ABSTRACT
Engineering of cellular biomolecules is an emerging landscape presenting creative therapeutic opportunities. Recently, several strategies such as biomimetic materials, drug-releasing scaffolds, stem cells, and dynamic culture systems have been developed to improve specific biological functions, however, have been confounded with fundamental and technical roadblocks. Rapidly emerging investigations on the bioengineering prospects of mammalian ribonucleic acid (RNA) is expected to result in significant biomedical advances. More specifically, the current trend focuses on devising non-coding (nc) RNAs as therapeutic candidates for complex neurological diseases. Given the pleiotropic and regulatory role, ncRNAs such as microRNAs and long non-coding RNAs are deemed as attractive therapeutic candidates. Currently, the list of non-coding RNAs in mammals is evolving, which presents the plethora of hidden possibilities including their scope in biomedicine. Herein, we critically review on the emerging repertoire of ncRNAs in neurological diseases such as Alzheimer’s disease, Parkinson’s disease, neuroinflammation and drug abuse disorders. Importantly, we present the advances in engineering of ncRNAs to improve their biocompatibility and therapeutic feasibility as well as provide key insights into the applications of bioengineered non-coding RNAs that are investigated for neurological diseases.
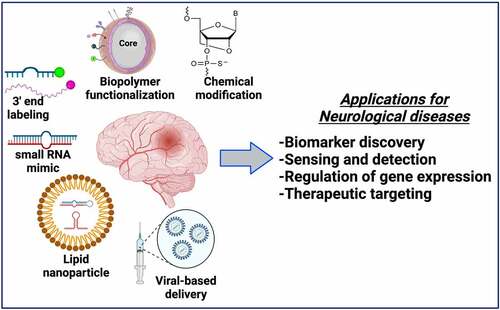
GRAPHICAL ABSTRACT
Introduction
Noncoding RNAs (ncRNAs) are a large cluster of non-protein coding nucleic acid species that have multiple functions in diverse cellular processes. Based on the biological functions and size (i.e., length of nucleotides) ncRNAs are divided into several types such as, ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), guide RNAs (gRNAs), microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and telomerase RNAs [Citation1–3]. Importantly, regulatory ncRNAs can be classified into microRNAs (miRNAs), small interfering RNAs (siRNAs), long noncoding RNAs (lncRNAs), Piwi-interacting RNAs (piRNAs), promoter-associated RNAs (PARs), and enhancer RNAs (eRNAs) [Citation1–3]. Several ncRNAs perform important roles in a wide range of biological and pathological processes. As such, interest in ncRNA biology has grown throughout biomedicine, biomedical informatics, and clinical sciences. In addition, the field of ncRNA research has witnessed significant progress in recent years due to amalgamation of new-age sequencing ‘omics’ technologies that have contributed to the growing repository of big-data in biomedicine [Citation4,Citation5].
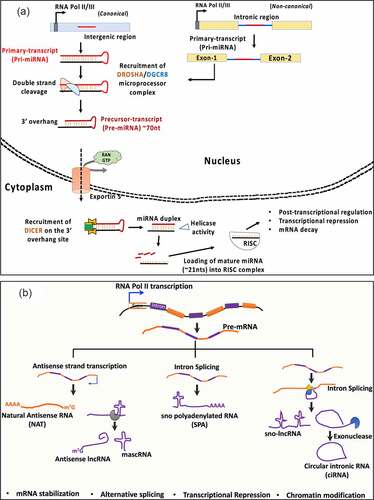
The dynamic expression and functionality across cell/tissue types has made ncRNAs attractive therapeutic targets. More recently, there have been intensive efforts for establishing siRNAs and miRNAs therapeutics against several diseases including cancer, cardiovascular, genetic disorders as well as to combat viral-induced pathogenesis [Citation6–11]. Currently, there are extensive developments in laboratory investigation approaches to test the therapeutic feasibility of ncRNAs. For instance, the therapeutic feasibility of ncRNAs is being extensively tested in cell and tissue engineering and models of human diseases including cancer, vascular diseases and neurodegeneration by strategies that include the use of antisense oligonucleotides (ASOs), small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs), anti-microRNAs (antimiRs), miRNA mimics, miRNA sponges, circular RNAs (circRNAs) whose details have been reviewed elegantly in previous studies [Citation12–14]. The mainstream approaches for ncRNA-based tissue regeneration therapy include altering endogenous cellular activity using ncRNAs to influence the behavior of resident stem/progenitor cells, cells incorporated into tissue engineered constructs, or modulating the fate of both implanted and endogenous cells with selected ncRNAs [Citation15–18]. MicroRNAs (miRNAs) are 18 ~ 21 nucleotides long, single stranded non-coding RNAs with pleiotropic functions [Citation19,Citation20]. MiRNAs are products of RNA Polymerase II driven transcription which target mature mRNAs in the cytoplasm based on the degree of sequences complementarity (seed, 2–8 nucleotides) between the 5ʹ end of the miRNA and 3ʹregion of the target mRNA (). MiRNA-dependent activity is primarily post-transcriptional that results in either mRNA degradation or halting of translation machinery, which in both cases represses target gene expression (). On the other hand, lncRNAs are defined as RNA transcripts longer than 200 nucleotides (nts) that are transcribed from intergenic and regions upstream of gene promoters, enhancer-coding regions including the anti-sense strands of protein-coding genes that can exist in both polyadenylated and non-polyadenylated states as illustrated in [Citation1,Citation21–23]. LncRNAs are associated with multiple mechanisms of action, such as chromatin modification, microRNA sequestration by sponging, scaffolding for protein interaction to regulate protein activity, transcriptional regulation by activation or repression via transcription factors, mRNA stabilization, splicing modulation, and modulation of translation and degradation of target mRNAs [Citation1,Citation21–23]. A brief description on the maturation of miRNA, lncRNA and other ncRNA species including the circular intronic RNAs (ciRNAs) along with their established cellular functions is illustrated in .
Figure 1. Cellular fates of miRNA and LncRNA maturation. (a) microRNAs, transcribed as primary transcripts from intergenic (pri-miRNA) or, intronic regions (mirtrons, non-canonical). DROSHA-DiGeorge syndrome critical region 8 (DGCR8) complex processes the pri-mRNA structures to produce precursor (pre-mRNA) sequence which is exported from the nucleus to the cytosol for further processing by DICER1 and helicase activity to produce a mature 18–22nts long sequence that undergoes ribonucleoprotein (RNP) assembly with the family of Argonaute proteins (AGO1-4), called as RNA-induced silencing complex (RISC) complex. (b) Long non-coding RNAs are products of Pol II transcription from intergenic regions. They are presumed to undergo processing events including capping, splicing and polyadenylation, in some cases. From left – Natural anti-sense transcripts are transcribed from the opposite(complementary) strands of mRNAs (usually coding in nature). Anti-sense lncRNA and MALAT-1 associated RNA (mascRNA) are produced because of RNAse P processing. U-A-U triple helix at the 3ʹends of mascRNA provides structural stability. sno polyadenylated RNAs (SPA) are 3ʹpolyadenylated and produced from read-through transcripts that are assembled with sno ribonuclear proteins (snoRNPs). In contrast, sno-lncRNAs lack a 5ʹm7G cap and 3ʹ poly (A) tail and are produced from excited introns post splicing events. Finally, the circular intronic RNAs (ciRNAs) are also a product of intron excision post splicing events often produced as an outcome of 3ʹexonuclease activity
It is no doubt that ncRNAs such as miRNAs, lncRNAs, circRNAs are abundant in the CNS. For instance, more than 40% of lncRNAs are expressed in the CNS whereas, several circRNAs and miRNAs are enriched in the neurons and their synaptic junctions. For example, studies have established several ncRNAs including miRNAs (miR-124, miR-125b, miR-21, miR-9), lncRNAs (Gomafu, Neat1) and circular RNAs (CDR1-AS) are highly expressed in the brain in a region or, cell-type-specific manner [Citation24–29]. For instance, miR-124 is abundantly expressed during early stages of embryonic development and neuronal differentiation during adulthood whereas, miR-21 is enriched in the microglia [Citation25]. Similarly, miR-383 is a neuronal enriched miRNA that is abundantly expressed in the brain stem and the cerebellum region [Citation25]. On the other hand, lncRNAs such as Gomafu, MALAT-1 and Neat 1 are implicated in glial specification and synaptic plasticity [Citation30]. Overall, these studies establish the importance of ncRNAs in the development and function of the mammalian brain and present the feasibility of therapeutic targeting of the enriched and abundantly expressed ncRNA species in neurological diseases. The improvement in genomic tools and advances in high power computational algorithms in amalgamation with ‘omics’ technologies has significantly advanced our knowledge on the role of several ncRNAs in phases of brain development to their alterations in diseased states. Neurological diseases or central nervous system (CNS) disorders are broadly defined by the loss of neurons featuring irreversible tissue injury that is manifested in dramatic phenotypic and behavioral changes. Extensive research and current discoveries have established that both miRNAs and lncRNAs are critical determinants of gene expression changes and phenotypic outcomes in neurological diseases. More recently, there is an increasing attention toward understanding the role of circular ncRNA species in healthy and diseased states of the mammalian brain [Citation29,Citation31,Citation32]. Recently there is a major drive in establishing circRNA species as effective stage-specific biomarkers for neuropsychiatric disorders [Citation33–36]. Collectively, the altered expression of ncRNAs such as miRNAs, lncRNAs and circRNAs in circulation (blood, serum, cerebrospinal fluid, CSF) in diseased conditions establishes their importance as biomarkers for disease diagnostics. Therefore, the development of ncRNA based therapeutic approaches could advance the existing repertoire of CNS therapeutics and could further our understanding of ncRNA biology in the healthy and diseased states of the mammalian brain. We acknowledge that the repertoire of ncRNAs in the brain is constantly emerging. However, our understanding of ncRNA regulation and function is primarily based on the evidence gained from miRNAs and lncRNAs. Hence, we critically review the role of ncRNAs (especially miRNAs, lncRNAs) in the major neurological diseases such as Alzheimer’s disease, Parkinson’s disease, along with neuroinflammation and drug abuse disorders that present a global healthcare burden and unmet clinical need. In this review we also discuss on the bioengineering approaches of these ncRNAs especially miRNAs and lncRNAs that are currently underway to improve their therapeutic and diagnostic feasibility for neurological diseases.
ncRNAs implicated in neurological disorders
ncRNAs confer a high level of regulation by governing gene expression changes at both transcriptional and post-transcriptional levels implicated in the development and function of the CNS. Novel high-throughput molecular and imaging technologies have established their function in neuronal development, differentiation, synapse function as well as in regulation of behavior and cognition. In the recent years an extensive line of evidence has underscored the critical role of ncRNAs in brain development, blood brain barrier formation and regulation, stress response, cellular homeostasis, and neuronal plasticity [Citation37–48]. In addition, loss of function studies have revealed the diseased/pathological outcomes associated with ncRNA function [Citation49–52]. While there are several elegant reviews that can be referred to gain insights into the role of ncRNA species in each of the neurological diseases, in this section we aimed to provide a general overview of the status and role of ncRNAs that are abundantly expressed in the CNS and are dramatically altered in Alzheimer’s disease, Parkinson’s disease and other neurovascular inflammatory conditions.
Alzheimer’s disease (AD)
Alzheimer’s disease is a complex neurodegenerative disorder and is the predominant cause of elderly dementia [Citation53,Citation54]. The symptoms mark a slow yet progressive memory loss and cognitive decline due to the impairment of region-specific neurons and cellular circuits, which eventually lead to neuronal death/loss. In this regard, several miRNAs and lncRNAs have been identified as key factors that are essential for the regulation of cognitive functions and memory processes in AD [Citation55,Citation56]. For example, in a mouse model of AD (Tg2576 mice), miR-124 was found to be dramatically induced in the hippocampus, which was directly associated with deficits in synaptic communication and memory dysfunction [Citation57]. Studies have shown that expression of several miRNAs was altered in AD pathology, such as miR-30a-5p and miR-128 [Citation58]. In the brains of transgenic animals of AD, a reduction of miR-298 and miR-328 was detected, which was associated to higher β-amyloid precursor protein converting enzyme (BACE1) protein [Citation59]. In related models the level of miR-195 has been reported to negatively correlate with BACE1 protein expression [Citation60]. miR-124 has also been shown to negatively correlate with BACE1 expression in mouse PC12 cells and primary cultured hippocampal neurons exposed to Aβ treatments [Citation61,Citation62]. Overexpression of miR-98 has been shown to induce Aβ production and tau phosphorylation in vivo, whereas inhibition of miR-98 exhibits opposing effects [Citation63]. In primary cultures of human brain cells and in brain specimens from AD patients, it has been shown that miRNAs are altered that may have effects on Aβ deposition [Citation64–67].
With regards to lncRNAs in AD pathology, BACE1-AS, a natural antisense transcript (NAT), which is a transcript that is endogenously transcribed from β-amyloid precursor protein cleaving enzyme 1 mRNA sequence has been shown to trigger production and aggregation of Aβ through BACE1 activity, leading to detrimental effects [Citation68]. In a recent study, BC200 RNA was discovered in quantifiable levels specifically in AD patients compared to a gradual decreasing trend seen in the normal age-matched control group [Citation69], and suppressing BC200 inhibited BACE1 expression [Citation70]. It is important to note that the direct role of BACE1 in AD pathology remains unclear. Moreover, the antisense transcript 17A expression has been found to be upregulated via the inflammatory signaling leading to increased secretion of Aβ in the cerebral tissue from AD patients [Citation71]. Another natural antisense transcript, NAT-Rad18 that acts by silencing Rad18 mRNA, has reported to be upregulated followed by Aβ-derived neurotoxic apoptosis [Citation72]. Studies have also shown that the complimentary strand of Glial cell-derived neurotrophic factor (GDNF) codes for GDNFOS lncRNA, which shows distinctive expression patterns in postmortem middle temporal gyrus from AD patients compared to age-matched normal group [Citation73].
Parkinson’s disease
Parkinson’s disease (PD) is considered a multifactorial and aging-related progressive disease, commonly characterized by movement-associated disabilities that depend on the extent of neurodegeneration [Citation74,Citation75]. It is established that motor dysfunction in PD underlies loss of dopaminergic neurons, specifically in midbrain’s substantia nigra [Citation74]. The motor deficits in PD primarily include bradykinesia, rigidity, and tremor [Citation75]. Using high throughput techniques such as RNA sequencing, microarray and microRNA qPCR profiling, studies have reported involvement of several microRNA and lncRNA species in PD pathology [Citation76]. For instance, in the postmortem human brain miR-30b in the substantia nigra and miR-30 c-2 and miR-30d in cingulate gyri were found to be induced in PD patients compared to healthy controls [Citation77,Citation78]. Moreover, induced miR-30b was detected in the exosomes isolated from the cerebrospinal fluid of PD patients [Citation79]. Differential expression levels of miR-30 family members have also been reported in the peripheral blood, plasma and serum of PD patients by several research groups [Citation80–82]. Similarly, miR-29a, miR-29b-1 and miR-29b-2 were upregulated in the anterior cingulate gyri of PD patients [Citation78] Moreover, miR-29b and miR-29 c were downregulated in the PBMCs of PD patients [Citation80] whereas, decreased miR-29a was observed in the blood sample of both drug-treated and untreated PD patients as compared to healthy controls [Citation83]. Based on these observations, it can be suggested that blood serum levels of miR-29a and miR-29 c decrease with the severity of PD pathology [Citation84]. miR-29 has been shown to regulate various processes that are important in PD development, such as apoptosis [Citation85–87], neuronal survival [Citation86], senescence [Citation88,Citation89], motor function [Citation86,Citation90], immune regulation [Citation91–93], and epigenetic modifications [Citation94,Citation95]. Studies have also reported alteration in expression of let-7 family of miRNAs i.e., let-7d-5p, let-7 f-5p and let-7 g in both the prefrontal cortex and blood of PD patients [Citation96]. In the central nervous system, miR-26a was observed to be upregulated in the substantia nigra tissues and in the exosomes isolated from the cerebrospinal fluid of PD patients as compared to healthy controls [Citation77,Citation79], and similar findings have been also reported in the striatal tissues of rodent PD models [Citation97].
On the other hand, recent studies have discovered the involvement of several lncRNA species in Parkinson’s pathology. For instance, NEAT1 levels were significantly upregulated in the peripheral blood of PD patients [Citation98] and has been shown to sponge miR-124 to accelerate Parkinson’s pathology [Citation99]. Homeobox (HOX) transcript antisense RNA (HOTAIR), ~2.2kb nucleotide long non-coding transcript found at the HOXC genetic locus has been reported in PD progression [Citation100]. Studies have revealed a notable decrease in MAPT-AS transcripts encoded from the antisense strand of microtubule-associated protein tau (MAPT) in brain regions of PD-derived tissue [Citation101].
Neurovascular inflammation
Both miRNAs and lncRNAs have been shown to play crucial roles in the control of brain function [Citation102–104]. Inflammation is one of the important factors that is involved during ischemia-induced brain damage. Pro-inflammatory secretory factors such as IL-1ß, IL-6, TNF-α are robustly induced in response to the damaged blood vessels, tissue injury and cellular apoptosis [Citation105,Citation106]. The combined effect exacerbates brain injury thus making it irreversible. One of the significantly affected signaling pathways is the NF- kß pathway sought to be a prime target for miRNA-mediated regulation [Citation107]. For instance, miR-146a has been shown to inhibit NF-kß signaling by targeting its adaptor proteins, tumor associated necrotic factor 6 (TRAF6) and receptor associated kinase-1 (RAK-1), involved in signal activation [Citation107,Citation108]. miR-146 and miR-9 regulate NF-kß signaling in rodent models of experimental stroke [Citation107,Citation109]. In another study miR-155 was shown to regulate inflammation by promoting TNF- α and IL-1 ß expression via toll-like receptor 4 (TLR4) response in transient middle cerebral artery occlusion (tMCAO) model [Citation110]. Stroke-induced ischemic damage induces oxidative stress mediated cell death causing irreversible tissue injury followed by secondary brain damage. Induced reactive oxygen and nitrogen species (ROS and RNS), hydrogen peroxide and free radicals are resultants that subsequently damage the neurovascular unit via apoptosis, cellular dysfunction and exacerbated pro-inflammatory signaling [Citation111,Citation112]. Clinically, the additive response of several molecular events underlying stroke pathology determines the infarct volume. Several microRNAs have been accounted to regulate an array of molecular events underlying pathology associated with stroke-induced ischemia [Citation113–116]. For instance, knockdown of miR-23a-3p in the cerebro-ventricular brain region of mice exposed to tMCAO reduced infarct volume in models of ischemia/reperfusion injury [Citation117]. On the other hand, microRNA miR-424 was shown to positively regulate superoxide dismutase activity where knockdown using antagomiRs in the cerebro-ventricular region rescued infarct volume via inhibition of cellular apoptosis [Citation118]. In the serum of acute ischemic stroke patients, miR-15a, miR-16, miR-17-5p, miR-125b-5p, miR-125a-5p, miR-143-3P and miR-106b-5p have been detected in elevated levels [Citation119,Citation120]. When miR-106b-5p was inhibited in rat brain post tMCAO, significant recovery was observed in neurological deficit scores and infarct volume [Citation121]. Stroke pathology significantly affects angiogenesis by altering the levels of angiogenic factors (for instance, vascular endothelial growth factor A, VEGF-A) essential for vascular remodeling and functional recovery post-stroke. Over the past few years, several miRNAs have been reported to regulate VEGF-A expression in models of experimental stroke. For instance, miR-140-5p, miR-377, miR-150, miR-107 and miR-210 have been shown to post-transcriptionally regulate VEGF-A mRNA levels by targeting its 3ʹ untranslated region (UTR). Ischemia has been shown to induce angiogenesis via the induction of HIF-1alpha dependent VEGF-A/Notch signaling axis and downregulation of miR-153-3p [Citation122].
Other species of small non-coding RNA include small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), endo-siRNA, vault RNA, yRNA, tRNA-derived stress-induced RNA, telomere small RNAs, centromere repeat-associated RNA, microRNA-offset RNA, tRNA-derived RNA fragments and splice-site RNAs that are involved in several aspects of development and function [Citation123,Citation124]. Although this diverse family of small non-coding RNAs, microRNAs are the most extensively studied in experimental models of neuroinflammation essentially due to their stability with a well-defined maturation cycle together with the access of current molecular tools and techniques. The depth of knowledge gained from the miRNAs and their function has served as a catalyst toward the development of efficient tools to study the molecular and cellular function of non-coding RNAs.
It is evident that tremendous advances have been made toward understanding the role of noncoding RNAs especially microRNAs. However, the focus is now steadily shifting toward understanding the role of lncRNAs in development and diseases of the mammalian brain. In general, the widespread use of RNA sequencing (RNA-seq) screens has led to the discovery of promising candidates that could pave the way for unveiling the therapeutic potential of ncRNAs. It can also be envisioned that lncRNAs can potentially serve as efficient tools for personalized medicine based on their specific expression patterns in each diseased condition or in a given population. Numerous challenges remain to be resolved before lncRNAs can reach clinical application. The evolving functional repertoire of lncRNAs highlights their involvement in more than one mechanism in diseased states adding layers of complexity to their molecular characteristics. For instance, single nucleotide polymorphisms (SNPs) in ANRIL and H19 have been associated with risks of cancer development and cardiovascular disease [Citation125,Citation126]. Additionally, the low conservation of lncRNAs across evolution of mammalian species is a bottleneck in their discovery and functional validation [Citation127–129]. Based on the knowledge gathered from microRNAs, it is often suggested that the conserved secondary structure of lncRNAs is also of higher importance than its full-length sequence. A key challenge to overcome constitutes the modes of tissue-specific delivery and ensuring the functionality in the targeted cell types of the anti-sense oligos targeting a specific lncRNA. There is a significant lack of in-depth understanding of lncRNA function. Moreover, the translation of lncRNA-based therapy into clinical applications is a long haul that is faced by challenges in understanding the molecular function as well as pharmacological features that include route of delivery, stability of RNA drug in both circulation and cells, duration of treatment, dosage adjustment, and off-target effects. Hence, extensive efforts are needed for a thorough characterization and functional validation of lncRNAs in neuronal and neurovascular pathology, both at the molecular and at the cellular level. A table with the list of miRNAs and lncRNAs well reported in the brain and CNS of patients with neurological disorders are listed in .
Table 1. List of non-coding RNAs (miRNAs, lncRNAs) detected in the brain regions and cerebrospinal fluid of patients suffering from neurological diseases
Disease-specific ncRNAs that have been reported with significantly altered expression in patients are listed in the table have been summarized Alzheimer’s disease, Parkinson’s disease, for Schizophrenia and Huntington’s disease.
It is known that ncRNAs are pleiotropic molecules that exhibit multiple functionalities. With regards to the therapeutic prospects of ncRNAs, it is also important to identify the genetic or molecular targets of ncRNAs that are implicated in diseased states. Therefore, given this pathophysiological importance we have summarized some of the widely reported miRNA and lncRNA species and their experimentally validated genetic targets in respective neurological diseases in .
Table 2. List of non-coding RNAs (miRNAs, lncRNAs) with their respective genetic targets reported in neurological disorders
Abbreviations of gene names: GSK-3β (Glycogen synthase kinase-3-beta); CAMKK2 (Calcium/Calmodulin Dependent Protein Kinase Kinase 2); SIRT1 (Sirtuin 1); ROCK1(Rho Associated Coiled-Coil Containing Protein Kinase 1); CFH (complement factor H); TRAF-6 (TNF Receptor Associated Factor 6); IL-1 (Interleukin-1); IRAK-1(Interleukin 1 Receptor Associated Kinase 1); FOXQ1 (Forkhead Box Q1), PTGS2(Prostaglandin-Endoperoxide Synthase 2); CDK5 (Cyclin Dependent Kinase 5); DUSP6 (dual-specificity phosphatase 6); PPP1CA (Protein Phosphatase 1 Catalytic Subunit Alpha); BACE-1 (beta-site amyloid precursor protein cleaving enzyme); PTPN (Protein Tyrosine Phosphatase Non-Receptor); NR3C1(Nuclear Receptor Subfamily 3 Group C Member 1); FGF-7(Fibroblast Growth Factor 7); SORL1 (Sortilin Related Receptor 1); BDNF (Brain-derived neurotrophic factor); GDNF (Glial cell-derived neurotrophic factor); EPHB2 (Ephrin type-B receptor 2); RAD18 (RAD1 Checkpoint DNA Exonuclease 18); LRP1 (LDL Receptor Related Protein 1); HMGB2 (High Mobility Group Box 2); SNCA(Alpha-synuclein); MAPK-JNK(Map kinase- Jun Kinase); PARK2 (Parkin2); LRRK2 (Leucine Rich Repeat Kinase 2); ATG5 (Autophagy Related 5); USP6 (Ubiquitin Specific Peptidase 6); NEDD4 (Neuronal precursor cell-expressed developmentally downregulated 4); USP3(Ubiquitin Specific Peptidase 3); DNMT3A (DNA methyltransferase 3 alpha); TET2 (Ten-Eleven Translocation 2); E2F1 (E2F Transcription Factor 1); PINK1 (PTEN Induced Kinase 1); TAK1(Transforming Growth Factor-Beta-Activated Kinase 1); TAB3(TGF-Beta Activated Kinase 1 (MAP3K7) Binding Protein 3); UCH-L1(ubiquitin carboxyl-terminal esterase L1); NLRP3 (Nod like receptor family pyrin domain containing 3); SOCS3 (Suppressor Of Cytokine Signaling 3); CASP1(Caspase 1); QK1(Quaking Homolog, KH Domain RNA Binding 1); SRSF (Serine And Arginine Rich Splicing Factor); SF1(Splicing factor 1); DISC1 (Disrupted-in-Schizophrenia 1); ERBB4 (Erb-B2 Receptor Tyrosine Kinase 4); GATA2 (GATA-binding factor 2); DPYSL3 (Dihydropyrimidinase Like 3); PTBP2 (Polypyrimidine Tract Binding Protein 2); MMP-9 (Matrix Metallopeptidase 9); STAT-4 (Signal Transducer And Activator Of Transcription 4); MeCP2 (Methyl-CpG Binding Protein 2); ZO-1(Zonula occludens-1); FZD3 (Frizzled 3); NRG2/3 (Neuregulin 2/3); SYN2/3 (Synapsis2/3); ATXN1(ataxin-1); HTT (Huntingtin); SOX9 (SRY-Box Transcription Factor 9); P53 (tumor suppressor protein coding gene 53); PGC1(Peroxisome Proliferator-Activated Receptor Coactivator 1).
Other neurological diseases
Dysregulated miRNA biogenesis and activity are directly implicated in the pathogenesis of several complex neurodegenerative and psychiatric diseases. Altered expression of miRNAs such as miR-181b, Let-7 g, miR-26b, miR-30b, miR29b, and miR-106b has been reported in the postmortem brain tissue of schizophrenia patients [Citation182–184]. Fragile X syndrome is a disease characterized by severe mental retardation which is caused by the loss of RNA-binding protein fragile X mental retardation protein (FMRP). Genetic knockdown of FMRP mitigates the effect of miR-125b and miR-132 on dendritic spine morphology [Citation185]. Studies have identified that miR-125b directly targets the NR2A subunit of the N-methyl-D-aspartate receptor (NMDAR), and that glutamate receptor NR2A mRNA which is associated with FMRP, supporting the idea that FMRP functions as a target engagement for miRNA activity [Citation185,Citation186]. Evidence also indicates a strong association between miRNA and Huntington’s disease (HD) pathology given the dysregulation in transcription and processing of microRNAs [Citation187]. In the brains of HD patients, miR-29a, miR-132, and miR-330 have been observed to be expressed in higher levels [Citation188,Citation189]. Significant downregulation of miR-22, miR-128, miR-29 c, miR-138, miR-132, miR-218; and miR-674, miR-344, and miR-222 has also been identified in the mouse models of HD [Citation190]. Several miRNAs such as miR-142-3p, miR-145, miR-146a/b, miR-22, miR-155, miR223/-3p, miR-584, and miR-326 have been also reported to be induced in multiple sclerosis (MS) patients implying their involvement in the pathogenic inflammatory process observed in MS. Dysregulation of various miRNAs targeting the inflammatory activity of various immune cells has also been extensively reported [Citation191–194].
Substance (drug) abuse and addiction are a debilitating disorder characterized by compulsive drug seeking behavior. At the molecular level, it is established that miRNAs play critical roles in synaptic plasticity, neuronal communication and signaling, which are significantly altered in the presence of drugs of abuse [Citation195,Citation196]. For example, multiple studies have demonstrated the involvement of miRNAs in cocaine addiction. Extended exposure to cocaine induces miR-212 concomitant with the increase of total and phosphorylated c-response element binding protein (CREB) in the dorsal striatum of rats, a key region involved in the development of compulsive cocaine use [Citation197]. In a miRNA profiling study of cocaine-induced plasticity genes, it was shown that the miRNAs let-7d and miR-124 are down-regulated, with concomitant induction of miR-181a in the mesolimbic dopaminergic system under chronic cocaine exposure [Citation198,Citation199]. Using next-generation miRNA sequencing method several miRNAs were identified that exhibit cocaine-induced expression changes, such as the miR-8 family members miR-429 and miR-200a/b in the nucleus accumbens and striatal synapses [Citation200]. Recently, the role of poly(ADP-ribose) polymerase-1 (PARP-1) was identified in plasticity, memory, and cocaine addiction [Citation201,Citation202]. Employing in vivo models, we reported that PARP-1 was modulated by both miR-125b and miR-124 that were significantly downregulated with cocaine exposure [Citation27,Citation203]. This dose-dependent nature of addiction was implicated in cocaine-induced activation of PARP-1-induced signaling for the maintenance of ATP levels in the dopaminergic neurons [Citation27,Citation203,Citation204]. Specifically, cocaine exposure downregulated both miR-125b and miR-124 in the nucleus accumbens (NAc), however only miR-124 expression was dramatically altered in the hippocampus and ventral tegmental area (VTA) [Citation27,Citation198,Citation203]. Other studies have also found that miR-495, let-7, and miR-212/132 were significantly downregulated in the NAc region upon cocaine treatments that could regulate several canonical reward pathways [Citation205]. Recent studies have also identified lncRNAs that are altered in key reward regions upon cocaine exposure. For instance, lncRNAs TRAF3IP2_AS1 (tumor necrosis factor receptor-associated factor 3-interacting protein 2_antisense 1) and PRKCQ_AS1 (protein kinase C theta antisense 1) are reported to be significantly altered in cocaine abusers [Citation206]. In another study using transcriptome profiling approach, lncRNA Gas5 was identified to be important in inducing cocaine-mediated effects in the mouse nucleus accumbens (NAc) [Citation207]. Using rodent model of methamphetamine abuse, research also reveals dramatic alterations in the lncRNA expression profile in the nucleus accumbens [Citation208].
Collectively, not only these ncRNAs are associated with drug abuse, but they are also associated with several brain diseases suggesting the involvement of novel miRNAs and lncRNAs whose molecular role and functional characterization demands further investigation.
Bioengineering of ncRNAs for neurological diseases
Bioengineering is the application of engineering principles to improve bio-molecular applications to address key challenges in medicine and biology. Bioengineering of ncRNAs can range from nanoparticle-based functionalization to engineering nucleoside chemistry to improve the delivery and pharmacological properties both in vitro and in vivo. In this regard, bioengineering of ncRNAs is a rapidly evolving field that is anticipated to revolutionize nucleic acid therapeutics for various human diseases including neurological and neurodegenerative disorders. Emerging pre-clinical and clinical evidence has established that miRNAs are secreted in bodily fluids suggestive of effective early-stage biomarkers and therapeutics [Citation209–211]. Therefore, the aim of selectively sensing, enriching, and capturing the changes in miRNA expression profile in a wide range of sample types is critical. Hence, bioengineering of miRNAs via magnetic nanoparticle (MNP) or polymer-based functionalization may provide enhanced therapeutic features when compared to traditional approaches [Citation212]. A summary of various bioengineering applications employed for miRNAs and lncRNAs are presented in . For instance, using assemblies of nano-(Fe2O3) particles in combination with Pt nanoparticles have been used for the detection of miR-21, polydopamine–functionalized (Fe3O4) core nanoparticles with carbon dots have been used for sensing of miR-167, and (Fe3O4) nanoparticles with carboxyl-modifications have been used in microfluidic devices to quantify miRNA-200a-3p [Citation212]. Moreover, Fe3O4-Ag (iron oxide-silver) core shell nanoparticles have been utilized for the capture and detection of miRNAs such as miRNA let-7b with an improved sensitivity in the lower femto-Molar (fM) range. Such proof-of-concept studies have been performed in a relatively homogeneous system without the interference of blood, cellular factors, complex tissue architecture etc., and thus it should be noted that the number of studies performed in natural body fluids is highly limited. The ability of concentrating circulating miRNAs on the MNPs by surface conjugation or functionalization approaches has resulted in a number of highly efficient miRNA sensing devices [Citation212,Citation213]. Cationic polyurethance (PU) short branch polyethylene imine (PEI) nanocomplexes have been developed as vehicles for miR-145 delivery to inhibit brain tumor by targeting genes such as Sox4 and Oct4, in vitro [Citation214]. Similarly, anti-miR-21 nanoparticles co-encapsulating the drug doxycycline have demonstrated enhanced cell apoptosis and reduced tumor growth in brain tumor models [Citation215]. Gold nanoparticles covalently functionalized with miR-182 have been tested to selectively deliver miR-182 to brain tumors that has resulted in reducing tumor size [Citation216]. Similarly, gold nanoparticles loaded with anti-miR-92b in apo-lipoprotein E coated liposomes have been shown to efficiently traverse through the blood brain barrier to reduce tumor size in mouse model of glioblastoma [Citation217]. Another advanced engineering approach includes ‘artificial miRNAs’, where the mature miRNAs sequence is replaced in the primary miRNA transcript for a complementary sequence to target the gene of interest [Citation218,Citation219]. When artificial miRNAs are transfected into mammalian cells, they undergo natural miRNA processing and produce the mature miRNA that directly targets the mRNA due to its sequence complementarity and it is postulated that this approach restricts the off-targets effects of a natural mature miRNA in cells [Citation218,Citation219]. These findings clearly establish that we are beginning to learn about the therapeutic aspects of bioengineering of miRNAs. Additionally, adeno-associated virus 5 (AAV5)-mediated delivery of miRNAs to the brain is currently being investigated as a ‘gene therapy’ in non-human primates in model of Huntington’s disease [Citation220]. The discussed proof-of-concept studies provide a foundation and present the feasibility for investigation of bioengineered miRNAs for neurological diseases such as Alzheimer’s disease, Parkinson’s disease as well as other neuropathologies.
Figure 2. Bioengineering of ncRNAs for biomedical applications. Both miRNAs and lncRNAs can be used for several therapeutic and diagnostic applications based on the bioengineering approaches used that include encapsulation of ncRNAs in functionalized nanoparticles, lipid/polymer based nanocarriers, terminal modifications including biotin or fluorophore tagging or chemical modifications by adding functional groups (i.e., 2ʹO-Methyl, P = S bonds) or by bridging the 2ʹoxygen with 4ʹcarbon as represented in the figure
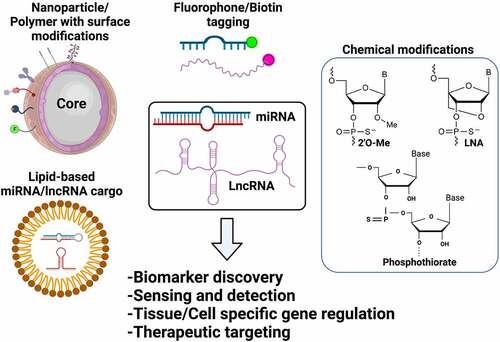
Based on current research trends, the future points toward development of cost-efficient functionalized approaches with improved biocompatibility for early capture, detection and delivery of miRNAs in disease systems [Citation212,Citation213]. Additional engineering of miRNAs includes nucleic acid modifications that improve the stability and target specificity of miRNAs when present in the cellular environment. RNA molecules are unstable due to the presence of a 2′ hydroxyl (OH) group [Citation221]. Other strategies include chemical modifications such as phosphothiorate group, phosphodiester group and lock nucleic acid modifications for miRNA and small RNA-based therapeutics (). Therefore, engineering of the nucleosides by incorporation of 2′-O-methyl (2′OMe)-modifications improves off-target effects without affecting basal immune response [Citation222,Citation223]. Similarly, 3ʹ cholesterol groups are also used to improve the delivery and bioavailability of small RNAs as cholesterol is a physiological and structural component of animal cell membranes [Citation224].
It must be noted that the diverse functionality of lncRNAs presents multiple ways in which these ncRNAs can be targeted. At the same time these diverse features pose a great deal of challenge in the therapeutic pipeline given the nonspecific events that could arise due to manipulation of a given lncRNA in the cell. However, reports suggest that lncRNAs can be targeted using engineered anti-sense oligos (ASOs) or by manipulating the levels of naturally occurring antisense transcripts (NATs) [Citation13,Citation17,Citation225].
Therapeutic feasibility of ncRNAs
The use of ncRNAs to treat human diseases is rapidly developing and so are the approaches to engineer ncRNAs for therapeutic and diagnostic compatibility. Several evidence point to the critical role of miRNAs and lncRNAs in disease models ranging from CNS pathologies, drug abuse to viral-induced nervous system disorders [Citation226–230]. As of now, we are beginning to understand the molecular role of lncRNAs whereas, with regards to miRNAs the field is witnessing a transition for small RNA therapeutics [Citation52,Citation231–235]. Several reports have elegantly described various RNA-based therapies that include antisense oligonucleotides (ASOs), short hairpin RNAs (shRNAs), ASO anti-microRNAs (anti-miRs) and small interfering RNAs (siRNAs), that have been developed for liver diseases, muscular disorders, CNS diseases and cancer [Citation12–14]. Currently, approximately 10 RNA-based therapeutics are approved by the FDA and multiple small RNA therapeutic approaches are in stages of clinical investigation for complex CNS and rare genetic disorders [Citation17,Citation225]. Importantly, we and others have discussed on the scope of ncRNA-based therapeutic applications in diseases such as COVID-19-induced neurological disorders, systemic inflammation and cardiovascular diseases [Citation6,Citation230,Citation236–240]. As an example, in 2018, the FDA approved ‘Patisiran’- the first therapy based on administration of RNAi (siRNA-based) for the treatment of rare progressive polyneuropathy caused by hereditary transthyretin-mediated amyloidosis. This drug works by targeting the 3ʹUTR region of transthyretin mRNA [Citation241,Citation242]. Neurological and neurovascular diseases present a global healthcare burden and an unmet clinical need [Citation243–247]. Hence, the observation that miRNAs modulate expression of candidate genes in various diseases has suggested researchers to develop miRNA-based therapeutic strategies [Citation13,Citation231,Citation243,Citation248]. Depending on the data available, a mimic or an antagonist of miRNAs could be explored as therapeutic agent. For example in the case of Alzheimer’s disease, it was shown that the injection of antisense oligonucleotides (ASO) into the CSF of nonhuman primates reduces target RNA (tau) expression in the brain regions analyzed, including the hippocampus [Citation249]. Based on these data, a clinical trial to test a tau ASO in patients with mild AD is currently under investigation by Biogen, IONIS Pharmaceuticals (NCT03186989). Similarly, miRNA silencing is thought to be an attractive therapeutic modality in many other neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis (ALS) [Citation250–253]
A recent study focused on identification of unique lncRNAs that could serve as targets for therapeutic agents [Citation254]. Abnormalities of lncRNAs that are involved in many fundamental neuronal cell properties are considered a hallmark of nervous system disorders [Citation24]. Oligonucleotide therapy using an ASO that is single stranded < 40nucleotides may modulate various mechanisms in consequence by binding to complementary targets such as mRNA, miRNA, and NATs [Citation255]. In this context, the drug potential of ASO against lncRNA in Angelman syndrome (AS) was postulated. Patients with genetic defects in AS present with malfunctioning nervous system features, caused by a maternally inherited defect in UBE3A, a gene that encodes for E3 ubiquitin ligase [Citation256]. ASO restored UBE3A mRNA that was silenced by lncRNA, an antisense transcript of small nucleolar RNA host gene 14 (SNHG14; previously known as UBE3A-ATS) in intact paternal UBE3A in vivo [Citation257] and in patients, showing a promising approach to clinical treatment [Citation256]. Moreover, ASOs that target naturally occurring antisense transcripts (NATs) have been shown to induce brain-derived neurotrophic factor (BDNF) in vivo to improve neurological outcomes [Citation258]. Importantly, using in vivo models it was discovered that these antisense-NATs targeting BDNF are reported to have higher degree of permeability through the blood brain barrier [Citation258]. In addition, lncRNA MALAT1 was suggested for application as a treatment strategy related to cancer [Citation259,Citation260]. The silencing of MALAT1 by siRNA or ASO has been reported to exhibit positive regulations in in vivo models of tumorigenesis by improving features such as tumor cell proliferation, migration, invasion, and apoptosis in various cancer types including lung cancer [Citation261Citation262], pancreatic cancer [262] and multiple myeloma [Citation259]. LncRNAs have drawn considerable attention in the past decade and currently there is a lack of lncRNA-based or targeting therapeutics in the pre-clinical or clinical domain. However at present most of the attention is diverted on claiming the value of lncRNAs as a diagnostic i.e., stage-specific biomarker. However, it is expected that the field of lncRNAs will witness rapid progress in both basic and clinical sciences.
Conclusion and outlook
Although we have gained insight into some of the key species of this evolving family of RNA, a vast majority of information is yet to be discovered. The field is currently innovating to overcome the several challenges in developing ncRNA-based therapeutics and diagnostics. It is important to acknowledge that bioengineering of ncRNAs is a relatively young field that needs extensive investigation. The feasibility of chemical modifications, genetic engineering approaches, compatibility of biopolymers, and oligonucleotide biochemistry are some of the key domains that can be advanced by the introduction of interdisciplinary approaches. Considering an example better and improved delivery modalities such as polyethylene glycol, hyaluronic acid or polymers with improved retention, biocompatibility and low degradability and low immunogenicity must be tested using disease relevant in vivo models. Another major problem for CNS therapeutics is the site or region-specific delivery of candidate ncRNAs such as, miRNAs to specific diseased sites in the brain. The development of effective miRNA delivery systems is vital as the delivery vehicle must allow miRNAs to cross the blood–brain barrier, which remains a major hurdle in neurodegenerative and neurovascular therapeutics. As miRNAs are easily degradable, the delivery and engineering systems can also be innovated to stabilize and extend the life of the miRNAs. Moreover, host cell endocytosis mechanisms must be exploited to improve the barrier permeability of engineered anti-sense oligos, small RNA mimics or delivery cargos. Similarly, lncRNAs can be investigated for their therapeutic prospects given their direct role in chromatin modifications as well as their direct impact in messenger RNA stabilization and post-transcriptional regulation. Furthermore, it is debated that using ncRNAs as a therapeutic modality may be a double-edged sword due to their multifaceted biological function and pleiotropic properties. Hence, effective methods of bioengineering ncRNAs are postulated as the future to improve our understanding on the therapeutic efficacy and feasibility of ncRNAs for neurological as well as related disorders. Often in vitro investigation provides clear cut results however are far from physiological relevance in contrast with in vivo approaches that are complex and include species variations and challenges in replication disease etiopathologies. Overall, given the tremendous advances have been made in nucleic acid modifications and delivery systems individually based on specific RNA species, a successful translation will require the integration of interdisciplinary expertise to improve the therapeutic index and feasibility of ncRNAs for neurological and neurodegenerative diseases.
Highlights
ncRNAs are critical regulators of gene function and play key roles in brain development and function.
Bioengineering improves the therapeutic feasibility of ncRNAs for pre-clinical and clinical testing.
Bioengineering of ncRNAs is key to development of novel diagnostics and therapeutics for neurological disorders.
Author Contributions
S.D. conceived the idea. S.D., A.K., T.D., T.K.D. wrote the original draft.; S.D., T.D., T.K.D reviewed and edited the draft; S.D., A.K., T.D., T.K.D. prepared the figures and summarized the table. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Statello L, Guo C-J, Chen -L-L, et al. Gene regulation by long non-coding RNAs and its biological functions. Nature Reviews. Molecular Cell Biology. 2021 Feb;22(2):96–118. DOI:10.1038/s41580-020-00315-9
- Cech TR, Steitz JA. The noncoding RNA Revolution—Trashing old rules to forge new ones. Cell. 2014 Mar;157(1):77–94. DOI:10.1016/j.cell.2014.03.008
- Beermann J, Piccoli M-T, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016 Oct;96(4):1297–1325. DOI:10.1152/physrev.00041.2015
- Dash S, Shakyawar SK, Sharma M, et al. Big data in healthcare: management, analysis and future prospects. J Big Data. 2019 Dec;6(1):54. doi: 10.1186/s40537-019-0217-0
- Liu CH, Wu D-Y, Pollock JD. Bioinformatic challenges of big data in non-coding RNA research. Front Genet. 2012;3. DOI:10.3389/fgene.2012.00178
- Huang C-K, Kafert-Kasting S, Thum T. Preclinical and clinical development of noncoding RNA therapeutics for cardiovascular disease. Circ Res. 2020 Feb;126(5):663–678. DOI:10.1161/CIRCRESAHA.119.315856
- Forterre A, Komuro H, Aminova S, et al. A comprehensive review of cancer MicroRNA therapeutic delivery strategies. Cancers (Basel). 2020 Jul;12(7):1852. DOI:10.3390/cancers12071852
- Grillone K, Riillo, C, and Scionti, F, et al. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic ‘dark matter. J Exp Clin Cancer Res. 2020 Dec;39(1):117. DOI:10.1186/s13046-020-01622-x
- Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy. J Pharm Pharmacol. 2020 Nov;72(12):1732–1749. DOI:10.1111/jphp.13351
- Finotti A, Fabbri E, Lampronti I, et al. MicroRNAs and Long Non-coding RNAs in Genetic Diseases. Mol Diagn Ther. 2019 Apr;23(2):155–171. DOI:10.1007/s40291-018-0380-6
- Ling H. Non-coding RNAs: therapeutic strategies and delivery systems Adv Exp Med Biol . 2016 937 ;229–237 doi:10.1007/978-3-319-42059-2_12.
- Ling H, Fabbri M, Calin Ga. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013 Nov;12(11):847–865. DOI:10.1038/nrd4140
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017 Mar;16(3):203–222. DOI:10.1038/nrd.2016.246
- van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012 Nov;11(11):860–872. DOI:10.1038/nrd3864
- Li S, Qian T, Wang X, et al. Noncoding RNAs and their potential therapeutic applications in tissue engineering. Engineering. 2017 Feb;3(1):3–15. DOI:10.1016/J.ENG.2017.01.005
- McElhinney JMWR, Hasan A, Sajini AA. The epitranscriptome landscape of small noncoding RNAs in stem cells. Stem Cells. Jun 2020. DOI:10.1002/stem.3233
- Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nature Reviews. Drug Discovery. 2021 Aug;20(8):629–651. DOI:10.1038/s41573-021-00219-z
- Duan Z, Yu A-M. Bioengineered non-coding RNA agent (BERA) in action. Bioengineered. 2016 Nov;7(6):411–417. DOI:10.1080/21655979.2016.1207011
- Dash S, Balasubramaniam M, Dash C, et al. Biotin-based pulldown assay to validate mRNA targets of cellular miRNAs. J Vis Exp. Jun 2018;136. doi:10.3791/57786
- Bartel DP. MicroRNAs. Cell. 2004 Jan;116(2):281–297. DOI:10.1016/S0092-8674(04)00045-5
- Yao R-W, Wang Y, Chen -L-L. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019 May;21(5):542–551. DOI:10.1038/s41556-019-0311-8
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017 Dec;18(1):206. DOI:10.1186/s13059-017-1348-2
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009 Jul;23(13):1494–1504. DOI:10.1101/gad.1800909
- Salta E, De Strooper B. Noncoding RNAs in neurodegeneration. Nat Rev Neurosci. 2017 Oct;18(10):627–640. DOI:10.1038/nrn.2017.90
- Pomper N, Liu Y, Hoye ML, et al. CNS microRNA profiles: a database for cell type enriched microRNA expression across the mouse central nervous system. Sci Rep. 2020 December;10(1):4921. DOI:10.1038/s41598-020-61307-5
- Isakova A, Fehlmann T, Keller A, et al. A mouse tissue atlas of small noncoding RNA. Proc Natl Acad Sci. 2020 October;117(41):25634–25645. DOI:10.1073/pnas.2002277117
- Dash S, Balasubramaniam, M, and Martínez-Rivera, F.J., et al. Cocaine-regulated microRNA miR-124 controls poly (ADP-ribose) polymerase-1 expression in neuronal cells. Sci Rep. 2020 December;10(1):11197. DOI:10.1038/s41598-020-68144-6
- Sun Y, Luo Z-M, Guo X-M, et al. An updated role of microRNA-124 in central nervous system disorders: a review. Front Cell Neurosci. 2015 May;9. DOI:10.3389/fncel.2015.00193
- Gokool A, Anwar F, Voineagu I. The landscape of circular RNA expression in the human brain. Biol Psychiatry. 2020 Feb;87(3):294–304. DOI:10.1016/j.biopsych.2019.07.029
- Barry G, Briggs, J, and Vanichkina, D, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014 April;19(4):486–494. DOI:10.1038/mp.2013.45
- Floris G, Zhang L, Follesa P, et al. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017 Sep;54(7):5156–5165. DOI:10.1007/s12035-016-0055-4
- Guerra B, Lima, J, Araujo, BHS, and Torres, LB, et al. Biogenesis of circular RNAs and their role in cellular and molecular phenotypes of neurological disorders. Semin Cell Dev Biol. 2021June;114:1–10. DOI:10.1016/j.semcdb.2020.08.003
- Shi Y, Song , R, Wang, Z, and Zhang, H, et al. Potential clinical value of circular RNAs as peripheral biomarkers for the diagnosis and treatment of major depressive disorder. EBioMedicine. 2021April;66:103337. DOI:10.1016/j.ebiom.2021.103337
- Ma Y, Liu Y, Jiang Z. CircRNAs: a new perspective of biomarkers in the nervous system. Biomed Pharmacother. 2020Aug;128:110251. DOI:10.1016/j.biopha.2020.110251
- Lu S, Yang , X, Wang, C, and Chen, S, et al. Current status and potential role of circular RNAs in neurological disorders. J Neurochem. 2019 Aug;150(3):237–248. DOI:10.1111/jnc.14724
- Zuo L, Zhang, L, and Zu, J, et al. Circulating circular RNAs as biomarkers for the diagnosis and prediction of outcomes in acute ischemic stroke. Stroke. 2020 January;51(1):319–323. DOI:10.1161/STROKEAHA.119.027348
- Qureshi IA, Mehler MF. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics. 2013 Oct;10(4):632–646. DOI:10.1007/s13311-013-0199-0
- Briggs JA, Wolvetang EJ, Mattick JS, et al. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015 December;88(5):861–877. DOI:10.1016/j.neuron.2015.09.045
- Mercer TR, Dinger ME, Mariani J, et al. Noncoding RNAs in long-term memory formation. Neurosci. 2008 October;14(5):434–445. DOI:10.1177/1073858408319187
- Bernard D, Prasanth, KV, and Tripathi , V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J. 2010 September;29(18):3082–3093. DOI:10.1038/emboj.2010.199
- Rajasethupathy P, Antonov, I, and Sheridan, R, et al. A role for neuronal piRNAs in the Epigenetic control of memory-related synaptic plasticity. Cell. 2012 April;149(3):693–707. DOI:10.1016/j.cell.2012.02.057
- Lin N, Chang , KY, and Li, Z, et al. An Evolutionarily Conserved Long noncoding RNA TUNA controls Pluripotency and neural lineage commitment. Mol Cell. 2014 March;53(6):1005–1019. DOI:10.1016/j.molcel.2014.01.021
- Bond AM, Vangompel, MJ, and Sametsky, EA, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009 August;12(8):1020–1027. DOI:10.1038/nn.2371
- Onoguchi M, Hirabayashi Y, Koseki H, et al. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci. 2012 October;109(42):16939–16944. DOI:10.1073/pnas.1202956109
- Ng S-Y, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. Embo J. 2012 Feb;31(3):522–533. DOI:10.1038/emboj.2011.459
- Mercer TR, Qureshi, IA, and Gokhan, S, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010 December;11(1):14. DOI:10.1186/1471-2202-11-14
- Tochitani S, Hayashizaki Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008 Aug;372(4):691–696. DOI:10.1016/j.bbrc.2008.05.127
- Rani N, Nowakowski, TJ, and Zhou, H, et al. A primate lncrna mediates notch signaling during neuronal development by sequestering miRNA. Neuron. 2016 June;90(6):1174–1188. DOI:10.1016/j.neuron.2016.05.005
- Sauvageau M, Goff, LA, and Lodato, S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013 December;2. DOI:10.7554/eLife.01749
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014 January;15(1):7–21. DOI:10.1038/nrg3606
- Hébert SS, Papadopoulou, AS, and Smith, P, et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010 October;19(20):3959–3969. DOI:10.1093/hmg/ddq311
- Goff LA, Groff, AF, and Sauvageau, M, et al Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci. 2015 June;112(22):6855–6862. DOI:10.1073/pnas.1411263112
- Long JM, Holtzman DM. Alzheimer disease: an update on pathobiology and treatment strategies. Cell. 2019 October;179(2):312–339. DOI:10.1016/j.cell.2019.09.001
- DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer’s disease. Mol Neurodegener. 2019 December;14(1):32. DOI:10.1186/s13024-019-0333-5
- Idda ML, Munk R, Abdelmohsen K, et al. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip Rev RNA. 2018 March;9(2):e1463. DOI:10.1002/wrna.1463
- Millan MJ. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: an integrative review. Progress in Neurobiology. 2017Sep;156:1–68. DOI:10.1016/j.pneurobio.2017.03.004
- Wang X, Liu, D, and Huang, HZ, et al. A novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in alzheimer’s Disease. Biol Psychiatry. 2018 March;83(5):395–405. DOI:10.1016/j.biopsych.2017.07.023
- Cosã-n-tomã¡s M, Alvarez-López, MJ, and Sanchez-Roige, S, et al. Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front Aging Neurosci. 2014 Mar;6. DOI:10.3389/fnagi.2014.00051
- Boissonneault V, Plante I, Rivest S, et al. MicroRNA-298 and MicroRNA-328 regulate expression of mouse β-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009 Jan;284(4):1971–1981. DOI:10.1074/jbc.M807530200
- Zhu H-C, Wang, L, and Wang, M, et al. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res Bull. 2012 Sep;88(6):596–601. DOI:10.1016/j.brainresbull.2012.05.018
- Makeyev EV, Zhang J, Carrasco MA, et al. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative Pre-mRNA splicing. Mol Cell. 2007 August;27(3):435–448. DOI:10.1016/j.molcel.2007.07.015
- Fang M, Wang, J, and Zhang, X, et al. The miR-124 regulates the expression of BACE1/β-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012 February;209(1):94–105. DOI:10.1016/j.toxlet.2011.11.032
- Hu Y-K, Wang X, Li L, et al. MicroRNA-98 induces an Alzheimer’s disease-like disturbance by targeting insulin-like growth factor 1. Neurosci Bull. 2013 December;29(6):745–751. DOI:10.1007/s12264-013-1348-5
- Long JM, Ray B, Lahiri DK. MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem. 2012 Sep;287(37):31298–31310. DOI:10.1074/jbc.M112.366336
- Long JM, Ray B, Lahiri DK. MicroRNA-339-5p down-regulates protein expression of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J Biol Chem. 2014 February;289(8):5184–5198. DOI:10.1074/jbc.M113.518241
- Jiang Y, Xu , B, and Chen, J, et al. Micro-RNA-137 inhibits tau hyperphosphorylation in Alzheimer’s disease and targets the CACNA1C gene in transgenic mice and human neuroblastoma SH-SY5Y Cells. Med Sci Monit. 2018Aug;24:5635–5644. DOI:10.12659/MSM.908765
- Fransquet PD, Ryan J. Micro RNA as a potential blood-based epigenetic biomarker for Alzheimer’s disease. Clin Biochem. 2018Aug;58:5–14. DOI:10.1016/j.clinbiochem.2018.05.020
- Das B, Yan R. Role of BACE1 in Alzheimer’s synaptic function. Transl Neurodegener. 2017 Dec;6(1):23. DOI:10.1186/s40035-017-0093-5
- Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci. 2007 June;104(25):10679–10684. DOI:10.1073/pnas.0701532104
- Li H, Zheng L, Jiang A, et al. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport. 2018 Sep;29(13):1061–1067. DOI:10.1097/WNR.0000000000001057
- Massone S, Vassallo, I, and Fiorino, G, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011 February;41(2):308–317. DOI:10.1016/j.nbd.2010.09.019
- Huang J, Huen, MS, and Kim, H, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009 May;11(5):592–603. DOI:10.1038/ncb1865
- Straten G, Eschweiler GW, Maetzler W, et al. Glial Cell-Line Derived Neurotrophic Factor (GDNF) Concentrations in Cerebrospinal Fluid and Serum of Patients with Early Alzheimer’s disease and normal controls. J Alzheimer’s Dis. 2009 August;18(2):331–337. DOI:10.3233/JAD-2009-1146
- Poewe W, Seppi, K, and Tanner, CM, et al. Parkinson disease. Nat Rev Dis Prim. 2017 Dec;3(1):17013. DOI:10.1038/nrdp.2017.13
- Dickson DW. Parkinson’s Disease and Parkinsonism: neuropathology. Cold Spring Harb Perspect Med. 2012 Aug;2(8):a009258–a009258. DOI:10.1101/cshperspect.a009258
- Goh SY, Chao YX, Dheen ST, et al. Role of MicroRNAs in Parkinson’s disease. Int J Mol Sci. 2019 Nov;20(22):5649. DOI:10.3390/ijms20225649
- Briggs CE, Wang Y, Kong B, et al. Midbrain dopamine neurons in Parkinson׳s disease exhibit a dysregulated miRNA and target-gene network. Brain Res. 2015 Aug;1618:111–121. DOI:10.1016/j.brainres.2015.05.021
- Tatura R, Kraus, T, and Giese, A, et al. Parkinson’s disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Parkinsonism Relat Disord. 2016December;33:115–121. DOI:10.1016/j.parkreldis.2016.09.028
- Gui Y, Liu H, Zhang L, et al. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget. 2015 November;6(35):37043–37053. DOI:10.18632/oncotarget.6158
- Martins M, Rosa, A, and Guedes, LC, et al. Convergence of miRNA expression Profiling, α-synuclein interacton and GWAS in Parkinson’s disease. PLoS One. 2011 Oct;6(10):e25443. DOI:10.1371/journal.pone.0025443
- Chen L, Yang J, Lü J, et al. Identification of aberrant circulating miRNAs in Parkinson’s disease plasma samples. Brain Behav. 2018 Apr;8(4):e00941. DOI:10.1002/brb3.941
- Burgos K, Malenica, I, and Metpally, R, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014 May;9(5):e94839. DOI:10.1371/journal.pone.0094839
- Margis R, Margis R, Rieder CRM. Identification of blood microRNAs associated to Parkinsońs disease. J Biotechnol. 2011 Mar;152(3):96–101. DOI:10.1016/j.jbiotec.2011.01.023
- Bai X, Tang , Y, and Yu, M, et al. Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci Rep. 2017 December;7(1):5411. DOI:10.1038/s41598-017-03887-3
- Kole AJ, Swahari V, Hammond SM, et al. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011 Jan;25(2):125–130. DOI:10.1101/gad.1975411
- Roshan R, Shridhar, S, and Sarangdhar, MA, et al. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA. 2014 Aug;20(8):1287–1297. DOI:10.1261/rna.044008.113
- Cao L, Zhang, Y, and Zhang, S, et al. MicroRNA-29b alleviates oxygen and glucose deprivation/reperfusion-induced injury via inhibition of the p53‑dependent apoptosis pathway in N2a neuroblastoma cells. Exp Ther Med. 2017 Oct. DOI:10.3892/etm.2017.5410
- Xu S, Wu, W, and Huang, H, et al. The p53/miRNAs/Ccna2 pathway serves as a novel regulator of cellular senescence: complement of the canonical p53/p21 pathway. Aging Cell. 2019 Jun;18(3):e12918. DOI:10.1111/acel.12918
- Martinez I, Cazalla D, Almstead LL, et al. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci. 2011 Jan;108(2):522–527. DOI:10.1073/pnas.1017346108
- Papadopoulou AS, Serneels, L, and Achsel, T, et al. Deficiency of the miR-29a/b-1 cluster leads to ataxic features and cerebellar alterations in mice. Neurobiol Dis. 2015Jan;73:275–288. DOI:10.1016/j.nbd.2014.10.006
- Steiner DF, Thomas, MF, and Hu, JK, et al. MicroRNA-29 Regulates T-Box Transcription Factors and Interferon-γ Production in Helper T Cells. Immunity. 2011 Aug;35(2):169–181. DOI:10.1016/j.immuni.2011.07.009
- Chandiran K, Lawlor, R, and Pannuti, A, et al. Notch1 primes CD4 T cells for T helper type I differentiation through its early effects on miR-29. Mol Immunol. 2018Jul;99:191–198. DOI:10.1016/j.molimm.2018.05.002
- Smith KM, Guerau-de-Arellano, M, and Costinean, S, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012 Aug;189(4):1567–1576. DOI:10.4049/jimmunol.1103171
- Lyu G, Guan, Y, and Zhang, C, et al. TGF-β signaling alters H4K20me3 status via miR-29 and contributes to cellular senescence and cardiac aging. Nat Commun. 2018 Dec;9(1):2560. DOI:10.1038/s41467-018-04994-z
- Morita S, Horii T, Kimura M, et al. miR-29 Represses the Activities of DNA Methyltransferases and DNA Demethylases. Int J Mol Sci. 2013 Jul;14(7):14647–14658. DOI:10.3390/ijms140714647
- Chatterjee P, Roy D. Comparative analysis of RNA-Seq data from brain and blood samples of Parkinson’s disease. Biochem Biophys Res Commun. 2017 Mar;484(3):557–564. DOI:10.1016/j.bbrc.2017.01.121
- Horst CH, Schlemmer, F, and de Aguiar Montenegro, N, et al. Signature of Aberrantly Expressed microRNAs in the Striatum of Rotenone-Induced Parkinsonian Rats. Neurochem Res. 2018 Nov;43(11):2132–2140. DOI:10.1007/s11064-018-2638-0
- Boros FA, Maszlag-Török R, Vécsei L, et al. Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain Res. 2020Mar;1730:146672. DOI:10.1016/j.brainres.2020.146672
- Chen M-Y, Fan K, Zhao L-J, et al. Long non-coding RNA nuclear enriched abundant transcript 1 (NEAT1) sponges microRNA-124-3p to up-regulate phosphodiesterase 4B (PDE4B) to accelerate the progression of Parkinson’s disease. Bioengineered. 2021 Jan;12(1):708–719. DOI:10.1080/21655979.2021.1883279
- Lin Q, Hou S, Dai Y, et al. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol Chem. 2019 Aug;400(9):1217–1228. DOI:10.1515/hsz-2018-0431
- Coupland KG, Kim WS, Halliday GM, et al. Role of the long non-coding RNA MAPT-AS1 in regulation of Microtubule Associated Protein Tau (MAPT) Expression in Parkinson’s disease. PLoS One. 2016 Jun;11(6):0157924. DOI:10.1371/journal.pone.0157924
- Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry. 2014 Apr;19(4):410–416. DOI:10.1038/mp.2013.196
- Guennewig B, and Cooper AA. The central role of noncoding RNA in the BRAIN Int Rev Neurobiol . 2014:153–194 doi:10.1016/B978-0-12-801105-8.00007-2.
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012 Aug;13(8):528–541. DOI:10.1038/nrn3234
- Rothwell N, Allan S, Toulmond S. The role of interleukin 1 in acute neurodegeneration and stroke: pathophysiological and therapeutic implications. J Clin Invest. 1997 Dec;100(11):2648–2652. DOI:10.1172/JCI119808
- Barone FC, Arvin, B, and White, RF, et al. Tumor Necrosis Factor-α. Stroke. 1997 Jun;28(6):1233–1244. DOI:10.1161/01.STR.28.6.1233
- Cheng HS, Njock M-S, Khyzha N, et al. Noncoding RNAs regulate NF-ÎoB signaling to modulate blood vessel inflammation. Front Genet. 2014 December;5. DOI:10.3389/fgene.2014.00422
- Walsh MC, Lee J, Choi Y. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol Rev. 2015 July;266(1):72–92. DOI:10.1111/imr.12302
- Cheng HS, Sivachandran, and Lau, A, et al. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med. 2013 Jul;5(7): DOI:10.1002/emmm.201202318
- Hunsberger JG, Fessler EB, Wang Z, et al. Post-insult valproic acid-regulated microRNAs: potential targets for cerebral ischemia. American Journal of Translational Research. 2012;4(3):316–332. [Online]. Available http://www.ncbi.nlm.nih.gov/pubmed/22937209
- Pizzino G, Irrera, N, and Cucinotta, M, et al. Oxidative stress: harms and benefits for human health. Oxid Med Cell Longev. 2017;1–13. DOI:10.1155/2017/8416763
- Mittal M, Siddiqui MR, Tran K, et al. Reactive Oxygen Species in Inflammation and Tissue Injury. Antioxid Redox Signal. 2014 Mar;20(7):1126–1167. DOI:10.1089/ars.2012.5149
- Zhong C, Yin C, Niu G, et al. MicroRNA miR-497 is closely associated with poor prognosis in patients with cerebral ischemic stroke. Bioengineered. 2021 Jan;12(1):2851–2862. DOI:10.1080/21655979.2021.1940073
- Xu W, Gao, L, and Zheng, J, et al. The Roles of MicroRNAs in stroke: possible therapeutic targets. Cell Transplant. 2018 Dec;27(12):1778–1788. DOI:10.1177/0963689718773361
- Li G, Morris-Blanco KC, Lopez MS, et al. Impact of microRNAs on ischemic stroke: from pre- to post-disease. Progress in Neurobiology. 2018Apr;163-164:59–78. DOI:10.1016/j.pneurobio.2017.08.002
- Tiedt S, Dichgans M. Role of Non-Coding RNAs in Stroke. Stroke. 2018 Dec;49(12):3098–3106. DOI:10.1161/STROKEAHA.118.021010
- Zhao H, Tao, Z, and Wang, R, et al. MicroRNA-23a-3p attenuates oxidative stress injury in a mouse model of focal cerebral ischemia-reperfusion. Brain Res. 2014Dec;1592:65–72. DOI:10.1016/j.brainres.2014.09.055
- Liu P, Zhao, H, and Wang, R, et al. MicroRNA-424 protects against focal cerebral ischemia and reperfusion injury in mice by suppressing Oxidative Stress. Stroke. 2015 Feb;46(2):513–519. DOI:10.1161/STROKEAHA.114.007482
- Wu J, Du K, Lu X. Elevated expressions of serum miR-15a, miR-16, and miR-17-5p are associated with acute ischemic stroke. International Journal of Clinical and Experimental Medicine. 2015;8(11):21071–21079. [Online]. Available http://www.ncbi.nlm.nih.gov/pubmed/26885038
- Tiedt S, Prestel, M, and Malik, R, et al. RNA-seq identifies circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res. 2017 Sep;121(8):970–980. DOI:10.1161/CIRCRESAHA.117.311572
- Wang S-W, Liu Z, Shi Z-S. Non-Coding RNA in acute ischemic stroke: mechanisms, biomarkers and therapeutic targets. Cell Transplant. 2018 Dec;27(12):1763–1777. DOI:10.1177/0963689718806818
- Yang R, Xu B, Yang B, et al. Non-coding RNAs: the extensive and interactive regulators of the blood-brain barrier permeability. RNA Biol. 2021 Jul;1–9. DOI:10.1080/15476286.2021.1950465
- Choudhuri S. Small noncoding RNAs: biogenesis, function, and emerging significance in toxicology. J Biochem Mol Toxicol. 2010 Feb;24(3):195–216. DOI:10.1002/jbt.20325
- Watson CN, Belli A, Di Pietro V. Small Non-coding RNAs: new class of biomarkers and potential therapeutic targets in neurodegenerative disease. Front Genet. 2019 Apr;10. DOI:10.3389/fgene.2019.00364
- Hennessy EJ. Cardiovascular Disease and long noncoding RNAs. Circ Cardiovasc Genet. 2017 Aug;10(4. DOI:10.1161/CIRCGENETICS.117.001556
- Hu W, Ding H, Xu Q, et al. Relationship between long noncoding RNA H19 polymorphisms and risk of coronary artery disease in a chinese population: a case-control study. Dis Markers. 2020May;2020:1–11. DOI:10.1155/2020/9839612
- Ruiz-Orera J, Albà MM. Conserved regions in long non-coding RNAs contain abundant translation and protein–RNA interaction signatures. NAR Genomics Bioinforma. 2019 Apr;1(1):e2–e2. doi: 10.1093/nargab/lqz002
- Kutter C, Watt, S, and Stefflova, K, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012 Jul;8(7):e1002841. DOI:10.1371/journal.pgen.1002841
- Guttman M, Garber, M, and Levin, JZ, et al. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010 May;28(5):503–510. DOI:10.1038/nbt.1633
- Angelucci F, Cechova K, Valis M, et al. MicroRNAs in Alzheimer’s disease: diagnostic markers or therapeutic agents?. Front Pharmacol. 2019 Jun;10. DOI:10.3389/fphar.2019.00665
- Wei W, Wang Z-Y, Ma L-N, et al. MicroRNAs in Alzheimer’s disease: function and potential applications as diagnostic biomarkers. Front Mol Neurosci. 2020 Aug;13. DOI:10.3389/fnmol.2020.00160
- Wang M, Qin L, Tang B. MicroRNAs in Alzheimer’s Disease. Front Genet. 2019 Mar;10. DOI:10.3389/fgene.2019.00153
- Yang S, Lim K-H, Kim S-H, et al. Molecular landscape of long noncoding RNAs in brain disorders. Mol Psychiatry. 2021 Apr;26(4):1060–1074. DOI:10.1038/s41380-020-00947-5
- Xin C, Liu J. Long non-coding RNAs in Parkinson’s disease. Neurochem Res. 2021 May;46(5):1031–1042. DOI:10.1007/s11064-021-03230-3
- Elkouris M, Kouroupi, G, and Vourvoukelis, A, et al. Long Non-coding RNAs associated with neurodegeneration-linked genes are reduced in Parkinson’s disease patients. Front Cell Neurosci. 2019 Feb;13. DOI:10.3389/fncel.2019.00058
- Hoss AG, Labadorf A, Beach TG, et al. microRNA Profiles in Parkinson’s disease prefrontal cortex. Front Aging Neurosci. 2016 Mar;8. DOI:10.3389/fnagi.2016.00036
- Nies YH, Mohamad Najib NH, Lim WL, et al. MicroRNA dysregulation in Parkinson’s disease: a narrative review. Front Neurosci. 2021 Apr;15. DOI:10.3389/fnins.2021.660379
- Cao T, Zhen X-C. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci Ther. 2018 Jul;24(7):586–597. DOI:10.1111/cns.12840
- Tian T, Wei, Z, and Chang, X, et al. The long noncoding RNA landscape in amygdala tissues from schizophrenia patients. EBioMedicine. 2018Aug;34:171–181. DOI:10.1016/j.ebiom.2018.07.022
- Li L, Zhuang Y, Zhao X, et al. Long Non-coding RNA in Neuronal Development and Neurological Disorders. Front Genet. 2019 Jan;9. DOI:10.3389/fgene.2018.00744
- Johnson R, Richter, N, and Jauch, R, et al. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genomics. 2010 May;41(3):269–274. DOI:10.1152/physiolgenomics.00019.2010
- Samadian M, Gholipour M, Hajiesmaeili M, et al. The Eminent Role of microRNAs in the Pathogenesis of Alzheimer’s Disease. Front Aging Neurosci. 2021 Mar;13. DOI:10.3389/fnagi.2021.641080
- Chang F, Zhang L-H, Xu W-P, et al. microRNA-9 attenuates amyloidβ-induced synaptotoxicity by targeting calcium/calmodulin-dependent protein kinase kinase 2. Mol Med Rep. 2014 May;9(5):1917–1922. DOI:10.3892/mmr.2014.2013
- Ramachandran D, Roy U, Garg S, et al. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic β-islets. FEBS J. 2011 Apr;278(7):1167–1174. DOI:10.1111/j.1742-4658.2011.08042.x
- Wang G, Huang , Y, and Wang, LL, et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci Rep. 2016 Jul;6(1):26697. DOI:10.1038/srep26697
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009 Aug;459(2):100–104. DOI:10.1016/j.neulet.2009.04.052
- Zhuang J, Chen, Z, and Cai, P, et al. Targeting MicroRNA-125b promotes neurite outgrowth but represses cell apoptosis and inflammation via Blocking PTGS2 and CDK5 in a FOXQ1-dependent way in alzheimer disease. Front Cell Neurosci. 2020 Dec;14. DOI:10.3389/fncel.2020.587747
- Banzhaf-Strathmann J, Benito E, May S, et al. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. The EMBO Journal. 2014 Aug;33(15):1667–1680. DOI:10.15252/embj.201387576
- An F, Gong G, Wang Y, et al. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget. 2017 Dec;8(69):114065–114071. DOI:10.18632/oncotarget.23119
- Hou TY, Zhou , Y, and Zhu, LS, et al. Correcting abnormalities in miR‐124/PTPN1 signaling rescues tau pathology in Alzheimer’s disease. J Neurochem. 2020 Aug;154(4):441–457. DOI:10.1111/jnc.14961
- Kozuka T, Omori, Y, and Watanabe, S, et al. miR-124 dosage regulates prefrontal cortex function by dopaminergic modulation.Sci Rep. 2019 Dec;9(1):3445. DOI:10.1038/s41598-019-38910-2
- Wang W-X, Rajeev, BW, and Stromberg, AJ, et al. The Expression of MicroRNA miR-107 decreases early in Alzheimer’s Disease and May Accelerate disease progression through regulation of -site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008 Jan;28(5):1213–1223. DOI:10.1523/JNEUROSCI.5065-07.2008
- Chen W, Wu, L, and Hu, Y, et al. MicroRNA-107 ameliorates damage in a cell model of Alzheimer’s disease by mediating the FGF7/FGFR2/PI3K/Akt pathway. J Mol Neurosci. 2020 October;70(10):1589–1597. DOI:10.1007/s12031-020-01600-0
- Faghihi MA, Zhang, M, and Huang, J, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010 May;11(5):R56. DOI:10.1186/gb-2010-11-5-r56
- Liu T, Huang, Y, and Chen, J, et al. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1-AS expression. Mol Med Rep. 2014 September;10(3):1275–1281. DOI:10.3892/mmr.2014.2351
- Li D, Zhang J, Li X, et al. Insights into lncRNAs in Alzheimer’s disease mechanisms. RNA Biol. 2021 Jul;18(7):1037–1047. DOI:10.1080/15476286.2020.1788848
- Parenti R, Paratore S, Torrisi A, et al. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during β-amyloid-induced apoptosis. Eur J Neurosci. 2007 Oct;26(9):2444–2457. DOI:10.1111/j.1460-9568.2007.05864.x
- Yamanaka Y, Faghihi MA, Magistri M, et al. Antisense RNA Controls LRP1 sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 2015 May;11(6):967–976. DOI:10.1016/j.celrep.2015.04.011
- Zhang L, Chen X, Chang M, et al. MiR-30c-5p/ATG5 axis regulates the progression of Parkinson’s Disease. Front Cell Neurosci. 2021 May;15. DOI:10.3389/fncel.2021.644507
- Swahari V, Nakamura, A, and Hollville, E, et al. MicroRNA-29 is an essential regulator of brain maturation through regulation of CH methylation. Cell Rep. 2021 Apr;35(1):108946. DOI:10.1016/j.celrep.2021.108946
- Wang R, Yao, J, and Gong, F, et al. miR‐29c‐3p regulates TET2 expression and inhibits autophagy process in Parkinson’s disease models. Genes Cells. 2021 Sep;26(9):684–697. DOI:10.1111/gtc.12877
- Shamsuzzama LK, Haque R, Nazir A. Role of MicroRNA Let-7 in modulating multifactorial aspect of neurodegenerative diseases: an overview. Mol Neurobiol. 2016 Jul;53(5):2787–2793. DOI:10.1007/s12035-015-9145-y
- Indrieri A, Carrella S, Carotenuto P, et al. The pervasive role of the miR-181 family in development, neurodegeneration, and Cancer. Int J Mol Sci. 2020 Mar;21(6):2092. DOI:10.3390/ijms21062092
- Liu Y, Song Y, Zhu X. MicroRNA-181a in Parkinson’s Disease by Inhibiting p38 Mitogen-Activated Protein Kinase (MAPK)/c-Jun N-Terminal Kinases (JNK) Signaling pathways. Med Sci Monit. 2017Apr;23:1597–1606. DOI:10.12659/MSM.900218
- Ye Y, He, X, and Lu, F, et al. A lincRNA-p21/miR-181 family feedback loop regulates microglial activation during systemic LPS- and MPTP- induced neuroinflammation. Cell Death Dis. 2018 Aug;9(8):803. DOI:10.1038/s41419-018-0821-5
- Scheele C, Petrovic, N, and Faghihi, MA, et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics. 2007 Dec;8(1):74. DOI:10.1186/1471-2164-8-74
- Carrieri C, Forrest, AR, and Santoro, C, et al. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci. 2015 Apr;9. DOI:10.3389/fncel.2015.00114
- Wang S, Zhang X, Guo Y, et al. The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget. 2017 Apr;8(15):24449–24456. DOI:10.18632/oncotarget.15511
- Cao B, Wang T, Qu Q, et al. Long Noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s Disease via Regulating miR-7/NLRP3 Pathway. Neuroscience. 2018Sep;388:118–127. DOI:10.1016/j.neuroscience.2018.07.019
- Ni C, Jiang, W, and Wang, Z, et al. LncRNA-AC006129.1 reactivates a SOCS3-mediated anti-inflammatory response through DNA methylation-mediated CIC downregulation in schizophrenia. Mol Psychiatry. 2021 Aug;26(8):4511–4528. DOI:10.1038/s41380-020-0662-3
- Shi Y, and Shang J. Long noncoding RNA expression profiling using arraystar LncRNA Microarrays Methods Mol Biol . 2016 1402 ;43–61 doi:10.1007/978-1-4939-3378-5_6.
- Miller BH, Zeier, Z, and Xi, L, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci. 2012 Feb;109(8):3125–3130. DOI:10.1073/pnas.1113793109
- Wanet A, Tacheny A, Arnould T, et al. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012 Jun;40(11):4742–4753. DOI:10.1093/nar/gks151
- Mellios N, Huang H-S, Baker SP, et al. Molecular Determinants of Dysregulated GABAergic gene expression in the prefrontal cortex of subjects with Schizophrenia. Biol Psychiatry. 2009 Jun;65(12):1006–1014. DOI:10.1016/j.biopsych.2008.11.019
- Merelo V, Durand, D, and Lescallette, AR, et al. Associating schizophrenia, long non-coding RNAs and neurostructural dynamics. Front Mol Neurosci. 2015 Sep;8. DOI:10.3389/fnmol.2015.00057
- Wright C, Turner JA, Calhoun VD, et al. Potential Impact of miR-137 and Its Targets in Schizophrenia. Front Genet. 2013;4. DOI:10.3389/fgene.2013.00058
- Chung DW, Rudnicki DD, Yu L, et al. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum Mol Genet. 2011 Sep;20(17):3467–3477. DOI:10.1093/hmg/ddr263
- Liu T, Im W, Mook-Jung I, et al. MicroRNA-124 slows down the progression of Huntington′s disease by promoting neurogenesis in the striatum. Neural Regen Res. 2015;10(5):786.
- Park S-Y, Lee JH, Ha M, et al. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat Struct Mol Biol. 2009 Jan;16(1):23–29. DOI:10.1038/nsmb.1533
- Chanda K, Das, S, and Chakraborty, J, et al. Altered levels of long NcRNAs Meg3 and neat1 in cell and animal models of huntington’s Disease. RNA Biol. 2018 Oct;15(10):1348–1363. DOI:10.1080/15476286.2018.1534524
- Khalil AM, Guttman, M, and Huarte, M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci. 2009 Jul;106(28):11667–11672. DOI:10.1073/pnas.0904715106
- Perkins DO, Jeffries, CD, and Jarskog, LF, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):27 doi:10.1186/gb-2007-8-2-r27.
- Gardiner E, Beveridge, NJ, and Wu, JQ, et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. 2012 Aug;17(8):827–840. DOI:10.1038/mp.2011.78
- Wang W, Kwon EJ, Tsai L-H. MicroRNAs in learning, memory, and neurological diseases. Learn Mem. 2012 Aug;19(9):359–368. DOI:10.1101/lm.026492.112
- Edbauer D, Neilson, JR, and Foster, KA, et al. Regulation of synaptic structure and function by FMRP-associated MicroRNAs miR-125b and miR-132. Neuron. 2010 Feb;65(3):373–384. DOI:10.1016/j.neuron.2010.01.005
- Kenny P, Ceman S. RNA secondary structure modulates FMRP’s Bi-Functional Role in the MicroRNA Pathway. Int J Mol Sci. 2016 Jun;17(6):985. DOI:10.3390/ijms17060985
- Bañez-Coronel M, Porta, S, and Kagerbauer, B, et al. A pathogenic mechanism in Huntington’s Disease Involves Small CAG-Repeated RNAs with neurotoxic activity. PLoS Genet. 2012 Feb;8(2):1002481. DOI:10.1371/journal.pgen.1002481
- Johnson R, Zuccato C, Belyaev ND, et al. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008 Mar;29(3):438–445. DOI:10.1016/j.nbd.2007.11.001
- Packer AN, Xing Y, Harper SQ, et al. The Bifunctional microRNA miR-9/miR-9* Regulates REST and CoREST and Is Downregulated in Huntington’s Disease. J Neurosci. 2008 Dec;28(53):14341–14346. DOI:10.1523/JNEUROSCI.2390-08.2008
- Lee S-T, Chu, K, and Im, WS, et al. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011 Jan;227(1):172–179. DOI:10.1016/j.expneurol.2010.10.012
- Piket E, Zheleznyakova GY, Kular L, et al. Small non-coding RNAs as important players, biomarkers and therapeutic targets in multiple sclerosis: a comprehensive overview. J Autoimmun. 2019Jul;101:17–25. DOI:10.1016/j.jaut.2019.04.002
- Amoruso A, Blonda, M, and Gironi, M, et al. Immune and central nervous system-related miRNAs expression profiling in monocytes of multiple sclerosis patients. Sci Rep. 2020 Dec;10(1):6125. DOI:10.1038/s41598-020-63282-3
- Zhou Z, Xiong H, Xie F, et al. a meta-analytic review of the value of mirna for multiple sclerosis diagnosis. front neurol. 2020 feb;11. DOI:10.3389/fneur.2020.00132
- Teuber‐Hanselmann S, Meinl E, Junker A. MicroRNAs in gray and white matter multiple sclerosis lesions: impact on pathophysiology. J Pathol. 2020 Apr;250(5):496–509. DOI:10.1002/path.5399
- Bali P, Kenny PJ. MicroRNAs and Drug Addiction. Front Genet. 2013;4. DOI:10.3389/fgene.2013.00043
- Gowen AM, Odegaard, KE, and Hernandez, J, et al. Role of microRNAs in the pathophysiology of addiction. WIREs RNA. 2021 May;12(3). DOI:10.1002/wrna.1637
- Hollander JA, Im, HI, and Amelio, A, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010 Jul;466(7303):197–202. DOI:10.1038/nature09202
- Chandrasekar V, Dreyer J-L. microRNAs miR-124, let-7d and miR-181a regulate Cocaine-induced Plasticity. Mol Cell Neurosci. 2009 Nov;42(4):350–362. DOI:10.1016/j.mcn.2009.08.009
- Saba R, Störchel, PH, and Aksoy-Aksel, A, et al. Dopamine-Regulated MicroRNA MiR-181a controls GluA2 Surface Expression in Hippocampal Neurons. Mol Cell Biol. 2012 Feb;32(3):619–632. DOI:10.1128/MCB.05896-11
- Eipper-Mains JE, Kiraly DD, Palakodeti D, et al. microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA. 2011 Aug;17(8):1529–1543. DOI:10.1261/rna.2775511
- Cohen-Armon M. Long-Term memory requires PolyADP-ribosylation. Science. 2004 Jun;304(5678):1820–1822. DOI:10.1126/science.1096775
- Scobie KN, Damez-Werno, D, and Sun, H, et al. Essential role of poly(ADP-ribosyl)ation in cocaine action. Proc Natl Acad Sci. 2014 Feb;111(5):2005–2010. DOI:10.1073/pnas.1319703111
- Dash S, Balasubramaniam, M, and Rana, T, et al. Poly (ADP-Ribose) Polymerase-1 (PARP-1) Induction by Cocaine Is Post-Transcriptionally Regulated by miR-125b. eneuro. 2017 Jul;4(4):ENEURO.0089–17.2017. DOI:10.1523/ENEURO.0089-17.2017
- Dash S, Dash C, Pandhare J. Activation of proline metabolism maintains ATP levels during cocaine-induced polyADP-ribosylation. Amino Acids. Aug 2021. DOI:10.1007/s00726-021-03065-w
- Bastle RM, Oliver, R, and Gardiner, A, et al. In silico identification and in vivo validation of miR-495 as a novel regulator of motivation for cocaine that targets multiple addiction-related networks in the nucleus accumbens. Mol Psychiatry. 2018 Feb;23(2):434–443. DOI:10.1038/mp.2016.238
- Bannon MJ, Savonen, CL, and Jia, H, et al. Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers. J Neurochem. 2015 Oct;135(1):50–59. DOI:10.1111/jnc.13255
- Xu H, Brown, AN, and Waddell, NJ, et al. Role of long noncoding RNA Gas5 in cocaine action. Biol Psychiatry. 2020 Nov;88(10):758–766. DOI:10.1016/j.biopsych.2020.05.004
- Zhu L, Zhu, J, and Liu, Y, et al. Methamphetamine induces alterations in the long non-coding RNAs expression profile in the nucleus accumbens of the mouse. BMC Neurosci. 2015 Dec;16(1):18. DOI:10.1186/s12868-015-0157-3
- Condrat CE, Thompson, DC, and Barbu, MG, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020 Jan;9(2):276. DOI:10.3390/cells9020276
- Gayosso-Gómez LV, Ortiz-Quintero B. Circulating MicroRNAs in blood and other body fluids as biomarkers for diagnosis, prognosis, and therapy response in lung cancer. Diagnostics. 2021 Mar;11(3):421. DOI:10.3390/diagnostics11030421
- Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as biomarkers and therapeutic targets in non-small cell lung cancer: current perspectives. Target Oncol. 2017 Apr;12(2):179–200. DOI:10.1007/s11523-017-0478-5
- Gessner I, Fries JWU, Brune V, et al. Magnetic nanoparticle-based amplification of microRNA detection in body fluids for early disease diagnosis. J Mater Chem B. 2021;9(1):9–22.
- Gessner I, Yu, X, and Jüngst, C, et al. Selective capture and purification of micrornas and intracellular proteins through antisense-vectorized magnetic nanobeads. Sci Rep. 2019 Dec;9(1):2069. DOI:10.1038/s41598-019-39575-7
- Lee SWL, Paoletti, C, and Campisi, M, et al. MicroRNA delivery through nanoparticles. J Control Release. 2019Nov;313:80–95. DOI:10.1016/j.jconrel.2019.10.007
- Lee TJ, Yoo, JY, and Shu, D, et al. RNA Nanoparticle-Based Targeted Therapy For Glioblastoma through Inhibition of Oncogenic miR-21. Mol Ther. 2017 Jul;25(7):1544–1555. DOI:10.1016/j.ymthe.2016.11.016
- Kouri FM, Hurley, LA, and Daniel, WL, et al. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015 Apr;29(7):732–745. DOI:10.1101/gad.257394.114
- Grafals-Ruiz N, Rios-Vicil, CI, and Lozada-Delgado, EL, et al. Brain targeted gold liposomes improve RNAi Delivery for Glioblastoma. Int J Nanomedicine. 2020Apr;15:2809–2828. DOI:10.2147/IJN.S241055
- Gangemi CMA, Alaimo, S, and Pulvirenti, A, et al. Endogenous and artificial miRNAs explore a rich variety of conformations: a potential relationship between secondary structure and biological functionality. Sci Rep. 2020 Dec;10(1):453. DOI:10.1038/s41598-019-57289-8
- Kotowska‐Zimmer A, Pewinska M, Olejniczak M. Artificial miRNAs as therapeutic tools: challenges and opportunities. WIREs RNA. 2021 Jul;12(4. DOI:10.1002/wrna.1640
- Keskin S, Brouwers, CC, and Sogorb-Gonzalez, V, et al. AAV5-miHTT Lowers Huntingtin mRNA and protein without off-target effects in patient-derived neuronal cultures and astrocytes. Mol Ther - Methods Clin Dev. 2019Dec;15:275–284. DOI:10.1016/j.omtm.2019.09.010
- Haussecker D. Current issues of RNAi therapeutics delivery and development. J Control Release. 2014Dec;195:49–54.DOI:10.1016/j.jconrel.2014.07.056
- Judge AD, Bola G, Lee ACH, et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006 Mar;13(3):494–505. DOI:10.1016/j.ymthe.2005.11.002
- Jackson AL. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006 May;12(7):1179–1187. DOI:10.1261/rna.25706
- Soutschek J, Akinc, A, and Bramlage, B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004 Nov;432(7014):173–178. DOI:10.1038/nature03121
- Kim Y-K. RNA therapy: current status and future potential. Chonnam Med J. 2020;56(2):87.
- Salvatori B, Biscarini S, Morlando M. Non-coding RNAs in Nervous System Development and Disease. Front Cell Dev Biol. 2020 May;8. DOI:10.3389/fcell.2020.00273
- Latowska J, Grabowska A, Zarębska Ż, et al. Non-Coding RNAs in brain tumors, the contribution of lncRNAs, circRNAs, and snoRNAs to cancer development—their diagnostic and therapeutic potential. Int J Mol Sci. 2020 Sep;21(19):7001. DOI:10.3390/ijms21197001
- Wu -Y-Y, Kuo H-C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J Biomed Sci. 2020 Dec;27(1):49. DOI:10.1186/s12929-020-00636-z
- Liu X, Shen L, Han B, et al. Involvement of noncoding RNA in blood-brain barrier integrity in central nervous system disease. Non-coding RNA Res. 2021 Sep;6(3):130–138. doi: 10.1016/j.ncrna.2021.06.003
- Dash S, Dash C, Pandhare J. Therapeutic Significance of microRNA-Mediated Regulation of PARP-1 in SARS-CoV-2 Infection. Noncoding RNA. 2021 Sep;7(4):60. DOI:10.3390/ncrna7040060
- Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem. 2014 Nov;6(17):1967–1984. DOI:10.4155/fmc.14.116
- Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011 Dec;18(12):1104–1110. DOI:10.1038/gt.2011.50
- Lam JKW, Chow MYT, Zhang Y, et al. siRNA Versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252.
- Liu SJ, Nowakowski, TJ, and Pollen, AA, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016 Dec;17(1):67. DOI:10.1186/s13059-016-0932-1
- Bitar M, Barry G. Multiple innovations in genetic and epigenetic mechanisms cooperate to underpin human brain evolution. Mol Biol Evol. 2018 Feb;35(2):263–268. DOI:10.1093/molbev/msx303
- Plowman T, Lagos D. Non-Coding RNAs in COVID-19: emerging Insights and Current Questions. Noncoding RNA. 2021 Aug;7(3):54. DOI:10.3390/ncrna7030054
- Greco S, Madè A, Gaetano C, et al. Noncoding RNAs implication in cardiovascular diseases in the COVID-19 era. J Transl Med. 2020 Dec;18(1):408. DOI:10.1186/s12967-020-02582-8
- Yang Q, Lin F, Wang Y, et al. Long Noncoding RNAs as Emerging Regulators of COVID-19. Front Immunol. 2021 Aug;12. DOI:10.3389/fimmu.2021.700184
- Natarelli L, Parca L, Mazza T, et al. MicroRNAs and Long Non-Coding RNAs as potential candidates to target specific motifs of SARS-CoV-2. Noncoding RNA. 2021 Feb;7(1):14. DOI:10.3390/ncrna7010014
- Perry RB-T, Ulitsky I. Therapy based on functional RNA elements. Science. 2021 Aug;373(6555):623–624. DOI:10.1126/science.abj7969
- Adams D, Gonzalez-Duarte, A, and O'Riordan, WD, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018 Jul;379(1):11–21. DOI:10.1056/NEJMoa1716153
- Wood H. FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat Rev Neurol. 2018 Oct;14(10):570. DOI:10.1038/s41582-018-0065-0
- Marsh J, Alifragis P. Synaptic dysfunction in Alzheimer’s disease: the effects of amyloid beta on synaptic vesicle dynamics as a novel target for therapeutic intervention. Neural Regen Res. 2018;13(4):616.
- Bužgová R, Kozáková R, Juríčková L. The unmet needs of patients with progressive neurological diseases in the czech republic: a qualitative study. J Palliat Care. 2019 Jan;34(1):38–46. DOI:10.1177/0825859718800489
- Scavone C, Di Mauro G, Mascolo A, et al. The new paradigms in clinical research: from early access programs to the novel therapeutic approaches for unmet medical needs. Front Pharmacol. 2019 Feb;10. DOI:10.3389/fphar.2019.00111
- Feigin VL, Vos, T, and Nichols, E, et al. The global burden of neurological disorders: translating evidence into policy. Lancet Neurol. 2020 Mar;19(3):255–265. DOI:10.1016/S1474-4422(19)30411-9
- Feigin VL, Vos, T, and Alahdab, F, et al. Burden of Neurological Disorders Across the US From 1990-2017. JAMA Neurol. 2021 Feb;78(2):165. DOI:10.1001/jamaneurol.2020.4152
- Hanna J, Hossain GS, Kocerha J. The Potential for microRNA therapeutics and clinical research. Front Genet. 2019 May;10. DOI:10.3389/fgene.2019.00478
- DeVos SL, Miller, RL, and Schoch, KM, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med. 2017 Jan;9(374):0481. DOI:10.1126/scitranslmed.aag0481
- Xia XG, Zhou H, Zhou S, et al. An RNAi strategy for treatment of amyotrophic lateral sclerosis caused by mutant Cu,Zn superoxide dismutase. J Neurochem. 2005 Jan;92(2):362–367. DOI:10.1111/j.1471-4159.2004.02860.x
- Do Carmo Costa M, Luna-Cancalon, K, and Fischer, S, et al. Toward RNAi therapy for the polyglutamine disease machado–joseph disease. Mol Ther. 2013 Oct;21(10):1898–1908. DOI:10.1038/mt.2013.144
- Liu Z, Li S, Liang Z, et al. Targeting β-secretase with RNAi in neural stem cells for Alzheimer’s disease therapy. Neural Regeneration Research. 2013 Nov;8(33):3095–3106. DOI:10.3969/j.issn.1673-5374.2013.33.003
- Nóbrega C, Nascimento-Ferreira, I, and Onofre, I, et al. Silencing Mutant Ataxin-3 Rescues Motor Deficits and Neuropathology in Machado-Joseph Disease Transgenic Mice. PLoS One. 2013 Jan;8(1):e52396. DOI:10.1371/journal.pone.0052396
- Yang H, Shang , D, and Xu, Y, et al. The LncRNA connectivity map: using LncRNA signatures to connect small molecules, LncRNAs, and diseases. Sci Rep. 2017 Dec;7(1):6655. DOI:10.1038/s41598-017-06897-3
- Wurster CD, Ludolph AC. Antisense oligonucleotides in neurological disorders. Ther Adv Neurol Disord. 2018Jan;11:175628641877693. DOI:10.1177/1756286418776932
- Buiting K, Williams C, Horsthemke B. Angelman syndrome — insights into a rare neurogenetic disorder. Nat Rev Neurol. 2016 Oct;12(10):584–593. DOI:10.1038/nrneurol.2016.133
- Meng L, Ward AJ, Chun S, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015 Feb;518(7539):409–412. DOI:10.1038/nature13975
- Modarresi F, Faghihi, M, and Lopez-Toledano, M, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012 May;30(5):453–459. DOI:10.1038/nbt.2158
- Amodio N, Stamato, MA, and Juli, G, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018 Sep;32(9):1948–1957. DOI:10.1038/s41375-018-0067-3
- Amodio N, Raimondi, L, and Juli, G, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018 Dec;11(1):63. DOI:10.1186/s13045-018-0606-4
- Gutschner T, Hämmerle, M, and Eissmann, M, et al. The Noncoding RNA MALAT1 Is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013 Feb;73(3):1180–1189. DOI:10.1158/0008-5472.CAN-12-2850
- Zhou Y, Shan, T, and Ding, W, et al. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating Hippo‐YAP signaling. J Cell Physiol. 2018 Aug;233(8):5805–5814. DOI:10.1002/jcp.26357
