ABSTRACT
Our study aimed to investigate the clinical significance and biological functions of Spindlin1 (SPIN1) in colorectal cancer (CRC) tumorigenesis and progression, as well as the mechanism underlying its upregulation. The expression of SPIN1 was detected by immunohistochemistry and western blotting assays. Bioinformatics prediction and dual-luciferase reporter assays were used to determine whether microRNA-381 (miR-381) could target SPIN1. A series of cell functional experiments were performed to investigate whether the miR-381-mediated regulation of SPIN1 is involved in the progression and aggressiveness of CRC cells via the Wnt/β-catenin pathway. Our results showed that SPIN1 is frequently overexpressed in CRC tissues and cell lines, and its upregulation is positively correlated with disease progression and lymph node metastasis. Moreover, SPIN1 depletion suppresses cell growth, migration, and invasion through inactivation of the Wnt/β-catenin signaling pathway, which recapitulates the effects of miR-381 upregulation. Moreover, SPIN1 is a target gene of miR-381, and miR-381 is downregulated in CRC. Furthermore, the reintroduction of SPIN1 partially abolished the miR-381-mediated inhibitory effects in CRC cells. In summary, our data revealed that the miR-381/SPIN1 axis greatly contributes to CRC tumorigenesis by orchestrating the Wnt/β-catenin pathway, thereby representing actionable therapeutic targets for colorectal cancer patients.
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer mortality worldwide, with a high incidence, rapid progression, and great metastatic potential [Citation1]. Despite the numerous improvements in surgery, chemotherapy, radiotherapy, and immunotherapy, the overall survival of patients with CRC remains dismal [Citation2]. At present, it is of great importance to uncover the complex underlying mechanisms responsible for CRC carcinogenesis and progression.
Spindlin1 (SPIN1), a member of the SPIN/SSTY family, was first identified as an overexpressed protein in ovarian cancer [Citation3]. The Chromatin reader SPIN1 recognizes trimethylated histone H3 lysine 4 to facilitate rRNA expression via protein-protein interaction [Citation4]. Recently, several studies have defined the oncogenic role of SPIN1 in tumorigenesis and progression in several cancers, such as non-small cell lung cancer (NSCLC), liver cancer, gastric cancer, and breast cancer [Citation5–8]. Our previous study identified SPIN1 as a novel and negative regulator of the uL18-MDM2-p53 pathway to control cancer cell proliferation and apoptosis [Citation9]. Additionally, the Wnt, PI3K/AKT, and RET pathways regulated by SPIN1 during tumorigenesis have been reported [Citation10]. Nevertheless, the cellular functions and clinical significance of SPIN1, along with the mechanism underlying its dysregulation in colorectal cancer, remain poorly characterized.
Emerging evidence has shown that miRNAs, a group of non-coding RNA molecules containing 20 to 22 nucleotides, were found to be involved in multiple physiological and pathologic processes [Citation11,Citation12], such as cell differentiation, invasion, and tumorigenesis [Citation13–15]. Specifically, the interaction between miRNAs and oxidative stress signals is involved in gastrointestinal carcinogenesis [Citation16]. Past studies have demonstrated that several miRNAs have been shown to contribute greatly to the development and progression of colorectal cancer [Citation17–20], among which miR-381 was discovered to exert tumor-suppressive functions. In addition, our preliminary bioinformatic predictions indicated that miR-381 could bind to the 3ʹ-UTR of SPIN1 mRNA. However, the exact correlation between miR-381 and SPIN1 in CRC has not been specifically reported.
Therefore, we hypothesized that the miR-381-mediated regulation of SPIN1 plays a significant role in the tumorigenesis and progression of CRC. In this study, our data showed that miR-381 inactivates the Wnt/β-catenin signaling pathway by targeting SPIN1, and therefore represents an actionable therapeutic target for colorectal cancer patients.
Materials and methods
Ethics statement and clinical tissues
Primary colorectal cancer tissues (from 2019 to 2020) and paraffin-embedded colorectal cancer samples (from 2016 to 2018) were obtained from the First Affiliated Hospital of Nanchang University. After resection, the fresh specimens were frozen and immediately stored in liquid nitrogen. Detailed clinicopathological features of the patients were collected and listed in Supplementary Table S1 and . None of the patients with colorectal cancer received radiotherapy, chemotherapy or immunotherapy before surgery. Our study was approved by the First Affiliated Hospital of the Nanchang University Ethics Review Board and complied with the Declaration of Helsinki. All CRC patients who participated in this study signed an informed consent document.
Table 1. The relationship between SPIN1 expression and clinical features of colorectal cancer patients
Cell culture
The human colon epithelial cell line, NCM460 (Cat. No. CRL-1831), CRC cell line DLD1(Cat. No. CCL-221), SW48 cells (Cat. No. CCL-231), SW480 (Cat. No. CCL-228), LOVO (Cat. No. CCL-229), HT-29 and HCT116 were obtained from the American Type Culture Collection (ATCC) or donated by Sun Yat-sen University. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, HyClone, Logan, UT, USA) supplemented with 12% fetal bovine serum (FBS; HyClone, USA) and penicillin-streptomycin antibiotics at 37°C and 5% CO2.
Cell transfection
HCT116, LOVO and DLD1 were selected for our further analysis due to the different expression levels of SPIN1. The negative control siRNA, SPIN1 siRNAs, NC mimics, miR-381 mimics, NC inhibitors, miR-381 inhibitors and SPIN1 overexpression plasmids were designed and synthesized by GenePharma (Suzhou, China). SPIN1 expression vector without 3′-UTR (SPIN1-no UTR) was constructed by inserting its CDS sequence into the vector (Promega, USA). All the transfection experiments were performed using Turbofect Reagent (Thermo Fisher, St Louis, MO, USA) according to the manufacturer’s instructions. The indicated cells were harvested 48 h upon transfection. Transfection efficiency was confirmed by qRT-PCR (quantitative real-time PCR) and western blotting.
Cell counting kit-8 (CCK-8) assay
Indicated cells were inoculated at 1 × 103 cells per well in 96-well plates for 1, 2, 3, 4, and 5 days. After adding 10 μL of the CCK-8 reagent post-transfection at the same time every day, the absorbance values of treated cells were detected daily at 450 nm using a Microplate Reader (Molecular Device, SpecrtraMax M5e, Sunnyvale, CA, USA) for five consecutive days.
Colony formation assays
In this study, 1 × 103 transfected CRC cells (LOVO, DLD1, and HCT116) were seeded into 6-well plates. After 10–15 days of incubation, the colonies were processed with paraformaldehyde and crystal violet for 60 min. ImageJ software was used to count the visible colonies. All experiments were performed at least twice.
Scratch wound and transwell invasion assays
Scratch wound and transwell invasion assays were performed to evaluate cell migration and invasion. The same number of indicated cells were seeded on a 6-well plate, and the cells were allowed to adhere to the plate and reach full confluency. A sterile 10 μL pipette tip was used to draw straight lines to make a wound. Microscopes were utilized to take images at the indicated times for further analysis.
Indicated cells were resuspended and plated into the upper chamber (8.0 μm pore size; Corning, USA). Upon incubation, the number of cells that invaded the bottom was estimated using the average number of cells over three microscopic fields. Biological experiments were performed more than twice.
Dual-luciferase reporter assay
Dual-luciferase reporter assays were carried out as reported previously [Citation21]. The binding sites between miR-381 and SPIN1 were mutated, and the SPIN1 mutant plasmid was constructed for the dual luciferase reporter assay. Briefly, the vector containing wild-type (WT) or mutated (MUT) 3ʹUTR of SPIN1 and miR-381 mimics/miR-381 inhibitor or negative control were transfected into CRC cells using TurboFect Reagent (Thermo Fisher, St Louis, MO, USA). Thirty-six hours post-transfection, the indicated cells were harvested and analyzed using a propriate luciferase detection kit (E1910, Promega, Madison, WI, USA). Luciferase activity was normalized to Renilla luciferase activity.
Western blotting analysis
Western blotting analysis was performed as previously described [Citation21,Citation22]. The antibodies and concentrations utilized in our study were shown as follows: SPIN1 (1:1500; #12,105-1-AP, Proteintech, Wuhan, China), β-catenin (1:1000; #8480, Cell Signaling Technology, MA, USA), cyclinD1 (1:1000; #55,506, Cell Signaling Technology, MA, USA), c-Myc (1:1000; #18,583, Cell Signaling Technology, MA, USA), β-actin (1:2000; #AF7018; Affinity, Jiangsu, China), and GAPDH (1:4000; #A2220; Affinity, Jiangsu, China). β-actin and GAPDH were used as internal controls, and the immunoblotting bands were visualized using ImageJ software. More than two independent experiments were performed.
Immunohistochemistry (IHC) assays
IHC was performed on 90 clinical CRC specimens. Briefly, the paraffin-embedded tissue was cut into 5 µm-thick tissue sections, which were then baked and fixed, deparaffinized with xylene, hydrated with ethanol, and heated in an antigen retrieval solution (EDTA, pH 9.0), after which endogenous peroxidase was quenched with 3% H2O2. After blocking with goat serum, the samples were incubated with anti-SPIN1 antibody (1:50, #19,531-1-AP, Proteintech, Wuhan, China) at 4°C overnight. On the second day, the tissue was covered with HRP-conjugated secondary antibody for 30 min and stained with 3,3ʹ-diaminobenzidine until brown particles appeared in the membrane, cytoplasm, or nucleus. Finally, the sections were counterstained with hematoxylin at room temperature. SPIN1 protein expression in CRC samples was determined using the staining index (SI). Samples with SI ≥ 6 were determined to have high SPIN1 expression, whereas those with SI < 6 were shown as low SPIN1 expression. SPIN1 immunostaining scores were evaluated by two experienced pathologists.
Quantitative real-time PCR (qRT-PCR) assays
Total RNA from transfected cells was collected using TRIzol (Invitrogen, Carlsbad, CA, USA), and qRT-PCR assays were performed using the SYBR Green PCR kit (BioRad, Hercules, USA) according to the manufacturer’s instructions. The fold changes in mRNA expression among the groups were assessed using the 2–∆∆Ct method. The sequences of the primers used are listed in . U6 and GAPDH were used to normalize the variation. All experiments were performed three times.
Table 2. The sequences of primers for qRT-PCR used in this study
Statistical analysis
All statistical analysis was performed using SPSS 20.0 software. These biofunctional experiments were repeated more than 2 times. Analysis of the correlations between SPIN1 or miR-381 and clinical features in CRC patients was performed using Chi-square tests. Statistical significance among groups was analyzed by one-way analysis of variance (ANOVA) or by the Student’s two-tailed t-test. Differences were considered statistically significant at P values lower than 0.05.
Results
SPIN1 is upregulated in CRC tissues and cell lines
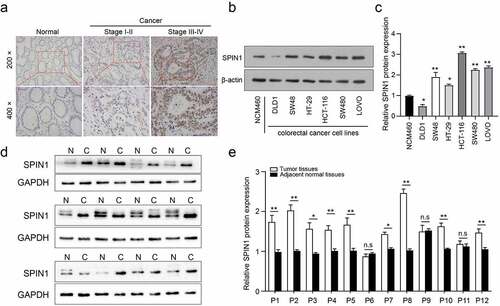
Our previous data have demonstrated that SPIN1 functions as a negative modulator of the uL18-MDM2-p53 signaling pathway, thus promoting tumorigenesis in human cancer [Citation9], but its role in CRC remains largely unknown. In this study, our immunohistochemical staining of 90 patients showed that SPIN1 was mainly localized in the nucleus of CRC cells, with higher SPIN1 expression in CRC specimens when compared to paired non-cancerous tissues (53/90, 58.9%, illustrated in )). Moreover, the protein expression of SPIN1 was higher in most CRC cell lines than in the normal colon epithelial cell line NCM460 ()). Furthermore, we analyzed the correlation between SPIN1 expression and clinicopathological factors in the CRC cohort (n = 90). As shown in , SPIN1 expression was strongly correlated with depth of invasion (p = 0.01), clinical TNM stage (p = 0.002), and lymph node metastasis (p = 0.004). Nevertheless, no statistically significant associations were found between SPIN1 expression and age, sex, or tumor size. Consistently, the protein expression of SPIN1 in fresh CRC tissues was also upregulated (9/12, 75%, )). Our data demonstrated that SPIN1 was markedly elevated and positively correlated with disease progression in CRC.
Figure 1. SPIN1 is upregulated in CRC tissues and cell lines. (a) Immunohistochemical staining images of SPIN1 in 90 pairs of paraffin-embedded colorectal cancer (CRC) tissues and adjacent non-cancerous tissues (magnification, 200× and 400×). (b, c) Representative western blotting images and quantitation of SPIN1 protein expression in six CRC cell lines and normal colon cells. (d) The SPIN1 protein expression in 12 pairs of CRC tissues and adjacent normal tissues were assessed by western blotting analysis. (e) The quantification analysis of the blots was analyzed by image J software. P, patient; Student’s t test, *p < 0.05; **p < 0.01.
SPIN1 depletion inhibits cell proliferation, migration and invasion
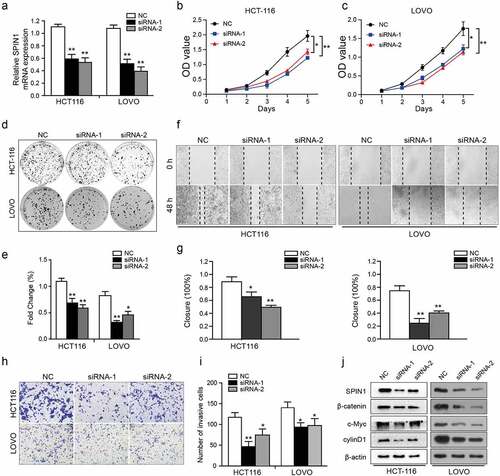
As SPIN1 overexpression was markedly correlated with aggressive clinical features of CRC patients, therefore, we further utilized a number of cellular functional experiments to investigate the biological role of SPIN1 in CRC cells. After transfection, the efficiency of siRNA transfection was assessed by qRT-PCR and western blotting simultaneously ()). Subsequently, CCK-8 and colony forming assays showed that SPIN1 depletion significantly suppressed the cell growth of CRC cells ()). Moreover, SPIN1 downregulation resulted in decreased cellular migration and invasion abilities ()). Mechanistically, SPIN1 knockdown reduced the protein expression of β-catenin, c-Myc and cyclinD1, which are the core components of the Wnt pathway ()). Collectively, our results indicate that SPIN1 plays a critical role in accelerating cell proliferation and invasion by positively regulating the Wnt/β-catenin pathway.
Figure 2. SPIN1 depletion inhibits cell proliferation, migration and invasion. (a) qRT-PCR assays were utilized to confirm the transfection efficiency of siRNAs in HCT-116 and LOVO cells. (b, c) Cell proliferation ability was tested by CCK-8 assays. (d, e) The colony forming assays were used to evaluate the colony formation ability. (f- i) The migration and invasion abilities were evaluated by scratch wound assays (f, g) and transwell invasion assays (h, i) upon transfection of negative control siRNAs and SPIN1 siRNAs, respectively. (j) Representative western blotting analysis of SPIN1 and Wnt/β-catenin pathway components in HCT-116 and LOVO cells upon indicated transfection. Student’s t test, *P < 0.05 and **P < 0.01.
SPIN1 is a target of miR-381 and miR-381 is downregulated in CRC
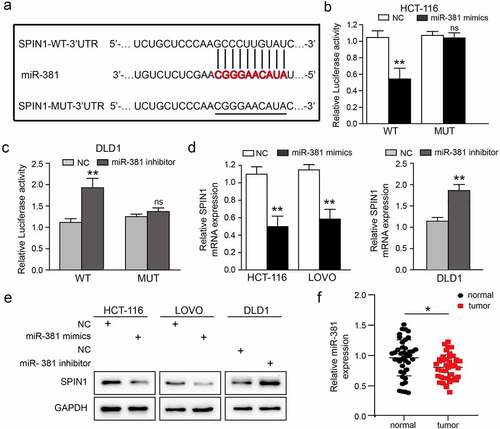
Bioinformatic tools (TargetScan and miRanda) were used to screen and identify potential miRNAs that could target SPIN1 in our study. We finally selected miR-381 for further investigation, as it was projected by these two prediction tools and found to be involved in tumorigenesis, including in CRC [Citation23,Citation24]. To validate the specific binding between miR-381 and SPIN1, a mutated luciferase reporter vector was constructed as the mutant-type (MUT) SPIN1 3ʹ-UTR ()). As clearly shown in ) and (c), miR-381 mimics or miR-381 inhibitor significantly affected the luciferase activity in CRC cells co-transfected with WT SPIN1 3ʹUTR, but not the MUT SPIN1 3ʹUTR. Moreover, qRT-PCR data showed that overexpression of miR-381 significantly inhibited the expression of SPIN1 mRNA in HCT116 and LOVO cells, while miR-381 inhibitors exerted the opposite effects ()). Furthermore, the western blotting analysis revealed that miR-381 upregulation remarkably repressed SPIN1 protein expression, while miR-381 downregulation increased SPIN1 expression in CRC cells ()). As well-documented, miRNAs often possess opposite expression patterns different from their target genes [Citation25]. Therefore, we detected the expression level of miR-381 in 42 pairs of CRC tissues. As shown in ), miR-381 was frequently downregulated in CRC tissues (25/42, 59.5%). After statistically analyzing the pathological characteristics of these 42 patients (), we found that downregulation of miR-381 was significantly related to undesirable characteristics, such as tumor size (p = 0.046) and TNM stage (p = 0.037). Taken together, our findings strongly support that SPIN1 is a target of miR-381, and miR-381 is downregulated and negatively correlated with poorer clinical characteristics in CRC.
Table 3. The association between miR-381 expression and clinicopathological factors in patients with colorectal cancer
Figure 3. SPIN1 is a target of miR-381 and miR-381 is downregulated in CRC. (a) Construction of the Wild-type (WT) and mutated (MUT) SPIN1 3ʹUTR luciferase reporter vectors. (b, c) The dual-luciferase reporter kit was used to detect the luciferase activity of WT and MUT SPIN1 3ʹUTR plasmids in HCT116 cells and DLD1 cells upon indicated transfection, respectively. (d) The expression of SPIN1 mRNA was assessed by qRT-PCR assays after indicated transfection in CRC cells. (e) Western blotting analysis of SPIN1 protein in CRC cells upon transfection by miR-381 mimics, miR-381inhibitor or negative controls. (f) Relative expression of miR-381 in colorectal cancer (CRC) tissues and paired adjacent tissues (n = 42). Student’s t test, *p < 0.05, **p < 0.01.
MiR-381 overexpression impedes CRC cell proliferation and invasion
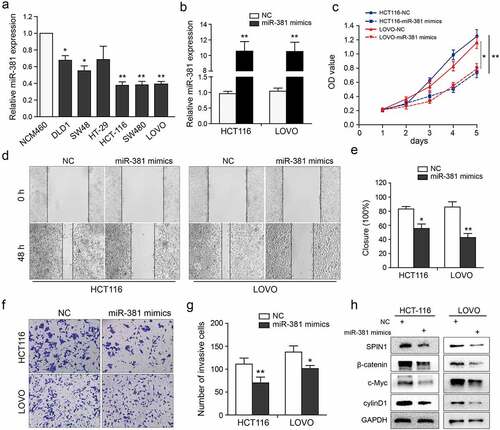
We further investigated the biological role of miR-381 in CRC cells. As clearly shown in ), miR-381 expression in CRC cells was markedly decreased when compared to NCM460 cells. Then, the qRT-PCR assays were used to validate the transfection efficiency of miR-381 mimics or negative controls ()). Clearly, miR-381 overexpression significantly suppressed cell growth, migration, and invasion abilities ()), which were similar to the effects of SPIN1 depletion. In addition, miR-381 overexpression inhibited the activity of Wnt/β-catenin pathway ()). Thus, these data indicate that miR-381 overexpression impedes cell proliferation and invasion of CRC cells.
Figure 4. MiR-381 overexpression impedes CRC cell proliferation and invasion. (a) The miR-381 expression in CRC cell lines was evaluated using qRT-PCR assays. (b) Transfection deficiency was validated by qRT-PCR assays in HCT116 and LOVO cells. (c) The well growth abilities of HCT-116 and LOVO were assessed using CCK-8 assays after relative transfection. (d-g) The role of miR-381 on migration and invasion was detected by wound healing and transwell invasion assays. Representative figures (magnification, ×200) and quantification analysis were formed by the indicated groups of cells. (h) Representative western blotting images of SPIN1 and Wnt/β-catenin pathway components following indicated transfection. GAPDH was used as an internal control. Student’s t test, *p < 0.05, **p < 0.01.
MiR-381 and SPIN1 are responsible for controlling the Wnt/β-catenin signaling pathway in CRC
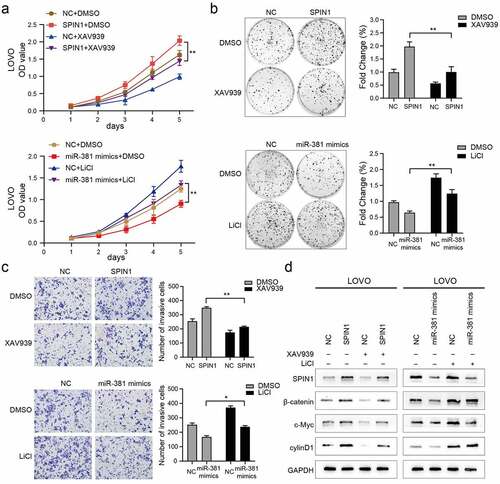
To further confirm that miR-381 and SPIN1 are critical in regulating cell proliferation and invasion by orchestrating the Wnt/β-catenin signaling pathway, the Wnt/β-catenin pathway inhibitor or activator was added for reverse assays. As shown in ), XAV-939 markedly rescued the proliferation and viability of SPIN1-overexpression CRC cells. Similarly, LiCl significantly suppressed the proliferation of CRC cells transfected with miR-381 mimics. The same phenomenon was observed in the colony formation assays and transwell invasion assays ()). More interestingly, we also observed that changes in Wnt/β-catenin pathway-related proteins (β-catenin, c-Myc, and cyclin D1) caused by SPIN1 overexpression plasmid or miR381 mimics can be recovered by XAV939 or LiCl, respectively ()). Collectively, these results demonstrated that MiR-381 and SPIN1 are responsible for controlling the Wnt/β-catenin signaling pathway, thereby affecting cell proliferation and invasion in CRC cells.
Figure 5. MiR-381 and SPIN1 are responsible for controlling the Wnt/β-catenin signaling pathway in CRC. (a-d) Rescue experiments of overexpressed SPIN1 and miR-381 mimics using with XAV939 or LiCl treatment for 24 h (XAV939,10 μmol/L; LiCl: 20 mmol/L). CCK-8 assays (a), Colony formation assays (b) and transwell invasion assay (c) were conducted to assess cell biological function upon indicated treatments. (d) The protein levels of SPIN1 and Wnt/β-catenin signaling pathway related targets (β-catenin, c-Myc and cyclin D1) were assessed by western blotting upon indicated treatments in LOVO cells. GAPDH was used as an internal control. Student’s t test, *p < 0.05, **p < 0.01.
Ectopic expression of SPIN1 attenuates the miR-381-induced effects on CRC cells
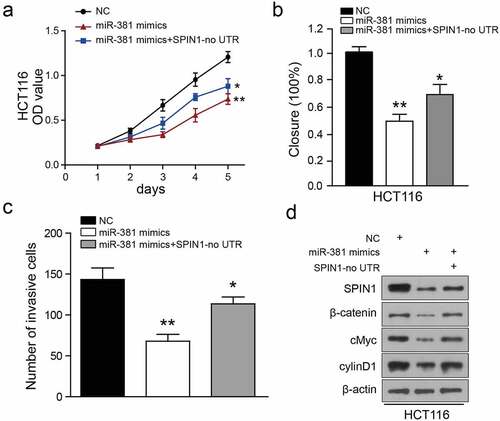
To investigate whether SPIN1 is a functional mediator of miR-381-induced effects, we synthesized a SPIN1 overexpression plasmid without 3ʹUTR and performed rescue assays. As illustrated in ), SPIN1 overexpression plasmid transfection restored SPIN1 expression, which was also partially suppressed by miR-381 overexpression in HCT116 cells. Similarly, functional experiments have demonstrated that overexpression of SPIN1 ameliorated the cancer-suppressive effects caused by miR-381 overexpression in CRC cells ()). Surprisingly, overexpression of SPIN1 could not completely abolish the effects of miR-381 on the Wnt/β-catenin pathway in HCT116 cells ()), suggesting that there may be other oncogenes or pathways involved in miR-381-mediated functions. In summary, these findings provide further evidence that SPIN1 functions as a main functional target of miR-381 in CRC.
Figure 6. Ectopic expression of SPIN1 attenuates the miR-381-induced effects on colorectal cancer cells. (a) CCK-8 assays performed on miRNA-381-overexpressing HCT-116 cells with SPIN1 overexpression plasmid. (b) Wound healing assays performed on miR-381-overexpressing HCT-116 cells with SPIN1 upregulation. (c) Quantification of the transwell invasion assays under the indicated transfection condition. (d) Representative western blotting images of lysates prepared from miRNA-381-overexpressing HCT-116 cells with SPIN1 upregulation. One-way ANOVA test, *p < 0.05, **p < 0.01.
Discussion
CRC is the third leading cause of cancer-related mortality globally, with a heavy burden on society [Citation26]. A detailed investigation of the underlying mechanism contributing to CRC development could provide new insights into the detection and treatment of CRC patients. Our previous data indicated that SPIN1 promotes tumorigenesis by negatively regulating the p53 pathway in multiple cancer cells [Citation9]. In the present study, our data revealed that the miR-381/ SPIN1 axis is critical in CRC proliferation and invasion via controlling the Wnt/β-catenin pathway.
Mounting evidence has demonstrated that dysregulation of SPIN1 contributes greatly to cancer tumorigenesis and progression in a series of tumors [Citation27–29]. Zhao et al. found that SPIN1 is highly expressed and triggers aberrant lipid metabolism in hepatocellular carcinoma patients [Citation6]. Moreover, increased SPIN1 expression enhanced cell growth, invasion and cell cycle progression in gastric cancer cells [Citation7]. Our study showed that SPIN1 was frequently upregulated in CRC tissues compared to adjacent noncancerous tissues. Additionally, SPIN1 upregulation was significantly associated with advanced clinical stage, depth of invasion, and metastasis, highlighting its critical role in the initiation and progression of CRC. Biological experiments have shown that SPIN1 depletion results in suppressed cell proliferation and invasion abilities in CRC cells, further implying that SPIN1 is a potential oncogene in CRC patients.
Numerous studies have documented that miRNAs are abnormally dysregulated in tumorigenesis, including CRC [Citation30–34]. Recently, SPIN1 was found to be negatively regulated by miR-148/152, leading to adriamycin resistance in breast cancer [Citation5]. Our luciferase reporter assays demonstrated that miR-381 could directly target the 3ʹ- UTR of SPIN1 mRNA. Several studies have suggested that miR-381 exerts its critical role in physical and pathological processes, including cell growth, apoptosis and tumorigenesis [Citation35–39]. For example, dendrobine suppressed the endoplasmic reticulum stress-induced apoptosis by increasing miR-381-3p expression in endothelial cells [Citation38]. Previously, miR-381 was reported to suppress cell proliferation and metastasis by targeting the BM1 and Rho/ROCK axis in bladder cancer [Citation40]. Additionally, miR-381 facilitates autophagy and apoptosis by suppressing the RELN-mediated PI3K/Akt pathway in prostate cancer cells [Citation41]. However, miR-381 may also exert oncogenic functions in some other cancers, including epithelioid sarcoma and glioma [Citation42,Citation43]. In the present study, we further strengthened the tumor suppressive role of miR-381 in CRC cells. Our data showed that miR-381 was frequently decreased in CRC clinical samples and cell lines. Moreover, functional assays revealed that overexpression of miR-381 repressed cancer cell proliferation and invasion, which recapitulated the impact of SPIN1 depletion. These data indicate that miR-381 exerts its tumor-suppressive roles by inhibiting SPIN1 expression, and this newly identified miR-381/SPIN1 regulatory axis greatly contributes to CRC tumorigenesis and progression.
It is well-established that the excessive activation of the Wnt/β-catenin pathway is involved in the development in CRC cells [Citation44]. Previous studies have shown that SPIN1 serves as a positive modulator of Wnt/β-catenin pathway [Citation4,Citation45]. In line with this, our study found that Wnt target genes, c-Myc and cyclinD1, were regulated by SPIN1. Moreover, β-catenin is also regulated by the level of SPIN1. We suspect SPIN1 overexpression might attract β-catenin translocates from the cytoplasm to the nucleus, thereby avoiding β-catenin degradation in the cytoplasm. In addition, our previous data showed that miR-381 delayed cancer progression in gastric cancer by suppressing the Wnt/β-catenin pathway [Citation35]. Consistent with it, we also found that miR-381 upregulation impeded the malignant behaviors of CRC cells and reintroduction of SPIN1 rescued the inhibitory effect of miR-381 on the Wnt/β-catenin pathway. More interestingly, the Wnt/β-catenin signaling-specific inhibitor XAV-939 and activator LiCl partially eliminated the effects of SPIN1 overexpression and miR-381 mimics, which further supports the notion that the miR-381/SPIN1 axis is critical for regulating the Wnt/β-catenin pathway. Besides, it is worth considering that SPIN1 overexpression could not completely abolish miR-381-induced effects, indicating that other mechanisms may also participate in miR-381 functions. In summary, our data indicate that miR-381 restrains the activation of the Wnt/β-catenin pathway by targeting SPIN1.
Conclusions
Collectively, our results showed that the dysregulated miR-381/SPIN1 axis greatly contributes to the initiation and progression of CRC cells by orchestrating the Wnt/β-catenin pathway, and therefore represents an actionable therapeutic target for CRC patients.
Highlights
SPIN1 is frequently upregulated in CRC, and SPIN1 overexpression facilitates the cell proliferation and invasion of CRC cells.
SPIN1 is a target of miR-381.
MiR-381 impedes colorectal cancer cell proliferation and invasion through inactivation of the Wnt/β-catenin pathway via targeting SPIN1.
Ethics approval and consent to participate
This study has been approved by the Medical Research Ethics Committee of the First Affiliated Hospital of Nanchang University.
Acknowledgements
We thank the National Natural Science Foundation of China (81860545, 82060493 and 82060566), the Natural Science Foundation of Jiangxi Province (20192BAB215040, 20192BAB215015 and 20202ACBL216011), the Outstanding Young Talents Project of Jiangxi Province (20192BCB23020), the Education Department of Jiangxi Province (701238001) and the Department of Health Project of Jiangxi Province (20201015 and 20203124) for their support.
Availability of data and materials
The datasets used or analyzed during the present study are available from the corresponding authors Ziling Fang and Xiaojun Xiang upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Keum N, Giovannucci EJ. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev . 2019;16(12):713–732.
- Dekker E, Tanis P, Vleugels J, et al. Colorectal cancer. Lancet (London, England). 2019;394:1467–1480.
- Yue W, Sun L, Li C, et al. [Screening and identification of ovarian carcinomas related genes]. Ai Zheng = Aizheng = Chin J Cancer. 2004;23:141–145.
- Su X, Zhu G, Ding X, et al. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev. 2014;28:622–636.
- Chen X, Wang Y, Gao PJ. SPIN1, negatively regulated by miR-148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res CR. 2018;37:100.
- Zhao M, Bu Y, Feng J, et al. SPIN1 triggers abnormal lipid metabolism and enhances tumor growth in liver cancer. Cancer Lett. 2020;470:54–63.
- Lv B, Ma R, Chen X, et al. E2F1-activated SPIN1 promotes tumor growth via a MDM2-p21-E2F1 feedback loop in gastric cancer. Mol Oncol. 2020;14:2629–2645.
- Jin Y, Su Z, Sheng H, et al. Circ_0086720 knockdown strengthens the radiosensitivity of non-small cell lung cancer via mediating the miR-375/SPIN1 axis. Neoplasma. 2021;68:96–107.
- Fang Z, Cao B, Liao J, et al. SPIN1 promotes tumorigenesis by blocking the uL18 (universal large ribosomal subunit protein 18)-MDM2-p53 pathway in human cancer. 2018;7:e31275.
- Li D, Guo J, Jia R. Histone code reader SPIN1 is a promising target of cancer therapy. Biochimie. 2021;191:78–86.
- Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science (New York, N.Y.). 2001;294(5543):853–858.
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23(1):175–205.
- Akbari A, Sedaghat M, Heshmati J, et al. Molecular mechanisms underlying curcumin-mediated microRNA regulation in carcinogenesis; Focused on gastrointestinal cancers. Biomed Pharmacother. 2021;141:111849.
- Cai L, Ye L, Hu X, et al. MicroRNA miR-330-3p suppresses the progression of ovarian cancer by targeting RIPK4. Bioengineered. 2021;12(1):440–449.
- Jinghua H, Qinghua Z, Chenchen C, et al. MicroRNA miR-92a-3p regulates breast cancer cell proliferation and metastasis via regulating B-cell translocation gene 2 (BTG2). Bioengineered. 2021;12:2033–2044.
- Akbari A, Majd HM, Rahnama R, et al. Cross-talk between oxidative stress signaling and microRNA regulatory systems in carcinogenesis: focused on gastrointestinal cancers. Biomed Pharmacother. 2020;131:110729.
- He X, Wei Y, Wang Y, et al. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther. 2016;9:1231–1239.
- Moody L, Dvoretskiy S, An R, et al. The efficacy of miR-20a as a diagnostic and prognostic biomarker for colorectal cancer: a systematic review and meta-analysis. 2019;11(8):1111.
- Sur D, Burz C, Sabarimurugan S, et al. Diagnostic and prognostic significance of MiR-150 in colorectal cancer: a systematic review and meta-analysis. J Pers Med. 2020;10. DOI:10.3390/jpm10030099
- Garo L, Ajay A, Fujiwara M, et al. MicroRNA-146a limits tumorigenic inflammation in colorectal cancer. Nat Commun. 2021;12:2419.
- Fang Z, Zhang L, Liao Q, et al. Regulation of TRIM24 by miR-511 modulates cell proliferation in gastric cancer. J Exp Clin Cancer Res CR. 2017;36:17.
- Fang Z, Deng J, Zhang L, et al. TRIM24 promotes the aggression of gastric cancer via the Wnt/β-catenin signaling pathway. Oncol Lett. 2017;13:1797–1806.
- He X, Wei Y, Wang Y, et al. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Oncotargets Ther. 2016;9:1231–1239.
- Liang Y, Zhao Q, Fan L, et al. Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomed Pharmacother. 2015;75:137–141.
- Gao F, Wu H, Wang R, et al. MicroRNA-485-5p suppresses the proliferation, migration and invasion of small cell lung cancer cells by targeting flotillin-2. Bioengineered. 2019;10(1):1–12.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clinicians. 2018;68:394–424.
- Du H, Liu BJ. MiR-1271 as a tumor suppressor in breast cancer proliferation and progression via targeting SPIN1. Eur Rev Med Pharmacol Sci. 2018;22:2697–2706.
- Song Q, Ji Q, Xiao J, et al. miR-409 inhibits human non-small-cell lung cancer progression by directly targeting SPIN1. Mol Ther Nucleic Acids. 2018;13:154–163.
- Li Y, Ma X, Wang Y, et al. miR-489 inhibits proliferation, cell cycle progression and induces apoptosis of glioma cells via targeting SPIN1-mediated PI3K/AKT pathway. Biomed Pharmacother Biomed Pharmacotherapie. 2017;93:435–443.
- Lee Y, Dutta AJ. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227.
- Hayes J, Peruzzi P, Lawler SJ. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469.
- Liu Y, Liu R, Yang F, et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16:53.
- Ren Y, Zhang H, Jiang PJ. MicroRNA-382 inhibits cell growth and migration in colorectal cancer by targeting SP1. Biol Res. 2018;51:51.
- Shuai F, Wang B, Dong SJ. MicroRNA-204 inhibits the growth and motility of colorectal cancer cells by downregulation of CXCL8. Oncol Res. 2018;26:1295–1305.
- Fang Z, Zhong M, Wang Y, et al. miR‑381 and miR‑489 suppress cell proliferation and invasion by targeting CUL4B via the Wnt/β‑catenin pathway in gastric cancer. Int J Oncol. 2019;54:733–743.
- Gu F, Zhang J, Yan L, et al. CircHIPK3/miR-381-3p axis modulates proliferation, migration, and glycolysis of lung cancer cells by regulating the AKT/mTOR signaling pathway. Open Life Sci. 2020;15:683–695.
- Ma H, Lv G, Zhang DH. miR-381 mediates the development of head and neck squamous cell carcinoma via targeting STC2. Oncotargets Ther. 2020;13:4485–4493.
- Meng J, Song X, Yan G, et al. Dendrobine suppresses endoplasmic reticulum stress-induced apoptosis through upregulating microRNA miR-381-3p to decrease caspase-4. Bioengineered. 2021;12:4452–4463.
- Sun X, Hu X, Wang X, et al. MiR-381-3p/RAB2A axis activates cell proliferation and inhibits cell apoptosis in bladder cancer. Cel Mol Biol. 2020;66:117–120.
- Chen D, Cheng L, Cao H, et al. Role of microRNA-381 in bladder cancer growth and metastasis with the involvement of BMI1 and the Rho/ROCK axis. BMC Urol. 2021;21:5.
- Liao W, Zhang YJ. MicroRNA-381 facilitates autophagy and apoptosis in prostate cancer cells via inhibiting the RELN-mediated PI3K/AKT/mTOR signaling pathway. Life Sci. 2020;254:117672.
- Tang H, Liu X, Wang Z, et al. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 2011;1390:21–32.
- Papp G, Krausz T, Stricker T, et al. chromosomes, cancer. SMARCB1 expression in epithelioid sarcoma is regulated by miR-206, miR-381, and miR-671-5p on both mRNA and protein levels. Genes, Chromosomes amp Cancer. 2014;53:168–176.
- Krishnamurthy N, Kurzrock RJ. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60.
- Drago-Ferrante R, Pentimalli F, Carlisi D, et al. Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget. 2017;8(17):28939–28958.
