ABSTRACT
Breast cancer (BC) is the most common cancer among women. LINC00675 and miR-513b-5p has been reported to be abnormally expressed in multiple types of cancers and modulate malignant phenotypes of cancer cells. However, to date, the functional role and underlying regulatory mechanism of LINC00675 and miR-513b-5p in BC remains largely unknown. Here, we found that LINC00675 was significantly downregulated in BC tissues and cell lines. Decrease of LINC00675 expression associated with higher tumor grade, lymphovascular invasion and shorter survival in BC patients. Functional experiments demonstrated that overexpression of LINC00675 suppressed BC cell proliferation, migration and invasion, whereas depletion of LINC00675 exerted opposite effects. Mechanistically, LINC00675 functioned as a competing endogenous RNA (ceRNA) to interact with miR-513b-5p and suppress its expression. Moreover, METTL3 increased the m6A methylation of LINC00675, which enhanced the association between LINC00675 and miR-513b-5p. Collectively, the central findings of our study suggest that LINC00675 represses BC progression through the inhibition of miR-513b-5p in a m6A-dependent manner.
KEYWORDS:
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer among women, and it is a serious threat for women’s health worldwide [Citation1]. Even through the great advances made in early diagnosis or treatment, including surgery, chemotherapy, radiotherapy and targeted therapy, resulted in substantial improvement in the therapeutic effects, the incidence and the mortality rate of BC remains high [Citation2,Citation3]. Therefore, it is necessary to reveal the mechanisms of BC initiation and development so that novel effective molecules and targets for diagnosis and treatment for BC could be developed.
The advancement of high-throughput sequencing technologies has provided new insights into the importance of long non-coding RNAs (lncRNAs) in gene regulation. LncRNAs are a class of non-protein-coding RNA transcripts that are longer than 200 nt. Increasing evidence has suggested that lncRNAs are involved in physiological and pathological processes at the epigenetic, transcriptional, post-transcriptional, translational, and post-translational levels. LncRNAs act as key regulators in cancer, which regulate oncogenes and tumor-suppressor genes to impact tumorigenesis, metastasis, drug resistance, angiogenesis and prognosis via interacting with DNA, mRNA, microRNA, and proteins [Citation4–6]. For instance, lncRNA HOTAIR interacts with polycomb repressive complex 2 (PRC2) and plays a pivotal role in the H3K27 methylation of many genes involved in cellular death, motility, and cycle progress [Citation7]. The involvement of lncRNAs in BC development has been explored. Overexpression of lncRNA BCRT1 correlates with poor prognosis of BC patients. LncRNA BCRT1 acts as a competing endogenous RNA (ceRNA) to competitively bind with miR-1303 and prevent the degradation of its target gene PTBP3, resulting in BC growth and metastasis in vitro and in vivo [Citation8]. LncRNA FGF13-AS1 expression is decreased in BC tissue. FGF13-AS1 functions as a tumor suppressor to inhibit BC cell proliferation, migration, and invasion by impairing glycolysis and stemness properties. FGF13-AS1 promotes the degradation of c-Myc mRNA via attenuating the interaction between IGF2BPs and c-Myc mRNA [Citation9]. LINC00675 locates in chromosome 17 and has been reported to be abnormally expressed in multiple types of cancers, such as pancreatic ductal adenocarcinoma, gastric cancer, glioma, cervical cancer, esophageal squamous cell carcinoma, and prostate cancer [Citation10–15]. However, there is no previous report about the functional role and underlying mechanism of LINC00675 in BC cells.
N6-Methyladenosine (m6A) modification is the most abundant eukaryotic RNA. m6A modification regulates essential biological processes of the target RNAs, including RNA splicing, decay, nuclear export, translation, microRNA processing, and the ceRNA activity of lncRNA [Citation16,Citation17]. Emerging evidence demonstrated that lncRNAs-regulated m6A modification of target RNAs is involved in tumor initiation and development [Citation18,Citation19]. However, the functional relevance between LINC00675 and m6A modification remains elusive.
In the present study, we sought to investigate the expression pattern and functions of LINC00675 in BC. Moreover, we provided mechanistic insights into the regulation of miR-513b-5p by LINC00675 in a m6A-dependent manner.
Materials and methods
Specimens
Seventy-four patients who were diagnosed with BC and underwent surgical resection at Hubei Cancer Hospital between 2015 and 2017 were randomly enrolled in this research. Fresh specimens of paired BC and adjacent non-tumor tissues were obtained from these 74 patients. All patients did not receive any treatment before surgery and signed the informed consent. Patients with other cancer or receiving chemotherapy, radiotherapy, or targeted therapy before surgery were excluded. All tissue samples were frozen in liquid nitrogen immediately and stored in −80°C until used. This study was approved in 2015 by the Ethics Committee of Hubei Cancer Hospital, followed the clinical research guidelines (ethics approval number: no. 2019–04-212), and complied the Declaration of Helsinki.
Cell culture
Five breast cancer cell lines (MCF7, T47D, BT474, MDA231, and BT549) and one normal breast cell line (MCF10A) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in an atmosphere containing 5% CO2.
Transfection
The mutant LINC00675 was constructed using TaKaRa MutanBEST Kit. The wild-type and mutant LINC00675 were cloned into the overexpression plasmid (pcDNA3.1), which was constructed by Genepharma Company (Shanghai, China). miR-513b-5p mimics and negative control were purchased from Genepharma (Shanghai, China). LINC00675 and METTL3 siRNAs and negative control were synthesized and purchased from Genepharma (Shanghai, China). The Lipofectamine 2000 kit (Invitrogen) was used for cell transfection according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were subjected for further experiments. The target sequence of siRNAs was listed as follow: LINC00675 siRNA#1 (KD1): CTGGGACTTCTTCATCTAC, LINC00675 siRNA#2 (KD2): GCTGCAATACTGAGGCTTT, siMETTL3: GCTACAGATCCTGAGTTAG.
Subcellular fractionation
Nuclear and cytoplasmic RNA was isolated using the PARIS Kit (Life Technologies, USA) according to the manufacturer’s instructions.
Cell counting kit-8 (CCK-8) and colony formation assay
3 × 103 cells per well were seeded into a 96-well plate and incubated for 1–4 days. After that, 10 μl CCK-8 solution was added into each well. The plate was incubated for another 1 h. The optical density (OD) value of each well was measured. For colony formation assay, 2× 103 cells per well were seeded into a 6-well plate and incubated for 7 days. Cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet and then counted
Transwell assay
Transwell assays were performed to detect cell migration and invasion. For the migration detection, cells in serum-free media were seeded into the upper Transwell chambers (Corning). For the invasion detection, cells in serum-free media were seeded into the upper Matrigel-coated Transwell chambers (BD Bioscience). The DMEM with 10% FBS was added to the lower chamber. After 24 h incubation, cells in the lower chamber were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The cells were counted in 10 random field under microscope.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA from cells or tissues was isolated using Trizol reagent (Invitrogen) according to the standard protocol. First-strand cDNA was synthesized using the EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen, Beijing). qRT-PCR was carried out in the Lightcycler 96 (Roche) using SYBR® Green (Takara, Dalian, China). The relative expression of RNAs was calculated using 2−ΔΔCT method. The sequence of gene-specific primers was listed as follow: LINC00675-forward: AAGGAGATGCCCTTCCTTTAC, LINC00675-reverse: GGACATCCTCGTGAGTACTTTG, METTL3-forward: CACTGATGCTGTGTCCATCT, METTL3-reverse: CTTGTAGGAGACCTCGCTTTAC.
RNA immunoprecipitation (RIP)
RIP assay was used to determine the association of LINC00675 and METTL3 or AGO2. Antibodies used for the RIP assay included anti-AGO2 (Millipore), METTL3 (Abcam) and control IgG (Millipore, USA). In brief, cells were lysed in RIP lysis buffer, and then incubated with indicated antibody and Magna beads. After wash, the coprecipitated RNAs were purified and then subjected to qRT-PCR analysis.
MS2bs (MS2-binding protein)-MS2bp (MS2-binding sequences)-based RIP (MS2-RIP)
MS2-RIP assay was used to determine the association of LINC00675 and miR-513b-5p. MS2-RIP was performed as previous studies described [Citation20,Citation21]. In brief, cells were co-transfected with pcDNA3.1-MS2, pcDNA3.1-MS2-LINC00675, or pcDNA3.1-MS2-LINC00675-mut and pMS2-GFP. After 48 hrs, cells were used to perform RNA immunoprecipitation (RIP) experiments using a GFP antibody (Roche) and the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) according to the manufacturer’s instructions. Antibodies used for the MS2-RIP assay included anti-GFP (Millipore) and control IgG (Millipore, USA). The coprecipitated RNAs were purified and then subjected to qRT-PCR analysis.
Methylated RNA immunoprecipitation (MeRIP)
MeRIP experiment was carried out to detect the m6A level of LINC00675 using Magna MeRIP™ m6A Kit (Millipore) according to the manufacturer’s instructions. In brief, total RNAs were first extracted from cells. RNAs were treated with DNase using TURBO DNA-freeTM Kit (Thermo) to avoid DNA contaminations. RNA concentration was adjusted to 1 μg/μl with nuclease-free water. RNA was chemically fragmented into ~100nt size and fragmented RNA was then incubated with m6A antibody for immunoprecipitation according to the standard protocol of Magna MeRIP™ m6A Kit (Merck Millipore).
RNA pull-down assay
LINC00675 or mutant LINC00675 (LINC00675-mut) were in vitro transcribed, respectively, and biotin-labeled with the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Roche), and purified with a RNeasy Mini Kit (Qiagen). Cell lysates were incubated with purified biotinylated transcripts for 1 hour; complexes were isolated with streptavidin agarose beads (Invitrogen). The miR-513b-5p pulled down by LINC00675 was detected by qRT-PCR analysis.
Luciferase reporter assay
Wild-type or mutant LINC00675 were cloned into luciferase reporter plasmid pmirGLO. pmirGLO, pmirGLO-LINC00675 or pmirGLO-LINC00675-mut was cotransfected with miR-513b-5p mimics or miR-NC into cells using the Lipofectamine 2000 kit. 48 hours later, the relative luciferase activity was detected and normalized to Renilla luciferase activity.
Statistical analysis
All experiments were performed with at least three biological replicates in triplicate. Data are expressed as the mean ± SD and analyzed by the SPSS software. Student t test (for two groups) or one-way ANOVA (for groups more than two) was used for the comparisons of different groups. Chi-square test was used to analyze the association between LINC00675 expression and clinicopathological features of BC patients. Kaplan-Meier method and log-rank test was performed for determine the relationship between LINC00675 expression and prognosis of BC patients. P less than 0.05 was considered statistically significant.
Results
Downregulation of LINC00675 is associated with poor prognosis of BC patients
To determine the expression pattern of LINC00675 in BC, the LINC00675 levels in 74 pairs of BC and adjacent non-tumor tissues were detected using qRT-PCR. The results showed that the BC tissues expressed obviously lower LINC00675 expression than non-tumor tissues did ()). Additionally, compared to normal breast cell line MCF10A, LINC00675 was significantly lower in BC cell lines, such as MCF7, T47D, BT474, MDA231, and BT549 ()). LINC00675 was high in luminal cell lines and low in basal-like breast cancer (BLBC) cell lines. Among these BC cells, BT549 cells expressed the lowest level of LINC00675 expression, while MCF7 cells exhibited the highest LINC00675 expression. These two cell lines were used for overexpression and knockdown experiments, respectively.
Figure 1. Downregulation of LINC00675 is associated with poor prognosis of BC patients
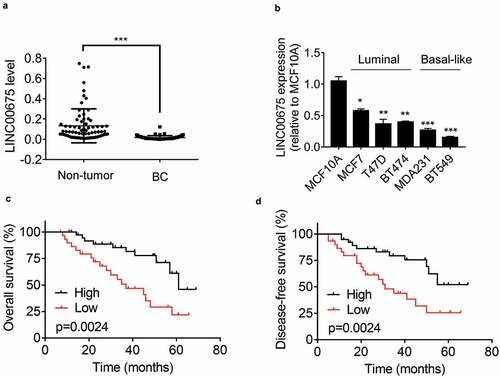
To reveal the clinical significance of LINC00675 expression in BC, we analyze the correlation between LINC00675 expression and clinicopathological features of BC patients. According to the median value of LINC00675 expression in BC tissues, enrolled BC patients were divided into high-expression and low-expression groups. The correlation analysis demonstrated that decreased LINC00675 expression was significantly associated with higher tumor grade and lymphovascular invasion (). Moreover, Kaplan-Meier survival analysis showed that low LINC00675 expression was correlated with poor overall survival of BC patients ()). Together, these data suggest that LINC00675 may act as a tumor suppressive lncRNA in BC.
Table 1. The correlation analysis between LINC00675 expression and clinicopathologic features of 74 BC patients
LINC00675 acts as a tumor suppressor in BC cells
To elucidate the functional role of LINC00675 in BC cells, LINC00675 was overexpressed in BT549 cells, but knocked down in MCF7 cells. The overexpression and knockdown efficiency was validated using qRT-PCR ()). We performed CCK-8 assay to evaluate the effect of LINC00675 on BC cell proliferation and found that overexpression of LINC00675 significantly suppressed the proliferation of BT549 cells ()). Conversely, the proliferative ability was enhanced by transfection of LINC00675 siRNAs in MCF7 cells ()). The results of colony formation assay further validated the suppressive effect of LINC00675 in BC cell proliferation ()).
Figure 2. LINC00675 suppresses the proliferation, migration and invasion of BC cells
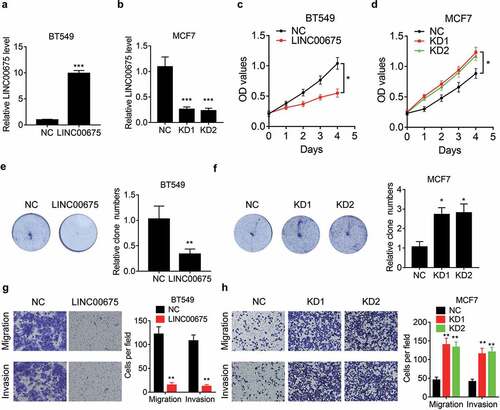
We then evaluated the function of LINC00675 in BC cell migration and invasion. Transwell assay results indicated that ectopic expression of LINC00675 obviously attenuated ()), while knockdown of LINC00675 enhanced, migration, and invasion in BC cells ()), supporting a metastasis-suppressive role of LINC00675 in BC.
LINC00675 directly interacts with miR-513b-5p
To investigate the underlying mechanism of LINC00675 inhibiting the malignant phenotypes of BC cells, we first detected the cellular distribution of LINC00675. The results of subcellular fractionation showed that LINC00675 mainly located in the cytoplasm of BC cells (). Cytoplasmic lncRNA usually acts as a ceRNA to sponge its target microRNA and exert its regulatory functions [Citation22]. Using an online prediction software LncBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php), we selected the top five microRNAs targeted by LINC00675 for verification. As evidenced by MS2-RIP assay, only miR-513b-5p could be significantly enriched by LINC00675 ()). LINC00675 contains a binding site for miR-513b-5p ()). We mutated the binding site of LINC00675 with miR-513b-5p, and found that mutation of LINC00675 failed to associate with miR-513b-5p ()). The interaction between LINC00675 and miR-513b-5p was also validated by affinity pull-down of endogenous miR-513b-5p using in vitro transcribed biotin-labeled LINC00675 ()). The AGO2-RIP showed that LINC00675 could be significantly enriched by AGO2 by ectopic expression of miR-513b-5p ()). For further confirmation, the wild-type and mutant LINC00675 were constructed into luciferase reporter plasmid pmirGLO. Transfection of miR-513b-5p mimics significantly decreased the luciferase activity of wild-type reporter, but did not affect the luciferase activity of mutant reporter ()). Moreover, we found that overexpression of LINC00675 was able to downregulate miR-513b-5p level ()), whereas knockdown of LINC00675 elevated its expression ()).
Figure 3. LINC00675 associates with miR-513b-5p
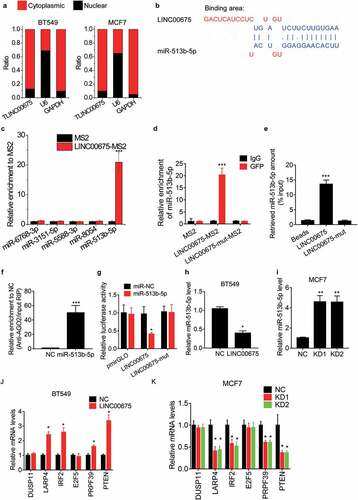
Previous studies have already identified the targets of miR-513b-5p, such as DUSP11, LARP4, IRF2, E2F5, PRPF39 and PTEN [Citation23–27]. Whether LINC00675 affected the expression of the targets of miR-513b-5p was then assessed. As demonstrated by qRT-PCR experiments, overexpression of LINC00675 significantly upregulated the expression of LARP4, IRF2, PRPF39, and PTEN ()), whereas knockdown of LINC00675 exerted an opposite function ()). However, LINC00675 could not influence the expression of DUSP11 and E2F5. All these findings suggest that LINC00675 functions as a ceRNA against miR-513b-5p.
LINC00675 exerts suppressive function via miR-513b-5p
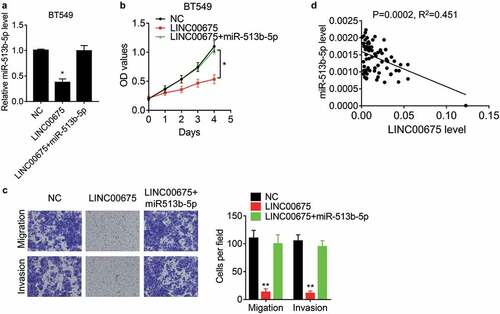
miR-513b-5p exerts oncogenic or suppressive function in different cancers [Citation23–27], but its effect on BC cells remains unknown. To elucidate whether LINC00675 functions via miR-513b-5p, rescue experiments were carried out. It was found that restoration of miR-513b-5p expression overcame the LINC00675-mediated suppression in BT549 cell proliferation, migration, and invasion ()). Moreover, the miR-513b-5p was detected in BC tissues and showed a negative correlation between LINC00675 and miR-513b-5p ()), supporting that miR-513b-5p is a bonafide downstream target of LINC00675.
Figure 4. LINC00675 exerts suppressive effects via miR-513b-5p
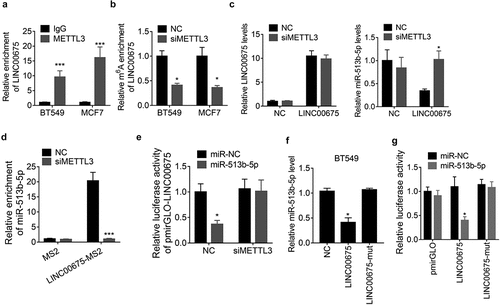
METTL3-mediated m6A modification of LINC00675 regulates its ceRNA activity
A recent study reported that N6-Methyladenosine (m6A) modification contributes to the ceRNA activity of lncRNA [Citation16]. To elucidate whether LINC00675 regulated miR-513b-5p in this manner, we performed a RIP assay to detect the association of LINC00675 with METTL3, the core component of m6A methylase complex, and observed that LINC00675 could be significantly pulled down by anti-METTL3 antibody compared to IgG ()). Moreover, knockdown of METTL3 resulted in a decrease of LINC00675 m6A level as demonstrated by MeRIP assay ()). However, knockdown of METTL3 did not affect the LINC00675 expression, but abolished the downregulation of miR-513b-5p expression induced by LINC00675 in BT549 cells ()), suggesting that METTL3 may influence ceRNA activity of LINC00675. To validate this hypothesis, we performed a MS2-RIP assay and found that depletion of METTL3 expression, LINC00675 failed to associate with miR-513b-5p ()). Furthermore, BT549 cells were co-transfected with the pmirGLO-LINC00675 reporter, METTL3 siRNAs and miR-513b-5p mimics. Similarly, the results of luciferase reporter experiment demonstrated that the suppressive effect of miR-513b-5p on pmirGLO-LINC00675 was abolished by deletion of METTL3 ()). The m6A sites of LINC00675 was predicted using the SRAMP online tool (http://www.cuilab.cn/sramp/) and the results showed that only one m6A site was predicted in LINC00675 transcript (Supplemental ). We mutated this site (A to G) and found that this mutation did not affect the miR-513b-5p expression ()). Moreover, miR-513b-5p was unable to decrease the activity of this mutant LINC00675 reporter ()), suggesting the importance of m6A sites of LINC00675 in regulating the expression of miR-513b-5p and its interaction with miR-513b-5p. Collectively, these findings suggest that m6A modification is critical for LINC00675 sponging miR-513b-5p.
Figure 5. m6A modification is involved in the ceRNA activity of LINC00675
Discussion
Here, we identified LINC00675 as a tumor-suppressive lncRNA significantly downregulated in BC. Gain- and loss-of-function experiments demonstrated that overexpression of LINC00675 attenuated the proliferation, migration, and invasion ability of BC cells, while knockdown of LINC00675 enhanced these cellular phenotypes. Moreover, decrease of LINC00675 expression was associated with advanced tumor grade and lymphovascular invasion. BC patients with high LINC00675 level exhibited better clinical outcome than those with low LINC00675 expression. Therefore, LINC00675 may serve as a prognostic factor for BC patients.
Previous studies have implicated the opposite function of LINC00675 in different specific types of cancer via diverse mechanism. For instance, in gastric cancer, LINC00675 was found to be downregulated in cancer tissues and inhibit metastasis through associating with vimentin and enhancing its phosphorylation [Citation11]. LINC00675 also competitively binds with LSD1 and promotes the binding of LSD1 and its target H3K4me2, thus suppressing SPRY4 transcription [Citation28]. Xu et al. reported that LINC00675 inhibits tumorigenesis and metastasis via suppressing Wnt/β-catenin signaling in esophageal squamous cell carcinoma [Citation14]. The same mechanism of LINC00675 was also found in cervical cancer and colorectal cancer [Citation12,Citation29]. In other cases, LINC00675 has been identified as oncogene. LINC00675 is upregulated in androgen-insensitive prostate cancer cell lines and castration-resistant prostate cancer patients. LINC00675 directly modulates androgen receptor’s (AR) interaction with MDM2 and blocks AR’s ubiquitination and stabilizes GATA2 mRNA which is a co-activator in the AR signaling pathway [Citation15]. In glioma, high-expression of LINC00675 was an independent unfavorable prognostic predictor. LINC00675 facilitates glioma cell proliferation, migration and invasion through regulating TRIP6 [Citation13]. Here, we verified that LINC00675 acted as a ceRNA against miR-513b-5p, suppressing BC cell proliferation, migration and invasion. Previous studies have reported that miR-513b-5p is involved in the proliferation, metastasis and drug resistance of cancer cells. The targets of miR-513b-5p includes DUSP11, LARP4, IRF2, E2F5, PRPF39, and PTEN [Citation23–27]. Here, our date demonstrated that LINC00675 significantly upregulated the expression of LARP4, IRF2, PRPF39 and PTEN, but could not influence the expression of DUSP11 and E2F5. LARP4 is an RNA binding protein which is involved in T cell activation-dependent mRNA stabilization [Citation30]. LARP4 also functions as a suppressor for motility of ovarian cancer cells [Citation31]. IRF2 upregulates β-catenin expression to modulate cellular survival [Citation32]. Loss of IRF2 leads to immune evasion through decreased MHC class I antigen presentation and increased PD-L1 expression in cancers [Citation33]. PRPF39 is a regulator of cisplatin sensitivity in liver cancer [Citation23]. PTEN is a well-known tumor suppressor. The loss of PTEN activity, identified in a series of primary and metastatic tumors, such as breast cancer, leads into uncontrolled transduction of the PI3K signal which is involved in a series of biological processes, such as cellular motility, invasion, proliferation, and survival [Citation34]. These findings suggested that LINC00675- miR-513b-5p may exerted different function via regulating specific target genes.
m6A modification is the most abundant eukaryotic RNA modification and involved in regulating essential biological processes of the transcriptome, such as RNA splicing, decay, nuclear export, translation, and microRNA processing [Citation17]. Dysregulation of the m6A regulators has increasingly been found in many neoplasms, including BC. For example, METTL3 upregulates the expression of onco-protein HBXIP via m6a modification, which promotes BC progression [Citation35]. METTL3 increases m6A level of the ITGA6 mRNA 3ʹUTR and promotes the translation of ITGA6 mRNA via binding of the m6A readers YTHDF1 and YTHDF3, inducing BC growth and metastasis [Citation36]. m6A demethylase FTO also promotes BC development via decreasing the m6A level of BNIP3 mRNA and inducing its degradation in an YTHDF2 independent manner [Citation37]. Recently, m6A modification has been found to regulate the ceRNA activity of lncRNA. linc1281 ensures embryonic stem cells identity by sponging let-7 family, and this lncRNA-microRNA interaction is regulated by m6A modification [Citation16]. Similarly, our present study revealed that METTL3 associated with LINC00675 and increased its m6A level, which did not affect the LINC00675 expression. Knockdown of METTL3 significantly attenuated the interaction between LINC00675 and miR-513b-5p. These findings suggest the importance of m6A modification of lncRNA in regulating microRNA.
Conclusions
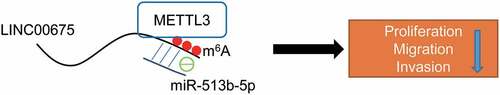
In summary, our study reveals that LINC00675 is downregulated in BC, and inhibits BC cell proliferation, migration and invasion by acting as a ceRNA to regulate miR-513b-5p, which is regulated by METTL3-mediated m6A modification ().
Authors’ contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, agreed to the submitted journal, and agree to be accountable for all aspects of the work.
Ethics statement
This study was approved by the Ethics Committee of Hubei Cancer Hospital and followed the clinical research guidelines (ethics approval number: no. 2019-04-212).
Supplemental Material
Download Zip (282.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used during this research are available.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021 Apr 5;149(4):778–789.
- Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev. 2016;17(S3):43–46.
- Fahad Ullah M. Breast cancer: current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51–64.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017 Aug 1;77(15):3965–3981.
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017 Oct 12;36(41):5661–5667.
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018 Jan 25;172(3):393–407.
- Kozlowska J, Kolenda T, Poter P, et al. Long intergenic non-coding RNAs in HNSCC: from “Junk DNA” to important prognostic factor. Cancers (Basel). 2021 Jun 12;13(12):2949.
- Liang Y, Song X, Li Y, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer. 2020 May 8;19(1):85.
- Ma F, Liu X, Zhou S, et al. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019;450:63–75.
- Li DD, Fu ZQ, Lin Q, et al. Linc00675 is a novel marker of short survival and recurrence in patients with pancreatic ductal adenocarcinoma. World J Gastroenterol. 2015 Aug 21;21(31):9348–9357.
- Zeng S, Xie X, Xiao YF, et al. Long noncoding RNA LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress gastric cancer progression. Cancer Lett. 2018;412:179–187.
- Ma S, Deng X, Yang Y, et al. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/beta-catenin signaling in cervical cancer. Biomed Pharmacothe. 2018 Dec;108:1686–1693.
- Li Z, Li Y, Wang Q. LINC00675 is a prognostic factor and regulates cell proliferation, migration and invasion in glioma. Biosci Rep. 2018;38(5). DOI:10.1042/BSR20181039.
- Zhong YB, Shan AJ, Lv W, et al. Long non-coding RNA LINC00675 inhibits tumorigenesis and EMT via repressing Wnt/beta-catenin signaling in esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018 Dec;22(23):8288–8297.
- Yao M, Shi X, Li Y, et al. LINC00675 activates androgen receptor axis signaling pathway to promote castration-resistant prostate cancer progression. Cell Death Dis. 2020 Aug 15;11(8):638.
- Yang D, Qiao J, Wang G, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018 May 4;46(8):3906–3920.
- Yang G, Sun Z, Zhang N. Reshaping the role of m6A modification in cancer transcriptome: a review. Cancer Cell Int. 2020;20:353.
- Lang C, Yin C, Lin K, et al. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 2021 Jun;11(6):e426.
- Wu J, Pang R, Li M, et al. m6A-induced LncRNA MEG3 suppresses the proliferation, migration and invasion of hepatocellular carcinoma cell through miR-544b/BTG2 signaling. Onco Targets Ther. 2021;14:3745–3755.
- Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014 May 12;25(5):666–681.
- Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015 Aug 1;75(15):3181–3191.
- Wang L, Cho KB, Li Y, et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22). DOI:10.3390/ijms20225758.
- Qin L, Zhan Z, Wei C, et al. HsacircRNAG004213 promotes cisplatin sensitivity by regulating miR513b5p/PRPF39 in liver cancer. Mol Med Rep. 2021 Jun;23(6). DOI:10.3892/mmr.2021.12060.
- Zhang Z, Wu X, Han Q, et al. Downregulation of long non-coding RNA UCA1 represses tumorigenesis and metastasis of osteosarcoma via miR-513b-5p/E2F5 axis. Anticancer Drugs. 2021 Feb 15. DOI:10.1097/CAD.0000000000001034.
- Wang X, Zhang X, Wang G, et al. Hsa-miR-513b-5p suppresses cell proliferation and promotes P53 expression by targeting IRF2 in testicular embryonal carcinoma cells. Gene. 2017 Aug 30;626:344–353.
- Lin W, Ye H, You K, et al. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020;20:5.
- Liu H, Zhang L, Ding X, et al. LINC00861 inhibits the progression of cervical cancer cells by functioning as a ceRNA for miR513b5p and regulating the PTEN/AKT/mTOR signaling pathway. Mol Med Rep. 2021 Jan;23(1). DOI:10.3892/mmr.2020.11662.
- Pan Y, Fang Y, Xie M, et al. LINC00675 suppresses cell proliferation and migration via downregulating the H3K4me2 level at the SPRY4 promoter in gastric cancer. Mol Ther Nucleic Acids. 2020;22:766–778.
- Shan Z, An N, Qin J, et al. Long non-coding RNA Linc00675 suppresses cell proliferation and metastasis in colorectal cancer via acting on miR-942 and Wnt/beta-catenin signaling. Biomed Pharmacothe. 2018 May;101:769–776.
- Tian Y, Zeng Z, Li X, et al. Transcriptome-wide stability analysis uncovers LARP4-mediated NFkappaB1 mRNA stabilization during T cell activation. Nucleic Acids Res. 2020 Sep 4;48(15):8724–8739.
- Egiz M, Usui T, Ishibashi M, et al. La-related protein 4 as a suppressor for motility of ovarian cancer cells. Tohoku J Exp Med. 2019 Jan;247(1):59–67.
- Guo Y, Xu J, Du Q, et al. IRF2 regulates cellular survival and Lenvatinib-sensitivity of hepatocellular carcinoma (HCC) through regulating beta-catenin. Transl Oncol. 2021 Jun;14(6):101059.
- Kriegsman BA, Vangala P, Chen BJ, et al. Frequent loss of IRF2 in cancers leads to immune evasion through decreased MHC class I antigen presentation and increased PD-L1 expression. J Immunol. 2019 Oct 1;203(7):1999–2010.
- Carbognin L, Miglietta F, Paris I, et al. Prognostic and predictive implications of PTEN in breast cancer: unfulfilled promises but intriguing perspectives. Cancers (Basel). 2019 Sep 19;11(9):1401.
- Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19.
- Jin H, Ying X, Que B, et al. N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019 Sep;47:195–207.
- Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019 Mar 28;18(1):46.