ABSTRACT
According to the United Nations Environment Programme (UNEP), soil health is declining over the decades and it has an adverse impact on human health and food security. Hence, soil health restoration is a need of the hour. It is known that microorganisms play a vital role in remediation of soil pollutants like heavy metals, pesticides, hydrocarbons, etc. However, the indigenous microbes have a limited capacity to degrade these pollutants and it will be a slow process. Genetically modified organisms (GMOs) can catalyze the degradation process as their altered metabolic pathways lead to hypersecretions of various biomolecules that favor the bioremediation process. This review provides an overview on the application of bioengineered microorganisms for the restoration of soil health by degradation of various pollutants. It also sheds light on the challenges of using GMOs in environmental application as their introduction may affect the normal microbial community in soil. Since soil health also refers to the potential of native organisms to survive, the possible changes in the native microbial community with the introduction of GMOs are also discussed. Finally, the future prospects of using bioengineered microorganisms in environmental engineering applications to make the soil fertile and healthy have been deciphered. With the alarming rates of soil health loss, the treatment of soil and soil health restoration need to be fastened to a greater pace and the combinatorial efforts unifying GMOs, plant growth-promoting rhizobacteria, and other soil amendments will provide an effective solution to soil heath restoration ten years ahead.
Graphical Abstract Schematic Representation Of Approaches Used To Restore Soil Health
1. Introduction
The existence and sustainable living of all organisms big and small greatly rely on the quality of various parameters that they interact within their ecological niche [Citation1]. Lithosphere, the home of various terrestrial living forms, has shaped itself from time to time in response to various extraneous and intrinsic polluting agents, resulting in variant levels of soil health in different parts of the world [Citation2]. Soil health plays a vital role in defining the members of a habitat, determining their longevity, productivity, and persistence. Regardless of their complexity, lower and higher living forms are equally affected by their resident soil health. The molding role of soil in controlling soil and human pathogens is a clear indication of that [Citation3].
Soil health, well-defined by its functionality and ecological equilibrium, relies on various physical factors such as porosity, moisture, texture, etc.; chemical factors viz, organic matter, nutrients, C, and N; and biological factors such as microbial diversity, soil respiration, and microbial biomass [Citation4]. The causes of soil damage are known to everyone including a long list of natural causes including rainfall, soil erosion, wind erosion, disasters like flood and landslide, etc. and anthropogenic activities of mining, deforestation, chemical fertilizer-based agriculture, urbanization, chemical-induced acidification [Citation5], alkalinization, salinization, oil spills, and many more [Citation6,Citation7]. However, the need of the hour is to find methods to find immediate solutions to overcome the damage caused by them.
Research and efforts to improve soil quality are relevant due to various reasons. First, the growth of autotrophic plants (the trophic level being a food source to all life forms directly or indirectly) solely depends on the soil health [Citation8]. Second, soil health directly contributes to the biodiversity and well-being of an ecosystem sustaining life of plants, animals, and humans [Citation9]. Agricultural productivity and the tolerance of soil to environmental stresses are good indicators of soil health, thereby implying that these targets could be achieved only by taking measures to restore soil health [Citation10]. Moreover, human health and soil health also remain entwined with each other as the latter prevents human exposure to pathogens, provides good nutrients and quality medicine, and enhances immunity [Citation11]. Furthermore, active soil biotic components aid to combat the drastic effects of climate changes and sequester more carbon dioxide, relieving the stress of global warming [Citation12]. The economy and agricultural productivity of a nation also depend on the soil resources, and its depletion would lead to the generation of barren nonproductive lands [Citation13].
Of the various factors aiding to build up the soil health, microbes play a pivotal role by degrading the pollutants. Soil health restoration is a cumulative outcome of indigenous microbes of the lithosphere [Citation14]. The advantages of relying on microbes are attributed to their versatility to detoxify a wide variety of pollutants [Citation15], eco-friendly nature [Citation16,Citation17], ability to enrich soil with nutrients [Citation18], survival in even harsh environments [Citation19,Citation20], production of plant growth-promoting substances [Citation21], ease of treatment [Citation22], and absence of toxic end products [Citation23].
Soil health can be addressed not only by removing the accumulated harmful chemicals but also by adding more nutrients to the soil to improves its health [Citation24]. Soil health restoration can be achieved by using microbes that are capable of adding fertility-contributing nutrients to the soil along with a dual role of removing or nullifying the effect of toxic xenobiotics. The current review discusses the prospects of utilizing the microbial detoxifying, biotransforming, and bioremediatory role in removing various xenobiotics in the soil, with a simultaneous role in improving soil fertility. Moreover, the use of bioengineered microbes to speed the process of detoxification or improvise their efficiency is also targeted in this paper to find effective solutions in timely soil health restoration.
2. Role of microorganisms in soil health improvement
Soil microbes are active engineers of soil where they condition the soil for plant growth by making the nutrients available and production of necessary growth regulators. They also help in the organic matter transformation and xenobiotic degradation in the soil [Citation25]. Natural microbial populations play distinct functional roles in adhering and desorbing inorganic nutrients to physical surfaces and degrading organic residues to make them a part of soil [Citation26–28]. The cumulative role of plants, as well as microbes, attributes to the fitness of soil for agriculture and farming [Citation29]. It is noted that even small human interventions such as the addition of sewage sludge enabled to increase the soil resident microbial population of Proteobacteria and Bacteroidetes in bauxite overloaded disposal areas and enhanced the process of soil formation [Citation30]. Apart from the soil formation, the process of nutrient cycling an essential part to maintain soil fertility is steered by microbes in the various biogeochemical cycles [Citation31].
The use of rhizosphere bacteria to improve soil fertility instead of chemical fertilizers has been encouraged to attain sustainable plant growth [Citation32]. It is now clear that the improvement of plant performance is a complex process involving interaction with specific microbes or consortiums. New approaches involve engineering symbiotic relationships to make nonlegumes and other staple crops to fix nitrogen [Citation33,Citation34], thereby converting them into soil fertility-contributing plants. This will significantly improve the global food supplies and help to meet sustainability goals.
The second contributory factor to soil health depends on the ability of soil microbes to detoxify and nullify the toxic pollutants that ultimately reach soil from various routes [Citation35]. Some of the microorganisms have superior ability in degradation of specific xenobiotics, for instance, pesticides viz. endosulfan, Lambda-cypermethrin, and deltamethrin, profenofos, and pyrethroid are degraded by microbes like Aspergillus sp. and Raoultella ornithinolytica [Citation36–38]. Similarly, a wide range of xenobiotics such as plastics [Citation22], hydrocarbons [Citation39], surfactants [Citation40], Polychlorinated biphenyls [Citation41], radioactive waste [Citation42], heavy metals [Citation43], etc. are effectively degraded by specific microbes. But there also do exist many multipotent microbes termed ‘Superbugs’ capable of degrading a wide range of xenobiotics [Citation44]. Compared to individual isolates, microbial consortiums are proven to be more effective in xenobiotic degradation as noted in several studies involving six isolates for tetrachlorvinphos degradation [Citation45] and an actinobacteria consortium composed of Pseudomonas, Enterobacter, Aspergillus, and Rhodotorula could effectively remove seven pesticides [Citation46].
Synthetic microbial consortia (SMC) were developed for improving plant growth and quality, which constituted the soil microbiome of high-quality crops. While formulating a microbial consortium, one must also consider the capability of these microbes in acclimatizing with the new environment. It is needless to say that the indigenous microbes are involved in ecological services to plants especially in their rhizosphere [Citation47]. Moreover, these indigenous microbes are also helpful in combating plant stress [Citation48,Citation49]. These rhizosphere-dwelling microbes are ideal for xenobiotic degradation and restoring soil quality [Citation50]. Reports are available on employing plant growth-promoting microbes in degradation of second-generation pesticides especially organophosphate pesticides [Citation51]. The microbes from the rhizosphere have multiple potentialities for degradation of pesticides and enhancement of plant growth [Citation52,Citation53].
The success of a selected microbial inoculum relies on its ability to thrive and act along with the autochthonous microbes and the abiotic components of that habitat [Citation26]. The survival and persistence of the microbe in the soil depend on how it interacts with other biotic components in the ecosystem, and quite often, plant interactions with microbial consortia are more effective than individual microbes [Citation54,Citation55]. Thus, the productivity of soil is undoubtedly dependent on microbial diversity and its growth-promoting qualities [Citation56].
3. Microbial xenobiotic degradation and plant growth promotion products to increase soil fertility
The concept of pollutant degradation of microbes serves as an effective factor in the removal of pollutants and the negative effects that such pollutants have on the soil and plants. However, there also exists another dimension of microbial activity in which they contribute to soil fertility through various products secreted by them amid their role as causatives of bioremediation. Many of these metabolites can be categorized as different classes including intermediates of xenobiotic degradation, biotransformed intermediates, and even plant growth-promoting factors produced by rhizobacteria. Reports on xenobiotic degradation products to serve as plant growth-promoting factors are still not clear, yet it is seen that microbes capable of conducting xenobiotic degradation are also found to be capable of expressing soil fertility-improving factors as in the case of Pseudomonas isolates performing hydrocarbon degradation [Citation57]. Pseudomonas in the former study also exhibited properties of phosphate solubilization, nitrogen fixation, and indole acetic acid production, which are key factors for plant growth promotion and soil fertility enhancement apart from its ability to degrade hydrocarbons. Another study on sodium doceyl sulfate (SDS) degradation also indicates that some dodecanol, a degradatory intermediate, could be biotransformed to rhamnolipids as a measure to overcome SDS stress and damage by the bioremediating microbe [Citation58].
The process of heavy metal detoxification by microbes also indicates that the presence of arbuscular mycorrhizal symbionts in plants enables them to bioaccumulate heavy metals in them, extends the tolerance level of plants to various stress such as drought or pollutants, improves nutrient availability or uptake by plants, and eventually promotes plant growth [Citation59]. The external application of plant growth regulators is also found to improve the plants’ ability to overcome the toxicity and stress caused by pesticides by triggering antioxidant mechanisms in another study [Citation60]. Thus the concept of Plant growth promoting rhizobacteria (PGPR) capable of degrading xenobiotics also gains relevance in efforts to improve soil health [Citation21].
Rhizoremediation, the so-called phenomenon of improving soil health using root-associated microbes, involves the participation of rhizobacteria that remediate xenobiotics in their root area and simultaneously produce plant growth-promoting factors to aid the plants. The principles of bioaugmentation and phytoremediation are visualized when plants provide nutrients to microbes, while rhizobacteria simultaneously remediate soil and increase nitrogen and phosphorus availability to plants, eventually contributing to soil health and plant growth [Citation61]. PGPR represent a group of bacteria resident in the rhizophere, in/around the root of plants directly or indirectly contributing to soil health, through various modes viz production of enzymes, hormones, and plant growth regulators, increasing bioavailability of nutrients, removal of antinutrient factors, protecting plants from antagonists, increased root growth, and many more mechanisms [Citation62,Citation63]. The use of plant growth-promoting rhizobacteria (PGPR) for the remediation of various soil pollutants such as petroleum [Citation64], heavy metals particularly mercury [Citation65], polychlorinated biphenyls [Citation66], etc. is found to be very effective.
briefly shows the role of rhizobacteria in improving soil health and the various mechanisms that they adopt to promote soil health and plant growth. Various enzymes such as aminocyclopropane‐1‐carboxylate (ACC) deaminase contributing to stressrelated ethylene production are reduced, whereas nitrogenases and phytases responsible for nitrogen fixation in soil and phytate removal are promoted in rhizobacteria when they are utilized in rhizoremediation [Citation67]. Phytates serve as an antinutrient factor to the availability of phosphorus in soil. Rhizobacteria-mediated pollutant removal and soil health improvement serves as a cost-effective, safe, and eco-friendly mechanism to deal with toxic substances in the soil [Citation68]. It is noted that rather than using individual microbes for soil health restoration, the use of consortium of bacteria to remove toxic pollutants and biofertilizer combinations to improve soil fertility give better results [Citation69]. Moreover, amendments of the soil with biowaste compost also provide an added advantage to nurture soil health and revive it [Citation70].
Table 1. Role of rhizobacteria in the improvement of soil health and plant growth
4. Bioengineering of microorganisms for soil health restoration by remediation
Owing to the disadvantage of indigenous microbes in acclimatizing in the new environment and performing degradation of pollutants efficiently, genetically engineered ones could be employed for better performance [Citation71]. These engineered microorganisms can efficiently remediate most of the contaminants, which cannot be degraded by normal indigenous microbes. A range of molecular tools are available for the construction of GMOs like biolistic transformation, electroporation, conjugation, horizontal transfer of bacterial DNA, molecular cloning, and transformation of protoplast. Transfer and expression of novel genes with high degradation capacity also minimize the remediation time. Engineered microbes could remediate a variety of compounds like toluene, octane, naphthalene, salicylate, and xylene by expressing genes encoded in the bacterial plasmid [Citation72]. There are four different approaches suggested by the researchers: a) manipulating the enzyme affinity and specificity; b) construction of gene and regulatory pathway modifications; c) process development, controlling, and monitoring of bioremediation; and d) employing sensor-based bioaffinity reporters to sense pollutants, reduce toxicity, and predict the end points [Citation72]. The ability to incorporate many genes contributing to xenobiotic degradation into a single microbe adds the potential to degrade a wide range of xenobiotics by a single microbe [Citation73]. shows the list of bioengineered microbes used to remove xenobiotics.
Table 2. List of recombinant microbes with different xenobiotic compounds
4.1. Heavy metal removal
Heavy metal removal using microbes follows the principles of biosorption and bioaccumulation to remove heavy metals from the polluted environments [Citation17]. The process of heavy metal biosorption involves the sorption and entrapping of heavy metals onto the outer lipid membrane and sometimes even on the exopolysaccharide secretions of the living or dead heavy metal sequester [Citation43]. On the other hand, bioaccumulation involves the use of various transporters such as porins, ion channels, primary active transporters, and secondary transporters that transport heavy metals from the environment to microbial cytoplasm to be further bound by metal-sequestering proteins within the microbial cytoplasm [Citation74].
Genetically engineered microbes for heavy metal removal adopt different strategies, viz. genetically engineering the transport proteins involved in metal transfer across microbial membrane as well as expressing various metal-binding proteins like ferritin, metallothionenin, polyphosphates, and phytoalexins that serve as storage proteins of metals in the microbial cytoplasm [Citation74]. Ferritins from worm Dendrorhynchus zhejiangensis aid in both transport and storage of heavy metals, making it a suitable candidate for metal detoxification [Citation75]. The recombinant expression of metal storage proteins such as metallothionenin in the surface layer proteins of Deinococcus improved the cadmium uptake approximately 3 times higher than normal metallothionenin expression in cytoplasm alone, whereas cell-free preparations of recombinant phosphatases were effective in uranium precipitation [Citation76].
Chromium (VI) remediation by a consortium of microbes indicated the presence of extracellular reductase rather than adsorption that converts them to reduced form of Cr (III) and further to Cr(OH)3, which is optimum at the pH of 8.0 and stable at a concentration of 50 mg/l [Citation77]. The microbes simultaneously produce various metabolites such as lactic acid during the heavy metal remediation to counteract the pH shift caused by formation of hydroxides. A decrease in the microbial diversity in the presence of Cr(VI) exposure clearly indicates the relevance of choosing Cr-resistant microbes in chromium and the concentration of chromium exposure. Similar studies of mercury remediation with mercury-resistant microbe Sphingobium SA2 indicate the complete detoxification of Hg2+ ions to nontoxic Hg0 ions by mercury reductases, which is yet another proof indicating the fate of heavy metals remediated by microbes [Citation78]. Apart from this, the microbial transformation of inorganic arsenic to volatile derivatives has known to play an important role in the biogeochemical cycling of arsenic [Citation79].
4.2. Pesticide degradation
Many genes have been discovered with a high ability to degrade pesticides, and this extends the possibility of developing a GMO suitable for the degradation of pesticides. As we move toward organic farming practices and the use of genetically engineered plants for enhanced yield, biological pesticides have become an important part of sustainable agricultural practices. However, the engineered microbe’s role is crucial in restoring soil health by simply degrading pesticide residues, which were otherwise recalcitrant and remain for years in the soil.
A commonly used pesticide atrazine, which poses a potential threat to other organisms, was degraded by gene atzA responsible for the production of atrazine chlorohydrolase [Citation80]. An engineered Escherichia coli with atrazine chlorohydrolase was proven to be successful in remediating soil polluted by atrazine in field-scale studies [Citation81]. In a similar study, gene tpd encoding for triazophos hydrolase obtained from Ochrobactrum sp. mp-4 was expressed in Pseudomonas putida KT2440 for degrading pesticides belonging to the organophosphorus group and other aromatic hydrocarbons [Citation82]. Hexachlorocyclohexane (HCH) and methyl parathion degradation was made efficient by expressing methyl parathion hydrolase gene (mpd) in a Sphingomonas sp. BHC-A [Citation83,Citation84].
For indigenous microbes, it may be harder to degrade a mixture of pesticides, and engineered microorganisms open a new possibility for the same. When a mixture of OP and OC was present, linA and mpd genes responsible for organochlorine and organophosphate degradation were expressed in E. coli that was more effective for simultaneous degradation of both pesticides [Citation85]. For expressing these novel genes for pollution control, protoplast fusion seems to be an ideal choice except for the related genes over expressions [Citation86].Organophosphate pesticides in soil are also degraded by organophosphorus hydrolase encoded by the OPH gene. Most of the enzyme secretions by microbes are intracellular and have low substrate diffusivity; hence, they are not efficient in the remediation of soil contaminated with pesticides. An engineered E. coli with the OPH gene that secretes OPH protein into the periplasm and with increased activity of 1.8 fold was more suitable for remediation of soil [Citation87].
4.3. Hydrocarbon degradation
Apart from other pollutants, oil pollution has become another major concern around the globe [Citation88]. Although it has a major impact on the marine environment, oil pollution of inland water and soil is also occurring due to spills during transportation. The severity and toxicity of the oil contamination may depend on the degree of spillage and exposure of other organisms [Citation89]. This oil spillage also damages soil and vegetation and, hence, needs to be cleaned up. Biological methods are advantageous when considering the soil sustainability, and they help in efficient soil restoration. Many indigenous strains from oil-contaminated sites with the ability to degrade these hydrocarbons were reported [Citation90,Citation91]. Since the oil is a complex mixture of hydrocarbons, genetically modified microorganisms are efficient in remediating these contaminated sites than indigenous strains. Superbug development by plasmids containing multiple genes with degrading enzymes may be introduced in an organism. An engineered Acinetobacter baumannii S30 pJES with the high efficiency to degrade total petroleum hydrocarbon (TPH) was developed with a reporter lux gene that allows bioremediation site monitoring [Citation92]. Similarly, Streptomyces coelicolor M145 was engineered to enhance the efficiency of n-hexadecane degradation by overexpressing alkB gene encoding for the enzyme alkane monooxygenase [Citation93].
In another study, Acinetobacter sp. BS3 was developed with insertion of xylE gene encoding for catechol 2,3-dioxygenase enzyme from Pseudomonas putida strain BNF1 responsible for biodegradation of hydrocarbons, which are aromatic in nature. This engineered strain expressed enzyme with broad substrate specificity, hence exhibiting a superior efficacy to degrade a variety of n-alkanes and other aromatic hydrocarbons when compared to its wild strain [Citation94]. Engineered psychrophilic recombinant Antarctic Pseudoalteromonas haloplanktis TAC125 successfully expressed toluene-o-xylene monooxygenase (capable of degrading a wide range of aromatics) along with its inherent laccase-like protein to address the remediation of cold and marine xenobiotic loaded effluents [Citation95]. Such solutions will enable the remediation of aromatics even in cold climate regions whenever necessary.
Another major threat is heterocyclic aromatic compounds (HACs) and polycyclic aromatic hydrocarbons (PAHs), which are essential raw materials in drug and pesticides manufacturing [Citation96]. These compounds are highly toxic, carcinogenic, and mutagenic to humans and for other living beings [Citation97,Citation98]. Bacteria belonging to Sphingobium and Sphingomonas genera were found to be efficient in biodegradation of such toxic compounds [Citation99]. Strains of these genera were also capable of degrading an array of hydrocarbon compounds like acridine, carbazole, dioxins, fluorene, m-xylene, phenanthrene, HCH, pentachlorophenol (PCP), etc., which are aromatic in nature [Citation96]. Genomes of about twenty-six bacteria of the genera Sphingobium and Sphingomonas were revealed [Citation96]. A bph and xyl gene cluster was identified in six strains with PAH-degrading ability and the major metabolic pathways involved were identified as homogentisate and β-ketoadipate pathways. A recombinant strain F14 was developed by combining a phenanthrene-degrading strain (Sphingomonas sp.) GY2B and a pyrene-degrading strain Pseudomonas sp. GP3A [Citation84]. Similarly, a recombinant P. putida strain ΔfadBA-phaZ was developed by overexpressing poly-3-hydroxy-n-phenylalkanoate (PHPhA) depolymerase encoding phaZ gene that helps in the degradation of different n-phenylalkanoic acids [Citation100].
5. Challenges of genetically engineered microbes for in situ applications
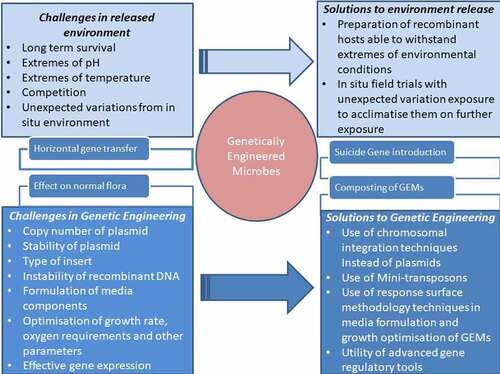
Ecological risk assessment is a crucial process in assessing the impact of microbial consortium or genetically modified microbe application in the field and thereby affecting indigenous soil microbiome [Citation101,Citation102]. Although the engineered microbes are efficient for bioaugmentation, their establishment and stable growth in the environment are quite difficult as they need to compete with the indigenous microorganisms [Citation103]. . depicts challenges and possible solutions in bioengineering microbes to remediate pollutants.
Moreover, there are many factors including copy number, growth rate, type of insert, oxygen availability, medium components, and environmental conditions influencing the stability of the recombinant plasmid. shows a comprehensive description of various challenges in the utility of GMOs in remediation of xenobiotics in the field.
Table 3. List of advantages and challenges associated with bioengineering microbes and their utility in bioremediation of xenobiotics
The use of bioengineered microbes in bioremediation faces a great challenge as many of them often fail to be effective in the natural environment for long term and combat the extremes of pH, salinity, temperature, etc. [Citation104]. The effective expression of biodegradatory genes would require their linking with chromosomal genetic elements rather than plasmids in many cases, but their effectiveness also needs to be verified [Citation105]. The instability of plasmids is a challenge in developing genetically modified microbes for bioremediation, and this can be overcome with the use of minitransposons. These minitransposons have a stable integration of recombinant genes with host chromosomes. Nonantibiotic resistance selection is better preferred for these systems in order to prevent gene transfer into the environment. A recombinant P. putida was developed with enhanced toluene degradation using minitransposons possessing antibiotic resistance markers [Citation106,Citation107].
There is also the chance of horizontal gene transfers between GEMs (genetically engineered microbes) and native microbes. The concept of transfer of hydrocarbon-degrading catabolic genes from recombinant Escherichia coli to indigenous bacteria of hydrocarbon-contaminated soil by mating experiments has proven to be effective in the removal of hydrocarbons and in the absence of hydrocarbons that the recombinant plasmids lost by selective pressure [Citation108]. Horizontal gene transfers from engineered Pseudomonas putida UWC3 to indigenous bacteria resulted in enhanced 2,4-D removal [Citation109]. In a similar study, a recombinant P. putida transferred PCB genes to indigenous microbes and showed a rapid disappearance [Citation110].
Suicide gene can play a role in the controlled release of these GEMs, as they get activated in the absence of pollutants and kill the GEMs [Citation111,Citation112]. Another strategy to eliminate the risk of horizontal gene transfer is composting where GEMs are exposed to lower pH and high temperature above 90°C, resulting in cell lysis and release of DNA that minimizes the horizontal gene transfer between GEMs and native microbes [Citation113].
Introducing GMO into agricultural land may have some effects on its normal soil microbiome structure. However, it is the same effect that occurs when a new species is introduced to the soil, even when there is no difference between the wild-type and genetic engineered strain [Citation114–116]. In a study, genetically modified Pseudomonas fluorescens when employed for polychlorinated biphenyl degradation, no effect was observed on the bacterial community structure and function [Citation117]. Moreover, changes in the structure of the microbial community as a result of introducing GEMs are insignificant compared to changes brought by other biotic and abiotic factors.
6. Future perspective and conclusion
Advanced sequencing techniques help to get a better understanding of the microbial flora of the soil [Citation118–121]. This approach could unveil the unrecognized microbial population and made it utilizable for the benefit of mankind. Engineering the soil microbiome is of great potential in improving agriculture. There are a set of microorganisms identified as keystone taxa that are associated with healthy plants [Citation122]. These microbial communities play a crucial role in the process of plant-microbial interaction that determines plant growth and health [Citation123].
Engineering microbes by advanced gene-editing tools such as CRISPR- Cas 9 provides a cheap and easy method for xenobiotic remediation and plant growth promotion to restore soil health. The bottleneck to soil health restoration using genetically engineered microbes is the lower expression levels of proteins than confer properties of relevance such as toxic xenobiotic remediation, higher resistance and accumulation of heavy metals, and faster degradation of a diverse range of pesticides. The use of CRISPR Cas-based systems in phytoremediation and endophytic microbes in pesticide remediation has been critically discussed [Citation124,Citation125]. Although the utility of such advanced gene-editing tools such as CRISPR Cas system, Zinc Finger nucleases (ZFN), and transcriptional activator-like effector nucleases (TALEN) has recently gained much attention, research activities using these molecular tools still need to be explored in the direction of more toxic waste remediatory research [Citation126]. Moreover, strict regulations on the applicability and field trials of genetically modified microbes are yet another factor that should be addressed to evaluate the success rates of further research in this direction. The effect of pollutants such as micro- and nanoplastics on soil and water health also needs to be addressed [Citation127].
The problem of soil health restoration is quite essential in every polluted country as the agricultural productivity is a direct indicator of the self-sustainability of every growing economy. The increasing population and necessity of more resources demand more fertile lands that could support our food and recovery of polluted landforms, which can never be neglected.
Microbes play a crucial role in the formation of soil and its fertility and ability to detoxify xenobiotics and maintain soil health. They act as double headed swords that can remove or detoxify pollutants from the soil and nourish the soil with minerals, metabolites, and growth regulatory compounds to enhance plant growth. Although microbes serve as efficient agents of soil remediation, the complexity in natural environments, selective detoxification of each type of pollutant, toxicity induced by high concentrations of pollutants, and optimization of xenobiotic remediation are some limiting factors in its enhanced applications to some extent. However, to speed up the process of soil health restoration and to tackle a wide variety of pollutants, the utility of genetically engineered microbes needs to be tried. The adoption of more reliable genetic tools, which would cause the least damage or no damage to the ecosystem, should be thus encouraged. Moreover, stable expression of biodegradatory chromosomally associated genetic elements instead of plasmids becomes essential for attaining the targeted remediation effects in long run. In such a scenario, combining the bioremediatory abilities of microbes along with their ability to enhance soil fertility will be promising to the sustainable development of soil.
Acknowledgements
Raveendran Sindhu acknowledges the Department of Science and Technology for sanctioning a project under the DST WOS-B scheme.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- OECD, 2014. Environmental quality of life.
- Jian J, Du X, Stewart RD. A database for global soil health assessment. Sci Data. 2020;7(1):16.
- Samaddar S, Karp DS, Schmidt R, et al. Role of soil in the regulation of human and plant pathogens: soils’ contributions to people. Philos Trans Royal Soc B. 2021;376(1834):20200179.
- Cardoso EJBN, Vasconcellos RLF, Bini D, et al. Soil health: looking for suitable indicators. What should be considered to assess the effects of use and management on soil health? Scientia Agricola. 2013;70(4):274–289.
- Fernandez IJ, Rustad LE, Norton SA, et al. Experimental acidification causes soil base‐cation depletion at the bear brook watershed in maine. Soil Sci Soc Am J. 2003;67(6):1909–1919.
- Hashim G, Coughlan K, Syers J, et al. On-site nutrient depletion: an effect and a cause of soil erosion. In Penning de Vries, F W T, Agus, F, and Kerr, J M(eds)., Soil erosion at multiple scales: principles and methods for assessing causes and impacts, (New York). 1998. p. 207–221.
- Maximillian J, Brusseau, ML, Glenn, EP, et al. Chapter 25 - pollution and environmental perturbations in the global system. In: Brusseau ML, Pepper IL, and Gerba CP, editors. Environmental and pollution science. Third. Amsterdam: Academic Press, 457–476, 2019.
- Parikh SJ, James BR. Soil: the foundation of agriculture. Nat Educ Knowledge. 2012;3(10):2.
- Maroun W, Atkins J. A practical application of accounting for biodiversity: the case of soil health. Social and Environmental Accountability Journal. 2021;41(1–2):37–65
- Rejesus RM, Aglasan S, Knight LG, et al. Economic dimensions of soil health practices that sequester carbon: promising research directions. J Soil Water Conserv. 2021;76(3):55A–60A.
- Brevik EC, Slaughter L, Singh BR, et al. Soil and human health: current status and future needs. Air Soil Water Res. 2020;13:1178622120934441.
- Girija Veni V, Srinivasarao, C, Sammi Reddy, K, et al. Chapter 26 - Soil health and climate change. In: Prasad MNV, and Pietrzykowski M, editors. climate change and soil interactions. San Diego, California: Elsevier; 2020. p. 751–767.
- Bunea A, Pacurar, I, Dulf, V et al, Study concerning the soil resources used in agriculture and forestry worldwide . ProEnvironment. 2020: 13; 39–43.
- Macdonald C, Singh B. Harnessing plant-microbe interactions for enhancing farm productivity. Bioengineered. 2014;5(1):5–9.
- Hernandez‐Raquet G, Durand E, Braun F, et al. Impact of microbial diversity depletion on xenobiotic degradation by sewage‐activated sludge. Environ Microbiol Rep. 2013;5(4):588–594.
- Bharadwaj A Bioremediation of xenobiotics: an eco-friendly cleanup approach. In Parmar, V S, Malhotra, P, and Mathur, D(eds).,: Green chemistry in environmental sustainability and chemical education. Singapore: Springer; 2018. p. 1–13.
- Rebello S, Sivaprasad MS, Anoopkumar AN, et al. Cleaner technologies to combat heavy metal toxicity. J Environ Manage. 2021;296:113231 doi:10.1016/j.jenvman.2021.113231.
- Rashid MI, Mujawar LH, Shahzad T, et al. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol Res. 2016;183:26–41.
- Rishad KS, Rebello S, Shabanamol PS, et al. Biocontrol potential of Halotolerant bacterial chitinase from high yielding novel Bacillus Pumilus MCB-7 autochthonous to mangrove ecosystem. Pestic Biochem Physiol. 2017;137:36–41.
- Nair AM, Rebello S, Rishad KS, et al. Biosurfactant facilitated biodegradation of quinalphos at high concentrations by Pseudomonas aeruginosa Q10. Soil Sediment Contaminat. 2015;24(5):542–553.
- Gunjal AB Role of plant growth–promoting rhizobacteria in degradation of xenobiotic compounds. In Verma, J P, Macdonald, C A, and Podile, A R. (eds)., New and future developments in microbial biotechnology and bioengineering. Amsterdam: Elsevier; 2021. p. 25–33.
- Cf SF, Rebello S, Mathachan Aneesh E, et al. Bioprospecting of gut microflora for plastic biodegradation. Bioengineered. 2021;12(1):1040–1053.
- Mishra S, Lin, Z, Pang, S, et al. Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front Bioeng Biotechnol. 2021;9(31);1–26.
- Docherty KM, Gutknecht JLM. Soil microbial restoration strategies for promoting climate-ready prairie ecosystems. Ecol Appl. 2019;29(3):e01858.
- Tejada M, Benítez C, Parrado J. Application of biostimulants in benzo (a) pyrene polluted soils: short-time effects on soil biochemical properties. Appl Soil Ecol. 2011;50:21–26.
- Finkel OM, Castrillo G, Herrera Paredes S, et al. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–163.
- Lakshmanan V, Selvaraj G, Bais HP. Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol. 2014;166(2):689–700.
- Kumar A, Dubey A. Rhizosphere microbiome: engineering bacterial competitiveness for enhancing crop production. J Adv Res. 2020;24:337–352.
- Lyu D, Msimbira LA, Nazari M, et al. The coevolution of plants and microbes underpins sustainable agriculture. Microorganisms. 2021;9(5):1036.
- Ke W, Zhang X, Zhu F, et al. Appropriate human intervention stimulates the development of microbial communities and soil formation at a long-term weathered bauxite residue disposal area. J Hazard Mater. 2021;405:124689.
- Basu S, Kumar , G, Chhabra, S et al, et al. Role of soil microbes in biogeochemical cycle for enhancing soil fertility. In Verma, J P, Macdonald, C A, and Podille, A R(eds)., New and future developments in microbial biotechnology and bioengineering. Amsterdam: Elsevier, 2021. 149–157.
- Nehal N, Rathore US, Sharma N Microbes and soil health for sustainable crop production. In Nath, Manoj, Bhatt, Deepesh, Bhargava, Prachi, and D.K., Choudhary(eds)., Microbial metatranscriptomics belowground, 1 . Switzerland: Springer. 2021. p. 581–613.
- Rogers C, Oldroyd GE. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot. 2014;65(8):1939–1946.
- Ryu M-H, Zhang J, Toth T, et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat Microbiol. 2020;5(2):314–330.
- Wu Q, Jiao S, Ma M, et al. Microbial fuel cell system: a promising technology for pollutant removal and environmental remediation. Environ Sci Pollut Res. 2020;27(7):6749–6764.
- Bhalerao TS, Puranik PR. Biodegradation of organochlorine pesticide, endosulfan, by a fungal soil isolate, Aspergillus Niger. Int Biodeterior Biodegrad. 2007;59(4):315–321.
- Malghani S, Chatterjee N, Yu HX, et al. Isolation and identification of profenofos degrading bacteria. Braz J Microbiol. 2009;40(4):893–900.
- Zhang X, Hao X, Huo S, et al. Isolation and identification of the Raoultella ornithinolytica-ZK4 degrading pyrethroid pesticides within soil sediment from an abandoned pesticide plant. Arch Microbiol. 2019;201(9):1207–1217.
- Kumari S, Mangwani N, Das S. Synergistic effect of quorum sensing genes in biofilm development and PAHs degradation by a marine bacterium. Bioengineered. 2016;7(3):205–211.
- Rebello S, Asok AK, Mundayoor S, et al. Surfactants: toxicity, remediation and green surfactants. Environ Chem Lett. 2014;12(2):275–287.
- Liu X, Germaine KJ, Ryan D, et al. Genetically modified Pseudomonas biosensing biodegraders to detect PCB and chlorobenzoate bioavailability and biodegradation in contaminated soils. Bioengineered Bugs. 2010;1(3):198–206.
- Vandana UK, Gulzar, A, Laskar, IH, et al. Role of microbes in bioremediation of radioactive waste. In Panpatte, D G, and Jhala, Y K(eds)., Microbial rejuvenation of polluted environment. Singapore: Springer, 2021. 329–352.
- Rebello S, Anoopkumar AN, Aneesh EM, et al. Hazardous minerals mining: challenges and solutions. J Hazard Mater. 2021;402:123474.
- Furukawa K. ‘Super bugs’ for bioremediation. Trends Biotechnol. 2003;21(5):187–190.
- Ortiz-hernández M, Sánchez-Salinas E. Biodegradation of the organophosphate pesticide tetrachlorvinphos by bacteria isolated from agricultural soils in México. Revista internacional de contaminación ambiental. 2010;26(1):27–38.
- Li J, Wu C, Chen S, et al. Enriching indigenous microbial consortia as a promising strategy for xenobiotics’ cleanup. J Clean Prod. 2020;261:121234.
- Qin Y, Druzhinina IS, Pan X, et al. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol Adv. 2016;34(7):1245–1259.
- Estrada B, Aroca R, Azcón-Aguilar C, et al. Importance of native arbuscular mycorrhizal inoculation in the halophyte Asteriscus maritimus for successful establishment and growth under saline conditions. Plant Soil. 2013;370(1):175–185.
- Armada E, Portela G, Roldán A, et al. Combined use of beneficial soil microorganism and agrowaste residue to cope with plant water limitation under semiarid conditions. Geoderma. 2014;232:640–648.
- Rani R, Kumar V, Usmani Z, et al. Influence of plant growth promoting rhizobacterial strains Paenibacillus sp. IITISM08, Bacillus sp. PRB77 and Bacillus sp. PRB101 using Helianthus annuus on degradation of endosulfan from contaminated soil. Chemosphere. 2019;225:479–489.
- Sundari SK, Prakash, A, Yadav, P, et al. Plant growth-promoting microbes as front-runners for on-site remediation of organophosphate pesticide residues in agriculture soils. In Arora, N K, and Kumar, N, Phyto and rhizo remediation. Singapore: Springer, 2019. 249–285.
- DalCorso G, Fasani E, Manara A, et al. Heavy metal pollutions: state of the art and innovation in phytoremediation. Int J Mol Sci. 2019;20(14):3412.
- Nathiya S, Janani R, Kannan VR. Potential of plant growth promoting Rhizobacteria to overcome the exposure of pesticide in Trigonella foenum-graecum (fenugreek leaves). Biocatal Agric Biotechnol. 2020;23:101493.
- Verbruggen E, Van Der HEIJDEN MARCELGA, Weedon JT, et al. Community assembly, species richness and nestedness of arbuscular mycorrhizal fungi in agricultural soils. Mol Ecol. 2012;21(10):2341–2353.
- Nemergut DR, Schmidt SK, Fukami T, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77(3):342–356.
- Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nature Rev Microbiol. 2014;12(4):263–273.
- Bello-Akinosho M, Makofane R, Adeleke R, et al. Potential of polycyclic aromatic hydrocarbon-degrading bacterial isolates to contribute to soil fertility. Biomed Res Int. 2016;2016:1–10.
- Rebello S, Joseph BV, Joseph SV, et al. Bioconversion of sodium dodecyl sulphate to rhamnolipids by transformed Escherichia coli DH5 α cells-a novel strategy for rhamnolipid synthesis. J Appl Microbiol. 2016;120(3):638–646.
- Begum N, Qin C, Ahanger MA, et al. Role of arbuscular mycorrhizal fungi in plant growth regulation: implications in abiotic stress tolerance. Front Plant Sci. 2019;10(1068). DOI:10.3389/fpls.2019.01068.
- Jan S, Singh R, Bhardwaj R, et al. Plant growth regulators: a sustainable approach to combat pesticide toxicity. 3 Biotech. 2020;10(11):466.
- Verma P, Rawat S Rhizoremediation of heavy metal-and xenobiotic-contaminated soil: an eco-friendly approach. In Shah, M P, Removal of emerging contaminants through microbial processes. Singapore: Springer; 2021. p. 95–113.
- Parewa HP, Meena, VS, Jain, LK, et al. Sustainable crop production and soil health management through plant growth-promoting rhizobacteria. In Meena, V J., Role of rhizospheric microbes in soil. Singapore: Springer, 2018. 299–329.
- Kumar G, Patel, JS, Maharshi, A, et al. 25 PGPR-mediated defence responses in plants under biotic and abiotic stresses. Advances in PGPR research. 2017; 427
- Hou J, Liu W, Wang B, et al. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere. 2015;138:592–598.
- Robas M, Jiménez PA, González D et al. Bio-mercury remediation suitability index: a novel proposal that compiles the PGPR features of bacterial strains and its potential use in phytoremediation. Int J Environ Res Public Health. 2021;18(8):4213.
- Asad SA Soil–PCB–PGPR interactions in changing climate scenarios. In Hashmi, M Z, Kumar, V, and Varma, A (eds)., Xenobiotics in the Soil Environment. Switzerland: Springer; 2017. p. 281–298.
- Turan M, Hashmi, M Z, Kumar, V, and Varma, A . Plant Growth Promoting Rhizobacteria’s (PGPRS) enzyme dynamics in soil remediation. In Larramendy, M L, and Soloneski, S (eds)., Soil contamination-current consequences and further solutions. Croatia: IntechOpen; 2016:209–231.
- Kaur J Kumar, Vivek, Prasad, Ram, and Kumar, Manoj. PGPR in management of soil toxicity. Rhizobiont in bioremediation of hazardous waste. Singapore: Springer; 2021. p. 317–344.
- Ajeng AA, Abdullah R, Malek MA, et al. The effects of biofertilizers on growth, soil fertility, and nutrients uptake of oil palm (Elaeis guineensis) under greenhouse conditions. Processes. 2020;8(12):1681.
- Chia WY, Chew KW, Le CF, et al. Sustainable utilization of biowaste compost for renewable energy and soil amendments. Environ Pollut. 2020;267:115662.
- Yuanfan H, Jin Z, Qing H, et al. Characterization of a fenpropathrin-degrading strain and construction of a genetically engineered microorganism for simultaneous degradation of methyl parathion and fenpropathrin. J Environ Manage. 2010;91(11):2295–2300.
- Kumar NM, Muthukumaran C, Sharmila G, et al. Genetically modified organisms and its impact on the enhancement of bioremediation. In Varjani, S J, Agarwal, A K, Gnansounou, E, and Gurunathan, B (eds)., Bioremediation, in bioremediation: applications for environmental protection and management. Singapore: Springer; 2018:53–76.
- Pant G, Garlapati D, Agrawal U, et al. Biological approaches practised using genetically engineered microbes for a sustainable environment: a review. J Hazard Mater. 2021;405:124631.
- Diep P, Mahadevan R, Yakunin AF. Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front Bioeng Biotechnol. 2018;6:157.
- Li C, Li Z, Li Y, et al. A Ferritin from Dendrorhynchus zhejiangensis with heavy metals detoxification activity. PLOS ONE. 2012;7(12):e51428.
- Misra CS, Sounderajan S, Apte SK. Metal removal by metallothionein and an acid phosphatase PhoN, surface-displayed on the cells of the extremophile, Deinococcus radiodurans. J Hazard Mater. 2021;419:126477.
- Ma L, Xu J, Chen N, et al. Microbial reduction fate of chromium (Cr) in aqueous solution by mixed bacterial consortium. Ecotoxicol Environ Saf. 2019;170:763–770.
- Mahbub KR, Krishnan K, Megharaj M, et al. Bioremediation potential of a highly mercury resistant bacterial strain Sphingobium SA2 isolated from contaminated soil. Chemosphere. 2016;144:330–337.
- Wang P, Sun G, Jia Y, et al. A review on completing arsenic biogeochemical cycle: microbial volatilization of arsines in environment. J Environ Sci. 2014;26(2):371–381.
- Neumann G, Teras R, Monson L, et al. Simultaneous degradation of atrazine and phenol by Pseudomonas sp. strain adp: effects of toxicity and adaptation. Appl Environ Microbiol. 2004;70(4):1907–1912.
- Strong LC, McTavish H, Sadowsky MJ, et al. Field‐scale remediation of atrazine‐contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ Microbiol. 2000;2(1):91–98.
- Zhang R, Cui Z, Zhang X, et al. Cloning of the organophosphorus pesticide hydrolase gene clusters of seven degradative bacteria isolated from a methyl parathion contaminated site and evidence of their horizontal gene transfer. Biodegradation. 2006;17(5):465–472.
- Zhongli C, Shunpeng L, Guoping F. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl Environ Microbiol. 2001;67(10):4922–4925.
- Lu J, Guo C, Li J, et al. A fusant of Sphingomonas sp. GY2B and Pseudomonas sp. GP3A with high capacity of degrading phenanthrene. World J Microbiol Biotechnol. 2013;29(9):1685–1694.
- Yang J, Liu R, Song W, et al. Construction of a genetically engineered microorganism that simultaneously degrades organochlorine and organophosphate pesticides. Appl Biochem Biotechnol. 2012;166(3):590–598.
- Dillon AJP, Camassola M, Henriques JAP, et al. Generation of recombinants strains to cellulases production by protoplast fusion between Penicillium echinulatum and Trichoderma harzianum. Enzyme Microb Technol. 2008;43(6):403–409.
- Kang DG, Choi SS, Cha HJ. Enhanced biodegradation of toxic organophosphate compounds using recombinant Escherichia coli with sec pathway‐driven periplasmic secretion of organophosphorus hydrolase. Biotechnol Prog. 2006;22(2):406–410.
- Carpenter A. Oil pollution in the North Sea: the impact of governance measures on oil pollution over several decades. Hydrobiologia. 2019;845(1):109–127.
- Ndimele PE. The political ecology of oil and gas activities in the Nigerian aquatic ecosystem. London, UK: Academic Press; 2017.
- Zheng C, Yu L, Huang L, et al. Investigation of a hydrocarbon-degrading strain, Rhodococcus ruber Z25, for the potential of microbial enhanced oil recovery. J Pet Sci Eng. 2012;81:49–56.
- Ismail S, Dadrasnia A. Biotechnological potential of Bacillus salmalaya 139SI: a novel strain for remediating water polluted with crude oil waste. PLoS One. 2015;10(4):e0120931.
- Mishra S, Sarma PM, Lal B. Crude oil degradation efficiency of a recombinant Acinetobacter baumannii strain and its survival in crude oil-contaminated soil microcosm. FEMS Microbiol Lett. 2004;235(2):323–331.
- Gallo G, Lo Piccolo L, Renzone G, et al. Differential proteomic analysis of an engineered Streptomyces coelicolor strain reveals metabolic pathways supporting growth on n-hexadecane. Appl Microbiol Biotechnol. 2012;94(5):1289–1301.
- Xie Y, Yu F, Wang Q, et al. Cloning of catechol 2, 3-dioxygenase gene and construction of a stable genetically engineered strain for degrading crude oil. Indian J Microbiol. 2014;54(1):59–64.
- Parrilli E, Papa R, Tutino ML, et al. Engineering of a psychrophilic bacterium for the bioremediation of aromatic compounds. Bioengineered Bugs. 2010;1(3):213–216.
- Zhao Q, Yue S, Bilal M, et al. Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. SciTotal Environ. 2017;609:1238–1247.
- Hu N-J, Huang P, Liu J-H, et al. Characterization and source apportionment of polycyclic aromatic hydrocarbons (PAHs) in sediments in the Yellow River Estuary, China. Environ Earth Sci. 2014;71(2):873–883.
- Nguyen TC, Loganathan P, Nguyen TV, et al. Polycyclic aromatic hydrocarbons in road-deposited sediments, water sediments, and soils in Sydney, Australia: comparisons of concentration distribution, sources and potential toxicity. Ecotoxicol Environ Saf. 2014;104:339–348.
- Aylward FO, McDonald BR, Adams SM, et al. Comparison of 26 sphingomonad genomes reveals diverse environmental adaptations and biodegradative capabilities. Appl Environ Microbiol. 2013;79(12):3724–3733.
- Sandoval Á, Arias-Barrau E, Bermejo F, et al. Production of 3-hydroxy-n-phenylalkanoic acids by a genetically engineered strain of Pseudomonas putida. Appl Microbiol Biotechnol. 2005;67(1):97–105.
- Hart MM, Antunes PM, Abbott LK. Unknown risks to soil biodiversity from commercial fungal inoculants. Nat Ecol Evol. 2017;1(4):1.
- Hart MM, Antunes PM, Chaudhary VB, et al. Fungal inoculants in the field: is the reward greater than the risk? Funct Ecol. 2018;32(1):126–135.
- Dechesne A, Pallud C, Bertolla F, et al. Impact of the microscale distribution of a Pseudomonas strain introduced into soil on potential contacts with indigenous bacteria. Appl Environ Microbiol. 2005;71(12):8123–8131.
- Hernández-Sánchez V, Wittich R-M. Possible reasons for past failures of genetic engineering techniques for creating novel, xenobiotics-degrading bacteria. Bioengineered. 2012;3(5):260–261.
- Gibson DG, Glass JI, Lartigue C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. science. 2010;329(5987):52–56.
- Sanchez-Romero JM, Diaz-Orejas R, De Lorenzo V. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl Environ Microbiol. 1998;64(10):4040–4046.
- Herrero M, De Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172(11):6557–6567.
- French KE, Zhou Z, Terry N. Horizontal ‘gene drives’ harness indigenous bacteria for bioremediation. Sci Rep. 2020;10(1):15091.
- Dejonghe W, Goris J, El Fantroussi S, et al. Effect of dissemination of 2, 4-dichlorophenoxyacetic acid (2, 4-D) degradation plasmids on 2, 4-D degradation and on bacterial community structure in two different soil horizons. Appl Environ Microbiol. 2000;66(8):3297–3304.
- Min MG, Kawabata Z, Ishii N, et al. Fate of a PCBS degrading recombinant pseudomonas putida AC30(PMFB2) and its effect on the densities of microbes in marine microcosms contaminated with PCBS. Int J Environ Stud. 1998;55(4):271–285.
- Pandey G, Paul D, Jain RK. Conceptualizing “suicidal genetically engineered microorganisms” for bioremediation applications. Biochem Biophys Res Commun. 2005;327(3):637–639.
- Paul D, Pandey G, Jain RK. Suicidal genetically engineered microorganisms for bioremediation: need and perspectives. Bioessays. 2005;27(5):563–573.
- Singh A, Billingsley K, Ward O. Composting: a potentially safe process for disposal of genetically modified organisms. Crit Rev Biotechnol. 2006;26(1):1–16.
- Amarger N. Genetically modified bacteria in agriculture. Biochimie. 2002;84(11):1061–1072.
- Viebahn M, Glandorf DCM, Ouwens TWM, et al. Repeated introduction of genetically modified pseudomonas putida WCS358r without intensified effects on the indigenous microflora of field-grown wheat. Appl Environ Microbiol. 2003;69(6):3110–3118.
- Johansen A, Olsson S. Using phospholipid fatty acid technique to study short-term effects of the biological control agent Pseudomonas fluorescens DR54 on the microbial microbiota in barley rhizosphere. Microb Ecol. 2005;49(2):272–281.
- De Cárcer DA, Martín M, Mackova M, et al. The introduction of genetically modified microorganisms designed for rhizoremediation induces changes on native bacteria in the rhizosphere but not in the surrounding soil. ISME J. 2007;1(3):215–223.
- Schweitzer JA, Bailey JK, Fischer DG, et al. Plant–soil–microorganism interactions: heritable relationship between plant genotype and associated soil microorganisms. Ecology. 2008;89(3):773–781.
- Panke-Buisse K, Poole AC, Goodrich JK, et al. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 2015;9(4):980–989.
- Mueller UG, Sachs JL. Engineering microbiomes to improve plant and animal health. Trends Microbiol. 2015;23(10):606–617.
- Sergaki C, Lagunas B, Lidbury I, et al. Challenges and approaches in microbiome research: from fundamental to applied. Front Plant Sci. 2018;9:1205.
- Banerjee S, Schlaeppi K, van der Heijden MG. Keystone taxa as drivers of microbiome structure and functioning. Nature Rev Microbiol. 2018;16(9):567–576.
- Ray P, Lakshmanan V, Labbé JL, et al. Microbe to microbiome: a paradigm shift in the application of microorganisms for sustainable agriculture. Front Microbiol. 2020;11:3323.
- Saxena P, Singh, NK, Harish,et al. 5 - Recent advances in phytoremediation using genome engineering CRISPR–Cas9 technology. In: Pandey VC, and Singh V, editors. Bioremediation of pollutants. Amsterdam: Elsevier; 2020. p. 125–141.
- Stein HP, Navajas-Pérez R, Aranda E, et al. Potential for CRISPR genetic engineering to increase xenobiotic degradation capacities in model fungi. In Prasad, Ram: Approaches in bioremediation. Switzerland: Springer; 2018. p. 61–78.
- Jaiswal S, Singh DK, Shukla P. Gene editing and systems biology tools for pesticide bioremediation: a review. Front Microbiol. 2019;10:87.
- Allouzi MMA, Tang DYY, Chew KW, et al. Micro (nano) plastic pollution: the ecological influence on soil-plant system and human health. SciTotal Environ. 2021;788:147815.
- Liu C, Lin H, Dong Y, et al. Identification and characterization of plant growth–promoting endophyte RE02 from Trifolium repens L. in mining smelter. Environ Sci Pollut Res. 2019;26(17):17236–17247.
- Ke T, Zhang J, Tao Y, et al. Individual and combined application of Cu-tolerant Bacillus spp. enhance the Cu phytoextraction efficiency of perennial ryegrass. Chemosphere. 2021;263:127952.
- He X, Xu M, Wei Q, et al. Promotion of growth and phytoextraction of cadmium and lead in Solanum nigrum L. mediated by plant-growth-promoting rhizobacteria. Ecotoxicol Environ Saf. 2020;205:111333.
- Abdelkrim S, Jebara SH, Saadani O, et al. In situ effects of Lathyrus sativus-PGPR to remediate and restore quality and fertility of Pb and Cd polluted soils. Ecotoxicol Environ Saf. 2020;192:110260.
- Tirry N, Kouchou A, El Omari B, et al. Improved chromium tolerance of Medicago sativa by plant growth-promoting rhizobacteria (PGPR). J Genet Eng Biotechnol. 2021;19(1):1–14.
- Jeyasundar PGSA, Ali A, Azeem M, et al. Green remediation of toxic metals contaminated mining soil using bacterial consortium and Brassica juncea. Environ Pollut. 2021;277:116789.
- Xu Z, Lei Y, Patel J. Bioremediation of soluble heavy metals with recombinant Caulobacter crescentus. Bioengineered Bugs. 2010;1(3):207–212.
- Kou S, Yang Z, Luo J, et al. Entirely recombinant protein-based hydrogels for selective heavy metal sequestration. Polym Chem. 2017;8(39):6158–6164.
- Si K, Ming T, Li Y, et al. Heavy metal detoxification by recombinant ferritin from Apostichopus japonicus. RSC Adv. 2017;7(66):41909–41918.
- Pazirandeh M, Chrisey LA, Mauro JM, et al. Expression of the Neurospora crassa metallothionein gene in Escherichia coli and its effect on heavy-metal uptake. Appl Microbiol Biotechnol. 1995;43(6):1112–1117.
- Sauge-Merle S, Lecomte-Pradines C, Carrier P, et al. Heavy metal accumulation by recombinant mammalian metallothionein within Escherichia coli protects against elevated metal exposure. Chemosphere. 2012;88(8):918–924.
- Bai F, Tian H. Recombinant Rhodococcus erythropolis expressing HAO and AMO genes promotes nitrogen and organic matter removal efficiency in the treatment of landfill leachate. Water and Environment Journal. 2021. doi:10.1111/wej.12743.