Flow cytometry was applied for the analysis of HT-22 cell apoptosis via the Annexin V-FITC Apoptosis Detection Kit (eBioscience, USA). HT-22 cells in each group were centrifuged at 400 × g for 5 min, rinsed 3 times with PBS, and then incubated with 5 μL Annexin V-FITC reagent and 10 μL PI reagent (Sigma-Aldrich, USA) for 15 min at room temperature in darkness. The apoptotic HT-22 cells were analyzed with a flow cytometer (Beckman Coulter, China) [Citation26].
ELISA
In order to determine TNF-α, IL-6, and IL-1β levels in HT-22 cells, ELISA assays were performed with corresponding ELISA assay kits (Mlbio, China) according to the standard protocol [Citation27].
RT-qPCR
Isolation of total RNA from HT-22 cells was performed via TRIzol (Invitrogen). Then, cDNA was generated with a reverse transcriptase kit (Takara, Japan) or Thermo Fisher’s K1622 kit (Thermo Fisher Scientific, USA). Afterward, qPCR was accomplished with SYBR-Green PCR Master Mix kit (Takara, China). Relative gene expression was evaluated by 2−ΔΔCt method, with GAPDH or U6 as the internal reference [Citation28]. The primers used were as follows: circ-CDR1as forward (F): 5′-GTGTCTCCAGTGTATCGGCG-3′ and reverse (R): 5′-TACTGGCACCACTGGAAACC-3′; TRAF3 F: 5′-CTTCCCGGGCTGTGATATTG-3′ and R: 5′-GGCTGTATCTGACCGCTAGG-3′; GAPDH F: 5′-ACCCACTCCTCCACCTTTGAC-3′ and R: 5′-TGTTGCTGTAGCCAAATTCGTT-3′; miR-28-3p F: 5′-CGCGCACTAGATTGTGAGCT-3′ and R: 5′-AGTGCAGGGTCCGAGGTATT-3′; U6 F: 5′-CTCGCTTCGGCAGCACATATACT-3′ and R: 5′-ACGCTTCACGAATTTGCGTGTC-3′.
Dual-luciferase assay
The 3′ UTR regions of TRAF3 and circ-CDR1as with binding or mutant sequences for miR-28-3p were cloned into pmirGLO luciferase vectors (Promega, USA) to synthesize wild-type plasmids (TRAF3-WT and circ-CDR1as-WT) or mutant-type plasmids (TRAF3-MUT and circ-CDR1as-MUT). The above plasmids were respectively transfected into HT-22 cells, together with NC mimics or miR-28-3p with Lipofectamine 2000 (Invitrogen). 48 h later, the luciferase activity was determined via the dual-luciferase reporter assay kit (Promega, USA) and normalized to Renilla luciferase activity.
Statistical analysis
The data were expressed as mean ± standard deviation (SD). Each experiment was conducted three times. Student’s t test or one-way ANOVA was applied to accomplish difference comparison between two or multiple groups. Statistical analysis was performed using GraphPad Prism 6.0. A difference with P < 0.05 was deemed statistically significant.
Results
In this work, we intended to investigate the role and molecular mechanism of DEX in H/R-challenged hippocampal neuronal dysfunction. Our results demonstrated that DEX exerted neuroprotective effects on hippocampal neurons against H/R treatment via the circ-CDR1as/miR-28-3p/TRAF3 regulatory network, providing novel therapeutic targets for DEX administration in CI/R treatment.
DEX relieves HT-22 cell dysfunction induced by H/R treatment
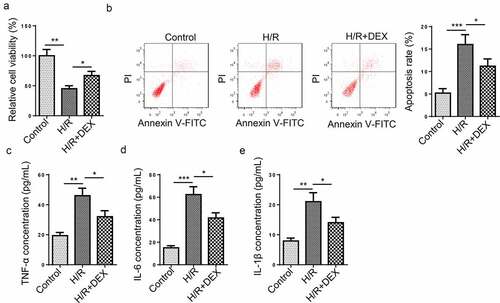
To discover the possible effects of DEX treatment on H/R-induced HT-22 cells, we treated HT-22 cells with DEX after H/R treatment. As shown in ), HT-22 cell viability prominently dropped after H/R treatment, while DEX treatment everted an ameliorative effect on cell viability. In addition, it was discovered that HT-22 cell apoptosis rate was significantly increased after H/R treatment, while DEX led to an opposite result ()), indicating that DEX reduced cell apoptosis. Moreover, the TNF-α, IL-6, and IL-1β levels were increased after H/R treatment, whereas DEX remarkably reversed such a phenomenon (). To sum up, DEX relieved H/R-induced apoptosis and inflammatory responses in HT-22 cells.
Figure 1. DEX relieves HT-22 cell dysfunction induced by H/R treatment. (a) Relative cell viability after H/R injury and DEX treatment was measured using CCK-8 assay. (b) Cell apoptosis rate after H/R injury and DEX treatment was measured by flow cytometry. (c-e) TNF-α (d), IL-6 (e), and IL-1β (f) levels were measured by ELISA. *P < 0.05; **P < 0.01; ***P < 0.001
Circ-CDR1as is down-regulated after DEX treatment and reverses the effects of DEX on H/R-treated HT-22 cell proliferation, apoptosis, and inflammation
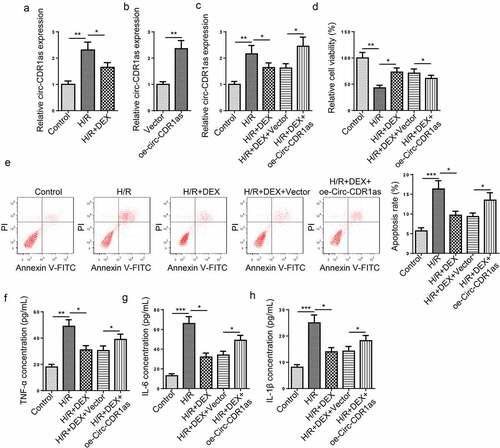
It was found that circ-CDR1as expression was markedly increased in HT-22 cells subject to H/R treatment; on the contrary, DEX led to down-regulated circ-CDR1as expression ()). To further discover the potential functions of circ-CDR1as in DEX-mediated neuroprotection against H/R treatment, cell viability, apoptosis, and inflammatory responses of HT-22 cells were detected in each group. Firstly, circ-CDR1as was overexpressed in HT-22 cells ()). As indicated by RT-qPCR assay, circ-CDR1as expression was evidently up-regulated by H/R, remarkably decreased by DEX, and increased again by circ-CDR1as overexpression ()). Functional assays exhibited that circ-CDR1as upregulation reversed the effect of DEX on cellular processes of HT-22 cells, as indicated by decreased cell viability, elevated apoptotic rate, as well as increased inflammatory responses ().
Figure 2. Circ-CDR1as is down-regulated after DEX treatment and reverses the effects of DEX on H/R-treated HT-22 cell proliferation, apoptosis, and inflammation. (a) Circ-CDR1as expression in HT-22 cells after H/R injury and DEX treatment was detected via RT-qPCR. (b) Circ-CDR1as overexpression efficiency was evaluated by RT-qPCR. (c) Circ-CDR1as expression in Control group, H/R group, H/R+ DEX group, H/R+ DEX+Vector group, or H/R+ DEX+oe-circ-CDR1as group was evaluated by RT-qPCR. (d) cell viability was measured using CCK-8 assay. (e) Cell apoptosis rate after H/R injury and DEX treatment was measured by flow cytometry. (f-h) TNF-α (g), IL-6 (h), and IL-1β (i) levels were measured by ELISA. *P < 0.05; **P < 0.01; ***P < 0.001
MiR-28-3p directly binds to circ-CDR1as
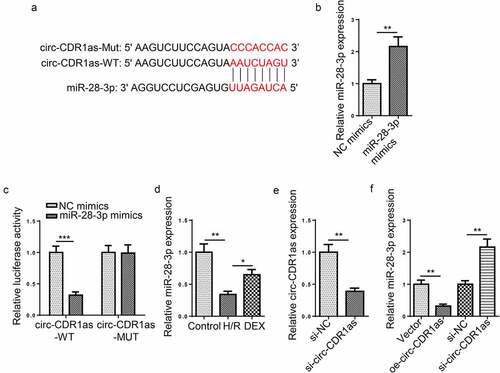
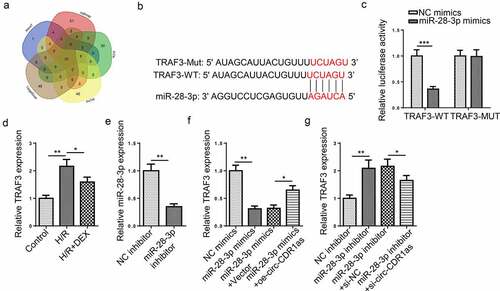
StarBase predicted that circ-CDR1as contains a latent binding region for miR-28-3p ()). The dual-luciferase reporter assay was performed to verify the binding condition between miR-28-3p and circ-CDR1as. Firstly, miR-28-3p was overexpressed in HT-22 cells ()). The results showed that miR-28-3p upregulation inhibited the luciferase activity of circ-CDR1as-WT but almost had no effect on the luciferase activity of circ-CDR1as-MUT ()). RT-qPCR results revealed that miR-28-3p was lowly expressed in H/R-challenged HT-22 cells, while DEX upregulated miR-28-3p expression ()). Next, circ-CDR1as was knockdown in HT-22 cells ()). Furthermore, miR-28-3p expression was downregulated or overexpressed in HT-22 cells after circ-CDR1as overexpression or knockdown, respectively ()). These data showed that circ-CDR1as negatively regulated miR-28-3p expression.
Figure 3. MiR-28-3p directly binds to circ-CDR1as. (a) Binding site between circ-CDR1as and miR-28-3p. (b) MiR-28-3p overexpression efficiency was evaluated by RT-qPCR. (c) The binding relationship between circ-CDR1as and miR-28-3p was verified by dual-luciferase activity assay. (d) MiR-28-3p expression in HT-22 cells after H/R injury and DEX treatment was detected via RT-qPCR. (e) Circ-CDR1as knockdown efficiency was evaluated by RT-qPCR. (f) MiR-28-3p expression in HT-22 cells transfected with Vector, oe-circ-CDR1as, si-NC, or si-circ-CDR1as was detected by RT-qPCR. *P < 0.05; **P < 0.01; ***P < 0.001
Circ-CDR1as expedited apoptosis and inflammatory responses of H/R-treated HT-22 cells via regulating miR-28-3p
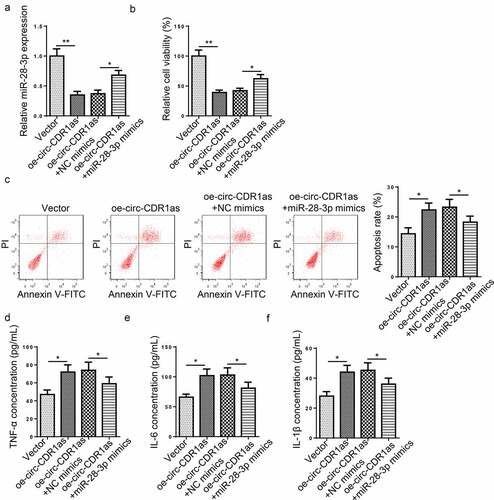
To explore the regulating role of circ-CDR1as and miR-28-3p in H/R-induced HT-22 cell dysfunction, H/R-treated HT-22 cells were transfected with Vector, oe-circ-CDR1as, oe-circ-CDR1as+NC mimics, or oe-circ-CDR1as+miR-28-3p mimics, respectively. RT-qPCR assay revealed that miR-28-3p expression was decreased after circ-CDR1as overexpression and then increased after miR-28-3p addition ()). Next, we measured cell viability, apoptotic rate, and levels of pro-inflammatory cytokines in each group. It was revealed that circ-CDR1as upregulation significantly inhibited proliferation and increased apoptosis of H/R-treated HT-22 cells; while miR-28-3p upregulation partly reversed such phenomena (). In addition, the TNF-α, IL-6, and IL-1β levels in H/R-treated HT-22 cells were remarkably lifted after circ-CDR1as amplification and declined after miR-28-3p overexpression (). Taken together, circ-CDR1as exacerbated H/R-induced apoptosis and inflammatory responses via miR-28-3p in HT-22 cells.
Figure 4. Circ-CDR1as expedited apoptosis and inflammatory responses of H/R-treated HT-22 cells via regulating miR-28-3p. (a) HT-22 cells were subject to H/R treatment and then transfected with Vector, oe-circ-CDR1as, oe-circ-CDR1as+NC mimics, or oe-circ-CDR1as+miR-28-3p mimics, with untreated HT-22 cells as the Control group. MiR-28-3p expression in each group was evaluated by RT-qPCR. (b) cell viability was measured using CCK-8 assay. (c) Cell apoptosis rate after H/R injury and DEX treatment was measured by flow cytometry. (d-f) TNF-α (e), IL-6 (f), and IL-1β (g) levels were measured by ELISA. *P < 0.05; **P < 0.01
MiR-28-3p targets TRAF3
Via 5 miRNA databases (microT, miRmap, PITA, PicTar, and TargetScan), 10 candidate downstream genes (MARCH6, RNF216, FAF2, ARF6, ZFP91, C15orf48, FUBP3, FAM168B, NR3C2, and TRAF3) were predicted for miR-28-3p ()). As Yao et al. disclosed that TRAF3 (tumor necrosis factor receptor-associated factor-3) expression was upregulated in neurons from hippocampus after cerebral ischemia [Citation29], TRAF3 was selected for subsequent experiments. The putative binding site between miR-28-3p and 3′UTR of TRAF3 was presented in ). Dual-luciferase activity assay manifested that miR-28-3p upregulation remarkably reduced the luciferase activity of TRAF3-WT, while the luciferase activity of TRAF3-MUT was nearly unchanged ()). RT-qPCR revealed that TRAF3 expression in HT-22 cells were significantly lifted after H/R and substantially declined after DEX treatment ()). Then, the efficiency of miR-28-3p inhibition was detected via RT-qPCR ()). TRAF3 expression were apparently decreased after miR-28-3p addition and distinctly increased after miR-28-3p inhibition; however, such phenomena were partly abrogated by circ-CDR1as amplification or circ-CDR1as silencing, respectively (). These results indicated that circ-CDR1as positively regulated TRAF3 expression in HT-22 cells via interaction with miR-28-3p, suggesting that circ-CDR1as exerting its effects via the miR-28-3p/TRAF3 pathway.
Figure 5. MiR-28-3p targets TRAF3. (a) Venn diagram of target mRNAs for miR-28-3p predicted by 5 databases (microT, miRmap, PITA, PicTar, and TargetScan). (b) Binding site between TRAF3 and miR-28-3p. (c) The binding relationship between TRAF3 and miR-28-3p was verified by dual-luciferase activity assay. (d) TRAF3 expression in HT-22 cells after H/R injury and DEX treatment were detected via RT-qPCR. (e) MiR-28-3p knockdown efficiency was evaluated by RT-qPCR. (f) TRAF3 expression in HT-22 cells transfected with NC mimics, miR-28-3p mimics, miR-28-3p mimics+Vector, or miR-28-3p mimics+oe-circ-CDR1as were detected by RT-qPC. (g) TRAF3 expression in HT-22 cells transfected with NC inhibitor, miR-28-3p inhibitor, miR-28-3p inhibitor+si-NC, or miR-28-3p inhibitor+si-circ-CDR1as were detected by RT-qPCR. *P < 0.05; **P < 0.01; ***P < 0.001
TRAF3 knockdown reverses the effect of circ-CDR1as overexpression on H/R-challenged HT-22 cells
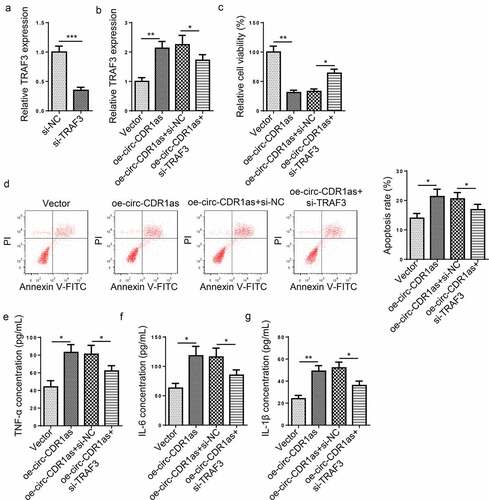
To further probe the role of TRAF3 in H/R-mediated damage to HT-22 cells, H/R-induced HT-22 cells were transfected with Vector, oe-circ-CDR1as, oe-circ-CDR1as+si-NC, and oe-circ-CDR1as+si-TRAF3, respectively. First of all, TRAF3 was knocked down in H/R-induced HT-22 cells ()). As indicated by RT-qPCR, TRAF3 expression was significantly elevated after circ-CDR1as addition, but declined after TRAF3 depletion ()). Functional assays exhibited that TRAF3 knockdown partly neutralized the effects of circ-CDR1as overexpression on viability, apoptosis, and inflammatory responses of HT-22 cells subject to H/R treatment ().
Figure 6. TRAF3 knockdown reverses the effect of circ-CDR1as overexpression on H/R-challenged HT-22 cells. (a) TRAF3 knockdown efficiency was evaluated by RT-qPCR. (b) HT-22 cells were subject to H/R treatment and then transfected with Vector, oe-circ-CDR1as, oe-circ-CDR1as+si-NC, and oe-circ-CDR1as+si-TRAF3, with untreated HT-22 cells as the Control group. TRAF3 mRNA and protein levels in HT-22 cells from each group were detected by RT-qPCR. (c) cell viability was measured using CCK-8 assay. (d) Cell apoptosis rate after H/R injury and DEX treatment was measured by flow cytometry. (e-g) TNF-α (f), IL-6 (g), and IL-1β (h) levels were measured by ELISA. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
DEX has not only effects on the hippocampus but also sensory processing, which involves the sensory cortex [Citation30]. Moreover, DEX significantly regulates the thalamus, the pulvinar nucleus in particular [Citation31], which plays a vital role in visual and auditory processing [Citation32,Citation33]. Moreover, accumulating evidence shows that DEX exerts significant neuroprotective effects in neurological diseases [Citation34–36]. In this work, the protective effect and regulatory mechanism of DEX on H/R-induced HT-22 cell dysfunction were further investigated. Our findings substantiated that DEX considerably attenuated H/R-induced apoptosis and inflammation in hippocampal neurons via the circ-CDR1as/miR-28-3p/TRAF3 cascade.
It has been widely recognized that circRNAs play crucial roles in the development and progression of neurological diseases, such as cerebral I/R [Citation37], Alzheimer’s disease (AD) [Citation38], Parkinson’s disease (PD) [Citation39], and Hirschsprung’s disease (HSCR) [Citation40]. Circ-CDR1as, also known as ciRS-7, is deeply involved in the biological processes of cells, including cell viability and apoptosis. For example, Mao et al. found that circ-CDR1as inhibited the proliferation of bone microvascular endothelial cells by regulating FIH-1 via interaction with miR-135b [Citation41]. Geng et al. revealed that circ-CDR1as contributed to apoptosis of hypoxia-treated mouse cardiac myocytes [Citation42]. Besides, Zhang et al. uncovered that circ-CDR1as also exacerbated inflammatory responses [Citation43]. In this study, we found that DEX exerted protective effects on hippocampal neuronal cells against H/R treatment, which was closely related to the downregulation of circ-CDR1as expression. However, circ-CDR1as overexpression could markedly weaken the protective effect of DEX on H/R-induced apoptosis and inflammation in HT-22 cells. Hence, DEX could attenuate H/R-induced dysfunctions in hippocampal neurons via down-regulating circ-CDR1as.
Emerging evidence has demonstrated that circRNAs can post-transcriptionally regulate the transcription and translation of messenger RNAs (mRNAs) as endogenous competitive RNAs for specific microRNAs (miRNAs) [Citation44]. Moreover, such a circRNA-miRNA-mRNA regulating network also exerts key effects in diverse neurological disorders, including cerebral I/R [Citation39,Citation45,Citation46]. Interestingly, our data demonstrated that circ-CDR1as sponged miR-28-3p as a ceRNA and miR-28-3p targeted TRAF3. Previous studies demonstrated that miRNAs also play important roles in CI/R-induced nerve damage. For instance, a report from Ma et al. showed that miRNA-589 protected against CI/R-induced inflammatory responses in primary cortical neurons via mediating TRAF6 [Citation47]. Xing et al. disclosed that miR-374 targeted WNT5A to alleviate CI/R injury [Citation48]. Besides, Liu et al. revealed that miR-211 exerted protective effects on OGD/R-challenged PC12 cells via reducing cell apoptotic rate [Citation49]. MiR-28-3p is differently regulated in human diseases and deeply involved in the regulation of cellular functions [Citation50]. In addition, Fan et al. disclosed that miR-28-3p expression was negatively related to IL-1β level in colorectal cancer [Citation51]. In the present study, we found that miR-28-3p was downregulated in H/R-induced HT-22 cells, while DEX increased miR-28-3p expression. Further experiments revealed that miR-28-3p could partially abolish the effects of circ-CDR1as overexpression on H/R-induced HT-22 cells by enhancing proliferation, reducing apoptosis, and impairing the secretion of pro-inflammatory cytokines. Therefore, circ-CDR1as aggravated cellular dysfunction of H/R-induced HT-22 cells via interaction with miR-28-3p.
As a member of the TRAF adaptor protein family, TRAF3 exerts critical functions in regulating cellular activities in multiple diseases [Citation52]. Sun et al. disclosed that TRAF3 upregulation substantially enhanced the inflammatory responses of caerulein-induced AR42J cells [Citation53]. Liu et al. revealed that TRAF3 promoted apoptosis and inflammatory responses of oxygen-glucose deprivation/reperfusion (OGD/R)-induced PC12 cells [Citation54]. Zhang et al. found that TRAF3 impaired the proliferation of MDA-MB-231 cells via inhibiting miR-29b-3p [Citation55]. Also, Liu et al. demonstrated that TRAF3 aggravated cardiac I/R-induced apoptosis and inflammation [Citation56]. Consistent with the above findings, TRAF3 expression was significantly increased in HT-22 cells after H/R treatment and remarkably after DEX administration. Rescue experiments showed that the promoting effects of circ-CDR1as upregulation on cellular dysfunction of H/R-treated HT-22 cells were partly abrogated by TRAF3 silencing. Taken together, circ-CDR1as upregulated TRAF3 expression, thereby promoting apoptosis and inflammatory responses in H/R-treated HT-22 cells.
Conclusion
In summary, this work demonstrated that DEX exerted neuroprotective effects against H/R-induced HT-22 cell dysfunction through regulating the circ-CDR1as/miR-28-3p/TRAF3 cascade. This study explored the neuroprotective effects and potential mechanisms of DEX in hippocampal neuron damage induced by cerebral I/R, providing a theoretical basis and certain targets for DEX application in cerebral I/R. In the future, in vivo experiments should be performed to further confirm the role of circ-CDR1as/miR-28-3p/TRAF3 axis in CI/R injury.
Research highlights
1. DEX attenuated H/R-induced dysfunctions of HT-22 cells by regulating circ-CDR1as
2.circ-CDR1as upregulated TRAF3 expression by targeting miR-28-3p
3.circ-CDR1as promoted dysfunction of H/R-induced HT-22 cells via miR-28-3p/TRAF3 axis
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Yao X, Yao R, Huang F, et al. LncRNA SNHG12 as a potent autophagy inducer exerts neuroprotective effects against cerebral ischemia/reperfusion injury. Biochem Biophys Res Commun. 2019;514(2):490–496.
- Clausen BH, Degn M, Martin NA, et al. Systemically administered anti-TNF therapy ameliorates functional outcomes after focal cerebral ischemia. J Neuroinflammation. 2014;11(1):203.
- Tobin MK, Bonds JA, Minshall RD, et al. Neurogenesis and inflammation after ischemic stroke: what is known and where we go from here. J Cereb Blood Flow Metab. 2014;34(10):1573–1584.
- Xu Q, Deng F, Xing Z, et al. Long non-coding RNA C2dat1 regulates CaMKIIdelta expression to promote neuronal survival through the NF-kappaB signaling pathway following cerebral ischemia. Cell Death Dis. 2016;7:e2173.
- Gomis M, Davalos A. Recanalization and reperfusion therapies of acute ischemic stroke: what have we learned, what are the major research questions, and where are we headed? Front Neurol. 2014;5:226.
- Chamorro ÁA, Dirnagl U, Urra X, et al. Neuroprotection in acute stroke: targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 2016;15(8):869–881.
- Jiang L, Hu M, Lu Y, et al. The protective effects of dexmedetomidine on ischemic brain injury: a meta-analysis. J Clin Anesth. 2017;40:25–32.
- Liu C, Fu Q, Mu R, et al. Dexmedetomidine alleviates cerebral ischemia-reperfusion injury by inhibiting endoplasmic reticulum stress dependent apoptosis through the PERK-CHOP-Caspase-11 pathway. Brain Res. 2018;1701:246–254.
- Kang L, Yayi H, Fang Z, et al. Dexmedetomidine attenuates P2X4 and NLRP3 expression in the spine of rats with diabetic neuropathic pain. Acta Cir Bras. 2019;34(11):e201901105.
- Li J, Chen Q, He X, et al. Dexmedetomidine attenuates lung apoptosis induced by renal ischemia-reperfusion injury through alpha2AR/PI3K/Akt pathway. J Transl Med. 2018;16(1):78.
- He Y, Yang Z, Li J, et al. Dexmedetomidine reduces the inflammation and apoptosis of doxorubicin-induced myocardial cells. Exp Mol Pathol. 2020;113:104371.
- Zhai Y, Zhu Y, Liu J, et al. Dexmedetomidine post-conditioning alleviates cerebral ischemia-reperfusion injury in rats by inhibiting high mobility group protein B1 group (HMGB1)/toll-like receptor 4 (TLR4)/nuclear factor kappa B (NF-kappaB) signaling pathway. Med Sci Monit. 2020;26:e918617.
- Wang Z, Zhou W, Dong H, et al. Dexmedetomidine pretreatment inhibits cerebral ischemia/reperfusioninduced neuroinflammation via activation of AMPK. Mol Med Rep. 2018;18:3957–3964.
- Zhao B, Li D, Zhang S, et al. Dexmedetomidine attenuates cerebral ischemia-reperfusion injury in rats by inhibiting the JNK pathway. Ann Palliat Med. 2021;10(6):6768–6778.
- Ou R, Mo L, Tang H, et al. circRNA-AKT1 sequesters miR-942-5p to upregulate AKT1 and promote cervical cancer progression, molecular therapy. Nucleic Acids. 2020;20:308–322.
- Han B, Zhang Y, Zhang Y, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142 -TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14(7):1164–1184.
- Sudhakara Prasad K, Cao X, Gao N, et al. A low-cost nanomaterial-based electrochemical immunosensor on paper for high-sensitivity early detection of pancreatic cancer. Sens Actuators B Chem. 2020;305:127516.
- Shin Low S, Pan Y, Ji D, et al. Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept. Sens Actuators B Chem. 2020;308:127718.
- Zhang ZH, Wang YR, Li F, et al. Circ-camk4 involved in cerebral ischemia/reperfusion induced neuronal injury. Sci Rep. 2020;10(1):7012.
- Yang X, Ji H, Yao Y, et al. Downregulation of circ_008018 protects against cerebral ischemia-reperfusion injury by targeting miR-99a. Biochem Biophys Res Commun. 2018;499:758–764.
- Liu C, Xu X, Huang C, et al. Circ_002664/miR-182-5p/Herpud1 pathway importantly contributes to OGD/R-induced neuronal cell apoptosis. Mol Cell Probes. 2020;53:101585.
- Quan H, Chen Q, Wang K, et al. Exendin-4 reversed the PC12 cell damage induced by circRNA CDR1as/miR-671/GSK3β signaling pathway. J Mol Neurosci. 2021;MN 71(4):778–789.
- Ryou MG, Liu R, Ren M, et al. Pyruvate protects the brain against ischemia-reperfusion injury by activating the erythropoietin signaling pathway. Stroke. 2012;43(4):1101–1107.
- Li H, Yao C, Shi K, et al. Astragaloside IV attenuates hypoxia/reoxygenation injury-induced apoptosis of type II alveolar epithelial cells through miR-21-5p. Bioengineered. 2021;12(1):7747–7754.
- Yao Y, Li X, Cheng L, et al. Circular RNA FAT atypical cadherin 1 (circFAT1)/microRNA-525-5p/spindle and kinetochore-associated complex subunit 1 (SKA1) axis regulates oxaliplatin resistance in breast cancer by activating the notch and Wnt signaling pathway. Bioengineered. 2021;12(1):4032–4043.
- Guohua H, Hongyang L, Zhiming J, et al. Study of small-cell lung cancer cell-based sensor and its applications in chemotherapy effects rapid evaluation for anticancer drugs. Biosens Bioelectron. 2017;97:184–195.
- Li J, Deng Q, Fan W, et al. Melatonin-induced suppression of DNA methylation promotes odontogenic differentiation in human dental pulp cells. Bioengineered. 2020;11(1):829–840.
- Yu L, Gong X, Sun L, et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PloS One. 2016;11(7):e0158347.
- Yao S, Tang B, Li G, et al. miR-455 inhibits neuronal cell death by targeting TRAF3 in cerebral ischemic stroke. Neuropsychiatr Dis Treat. 2016;12:3083–3092.
- Ibrahim LA, Mesik L, Ji XY, et al. Cross-modality sharpening of visual cortical processing through layer-1-mediated inhibition and disinhibition. Neuron. 2016;89:1031–1045.
- Coull JT, Jones ME, Egan TD, et al. Attentional effects of noradrenaline vary with arousal level: selective activation of thalamic pulvinar in humans. Neuroimage. 2004;22(1):315–322.
- Fang Q, Chou XL, Peng B, et al. Circuit via retino-colliculo-pulvinar pathway enhances feature selectivity in visual cortex through surround suppression. Neuron. 2020;105(2):355–369 e356.
- Chou XL, Fang Q, Yan L, et al. Contextual and cross-modality modulation of auditory cortical processing through pulvinar mediated suppression. Elife. 2020;9. DOI:10.7554/eLife.54157
- Sun W, Zhao J, Li C. Dexmedetomidine provides protection against hippocampal neuron apoptosis and cognitive impairment in mice with Alzheimer’s Disease by mediating the miR-129/YAP1/JAG1 axis. Mol Neurobiol. 2020;57(12):5044–5055.
- Chen L, Cao J, Cao D, et al. Protective effect of dexmedetomidine against diabetic hyperglycemia-exacerbated cerebral ischemia/reperfusion injury: an in vivo and in vitro study. Life Sci. 2019;235:116553.
- Zhang P, Li Y, Han X, et al. Dexmedetomidine regulates 6-hydroxydopamine-induced microglial polarization. Neurochem Res. 2017;42(5):1524–1532.
- Zhang Z-H, Wang Y-R, Li F, et al. Circ-camk4 involved in cerebral ischemia/reperfusion induced neuronal injury. Sci Rep. 2020;10(1):7012.
- Yang H, Wang H, Shang H, et al. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer’s disease. Cell Cycle. 2019;18(18):2197–2214.
- Feng Z, Zhang L, Wang S, et al. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun. 2020;522(2):388–394.
- Wen Z, Shen Q, Zhang H, et al. Circular RNA CCDC66 targets DCX to regulate cell proliferation and migration by sponging miR-488-3p in Hirschsprung’s disease. J Cell Physiol. 2019;234(7):10576–10587.
- Mao Z, Liu G, Xiao G-Y, et al. CircCDR1as suppresses bone microvascular endothelial cell activity and angiogenesis through targeting miR-135b/ FIH-1 axis. Orthop Surg. 2021;13:573–582.
- Geng -H-H, Li R, Su Y-M, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PloS One. 2016;11(3):e0151753.
- Zhang W, Zhang C, Hu C, et al. Circular RNA-CDR1as acts as the sponge of microRNA-641 to promote osteoarthritis progression. J Inflamm (Lond). 2020;17(1):8.
- Chen H, Gu B, Zhao X, et al. Circular RNA hsa_circ_0007364 increases cervical cancer progression through activating methionine adenosyltransferase II alpha (MAT2A) expression by restraining microRNA-101-5p. Bioengineered. 2020;11(1):1269–1279.
- Lu Y, Tan L, Wang X. Circular HDAC9/microRNA-138/Sirtuin-1 pathway mediates synaptic and amyloid precursor protein processing deficits in Alzheimer’s Disease. Neurosci Bull. 2019;35(5):877–888.
- Chen W, Wang H, Feng J, et al. Overexpression of circRNA circUCK2 attenuates cell apoptosis in cerebral ischemia-reperfusion injury via miR-125b-5p/GDF11 signaling. Mol Ther Nucleic Acids. 2020;22:673–683.
- Ma GP, Yang BZ, Zhang YS, et al. Protective effects of miRNA-589 on cerebral ischemia-reperfusion injury. J Biol Regul Homeost Agents. 2020;34:1269–1275.
- Xing F, Liu Y, Dong R, et al. miR-374 improves cerebral ischemia reperfusion injury by targeting Wnt5a. Exp Anim. 2021;70(1):126–136.
- Yan L, Zhang Y, Li K, et al. miR-593-5p inhibit cell proliferation by targeting PLK1 in non small cell lung cancer cells. Pathol Res Pract. 2020;216(2):152786.
- Zhao X, Wang S, Sun W. Expression of miR-28-3p in patients with Alzheimer’s disease before and after treatment and its clinical value. Exp Ther Med. 2020;20:2218–2226.
- Fan HN, Liao XH, Zhang J, et al. Macrophages promote cell proliferation in colorectal cancer via IL-1β-mediated downregulation of miR-28-3p. J Biol Regul Homeost Agents. 2020;34:1657–1668.
- Wang P-X, Zhang X-J, Luo P, et al. Hepatocyte TRAF3 promotes liver steatosis and systemic insulin resistance through targeting TAK1-dependent signalling. Nat Commun. 2016;7(1):10592.
- Sun H, Tian J, Li J. MiR-92b-3p ameliorates inflammation and autophagy by targeting TRAF3 and suppressing MKK3-p38 pathway in caerulein-induced AR42J cells. Int Immunopharmacol. 2020;88:106691.
- Liu E, Sun H, Wu J, et al. MiR-92b-3p regulates oxygen and glucose deprivation-reperfusion-mediated apoptosis and inflammation by targeting TRAF3 in PC12 cells. Exp Physiol. 2020;105:1792–1801.
- Zhang B, Shetti D, Fan C, et al. miR-29b-3p promotes progression of MDA-MB-231 triple-negative breast cancer cells through downregulating TRAF3. Biol Res. 2019;52(1):38.
- Liu X, Zhang L, Qin H, et al. Inhibition of TRAF3 expression alleviates cardiac ischemia reperfusion (IR) injury: a mechanism involving in apoptosis, inflammation and oxidative stress. Biochem Biophys Res Commun. 2018;506(1):298–305.
