ABSTRACT
Circular RNAs (CircRNAs) gain importance as regulatory molecules in prostate cancer (PCa), but molecular mechanism of most circRNAs in pathogenesis of PCa remains to be studied. This study aimed to explore the role of hsa_circ_0030586 in PCa. Gene Expression Omnibus database (GSE77661) was used to screen out candidate circRNAs. Quantitative real-time PCR was used to verify the relative expressions of circRNAs, miRNAs, and genes in PCa cells. A CCK-8 assay was used to evaluate the cells’ proliferation. Transwell and wound healing assay were used to determine the cells’ migration and invasion. Western blotting and immunohistochemistry were used to detect the protein expression of PI3K/AKT signaling proteins and epithelial–mesenchymal transition (EMT) markers. Furthermore, a nude mice tumorigenesis experiment in vivo was conducted to determine the function of hsa_circ_0030586 on PCa. Our results showed that hsa_circ_0030586 is significantly upregulated in PCa cells (p < 0.05). Its circular structure was confirmed via agarose gel electrophoresis and Sanger sequencing. Interfering with hsa_circ_0030586 in PC3 cells inhibited cell proliferation, migration, and invasion and led to the significant upregulation of E-cadherin and the significant downregulation of p-AKT/AKT, IKKα, PIK3CB, and Twist (all p < 0.05). Conversely, the hsa_circ_003058 interference fragment combined with the transfection of a miR-145-3p inhibitor could reverse the above effects. In vivo tumorigenesis of the xenograft model confirmed that interfering with hsa_circ_0030586 suppressed tumor cell proliferation and inhibited PI3K-AKT signaling and EMT in PC3 cells. Hsa_circ_0030586 is significantly upregulated in PCa cells and may promote EMT via PI3K-AKT signaling.
Introduction
Prostate cancer (PCa) is one of the most common malignant tumors in males worldwide, especially the elderly. Meanwhile, its incidence and mortality rates are substantially high [Citation1]. About 1.6 million men are diagnosed with PCa each year [Citation2]. Age and heredity are the main risk factors associated with PCa development [Citation3]. It is a heterogeneous disease and can be indolent or aggressive [Citation2]. Although most patients are diagnosed with localized PCa and could be cured via surgery and radiotherapy, up to 30% of men will relapse within 5–10 years, and a rapid biochemical relapse will lead to specific deaths in 10% of PCa patients [Citation4]. The main PCa treatments are prostatectomy for primary PCa and androgen deprivation therapy for metastatic PCa. However, treatment options are limited for castration-resistant PCa (CRPC). Hence, it is important to discern who needs treatment to avoid unnecessary procedures that may lead to serious complications and affect the patients’ quality of life [Citation5]. Thus, an in-depth understanding of PCa’s pathogenesis and potential therapeutic targets is urgently required.
Circular RNAs (circRNAs) are noncoding RNAs with covalently closed loop structures. They play an important role in different biological progress by acting as micro-RNA (miRNA/miR) sponges [Citation6–8]. Recently, circRNAs have become an important regulatory molecule in PCa. For instance, the knockdown of circ_CCNB2 was shown to promote PCa radiosensitivity through autophagy repression via the miR-30b-5p/kinesin family member 18A axis [Citation9]. Downregulated hsa_circ_0000735 reduced tumor growth in PCa via sponging miR-7 and boosted PCa’s sensitivity to docetaxel [Citation10]. The circular RNA_LARP4 suppressed migration and invasion by upregulating forkhead box O3 alpha in PCa [Citation11]. However, the molecular mechanisms of most circRNAs in the pathogenesis, invasion, and metastasis of PCa remain unknown and need to be further studied [Citation12–14].
miRNAs are noncoding RNAs that are 20–25 nucleotides long and can regulate gene expression at the transcriptional or posttranscriptional level by binding to the 3′ untranslated regions [Citation15] of mRNA. miRNAs act as regulators in various physiological processes, including proliferation, tumorigenesis, and apoptosis. A previous study has shown that miR-145 could inhibit the proliferation and invasion of LNCaP cells and was involved in the regulation of lncRNAPCGEM1 [Citation16]. It has also been shown to affect circRNA expression in LNCaP cells [Citation17]. A recent study showed that circCOL1A1 promotes the progression of gastric cancer cells by sponging miR-145 [Citation18]. However, the role of miR-145-3p in PCa and its relationship with hsa_circ_003058 remain to be studied.
In this study, we intended to explore the biological role and molecular mechanism of hsa_circ_0030586 in PCa progression, which may provide new targets for the diagnosis and treatment of PCa.
Materials and Methods
Cell culture and transfection
Human PCa cell lines (RWPE-1, DU145, PC3, 22Rv1, LNCaP, PC-3 M-2B4, and PC-3 M-IE8) were purchased from the CellCook Company (Guangzhou, China). Cells were cultured in F-12 K medium supplemented with 10% fetal bovine serum (FBS), 100 units/mL of penicillin, and 100 μg/mL of streptomycin (Sigma-Aldrich), in 5% CO2 at 37°C. The circ_0030586 siRNA (5′-ATAGAAATCCAATAGGCATCA-3′) and siRNA negative control (siRNA NC: 5′-TGATGCCTATTGGATTTCTAT-3′) were synthesized by Shanghai GenePharma Co., Ltd. Additionally, the lentiviral of shRNA and shRNA negative control (shRNA-NC) was also purchased from GenePharma.
RNA sequencing and bioinformatics analysis
The total RNA of cells was isolated using Trizol reagent (Invitrogen, USA) following the manufacturer’s instructions. The quantity and quality of total RNA were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The sequencing library was constructed according to the protocol of the NEB Next Ultra™ RNA Library prep kit (Illumina, San Diego, USA). Next, the differentially expressed mRNAs were analyzed and screened using R software according to their log2 ratio (siRNA/NC), absolute value of ≥1, and FDR value of ≤0.001.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted by using the Database for Annotation, Visualization, and Integrated Discovery (http://david.abcc.ncifcrf.gov). The circRNA–miRNA–mRNA network was constructed and visualized using Cytoscape software.
Quantitative real-time PCR and real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen, USA) and reverse transcribed into complementary DNA following the manufacturer’s instructions in the PrimeScript RT reagent kit (TaKaRa, Dalian, China). Then, TB GreenPremix Ex Taq II (TaKaRa, Dalian, China) was used for real-time PCR on an ABI 7500 system (Applied Biosystems, Foster City, CA, USA). Relative RNA levels were calculated using the standard 2−ΔΔCt method, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was used as an internal standard. Additionally, gel electrophoresis and Sanger sequencing were conducted to identify the cyclization site. lists the primer sequences.
Table 1. Primers used in the study
CCK-8 assay
The proliferation ability of PC3 cells was tested using a cell counting kit-8 (CCK-8, Beyotime Biotech, Haimen, China). First, 3 × 103 cells were seeded per well in a 96-well plate. Then, 10 μL of the CCK-8 solution was added to each well. After incubation for 2 h at 37°C, absorbance at 450 nm was measured using a Spectra Max 250 spectrophotometer (ELx800, BioTek, Inc., IL, USA). The experiment was repeated three times.
Cell migration and invasion assays
Cell migration and invasion assays in the absence and presence of Matrigel were conducted using transwell chambers (Corning, NY, USA) according to the manufacturer’s instructions. PC3 cells were transfected with 200 pmol of si-circRNA or with negative controls. After 24 h, 200 μL of the cell suspension in serum-free medium was seeded into the upper chamber, and 700 μL of culture medium supplemented with 20% FBS was added to the lower compartment. After incubation for 24 h, cotton swabs were used to wipe the cells located on the upper surfaces of the chamber, and the cells located on the lower surfaces were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet for 30 min at room temperature, respectively. The stained cells were counted in six randomly selected fields under a microscope (Leica, Wetzlar, Germany).
Wound healing assay
PC3 cells were transfected with si-circRNA or negative controls for 24 h. Then, they were evenly inoculated into a 24-well cell culture plate. PC3 cells were trypsinized and then seeded into a 24-well plate. After the cells were completely attached, the medium was changed and mitomycin C was added to a final concentration of 1 μg/mL for 1 h to inhibit cell division. Then, a 200 μL pipette tip was used to scratch the bottom of the plate vertically against the edge of the lid. After washing three times with phosphate-buffered saline (PBS) to remove the dead cells, a basal medium was added, and cells were placed in an incubator to continue culturing. The samples were imaged under a microscope and recorded after 0, 24, and 48 h. This experiment was repeated three times.
Western blot analysis
For Western blot (WB) analysis, the total protein of PCa cells was extracted using a RIPA lysis buffer (#P0013J; Beyotime, Beijing, China) according to the protocol, and protein concentration was measured through the BCA method. A total of 30 μg of protein was loaded and separated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane. After blocking with 5% milk at room temperature for 2 h, primary antibodies targeting E-cadherin (dilution 1:5000; Proteintech, 20,874–1AP), Twist (dilution 1:1000; Proteintech, 25,465-1-AP), p-AKT (dilution 1:2000; CST, 4060s), AKT (dilution 1:2000; CST, 2920s), IKKα (dilution 1:10,000; Abcam, ab32041), PIK3CB (dilution 1:1000; Abcam, ab151549), and GAPDH (dilution 1:8000; Proteintech, 60,004-1-1 g) were incubated at 4°C overnight. HRP-conjugated horseradish peroxidase secondary antibodies were incubated at room temperature for 2 h. Finally, blots were developed with ECL substrates. Image J software (National Institutes of Health, Bethesda, MD, USA) was used for a semiquantitative analysis, and GAPDH was used as an internal control.
Immunohistochemistry
Immunohistochemistry (IHC) was conducted to assess the expression of epithelial–mesenchymal transition (EMT) markers (E-cadherin and Twist). The samples were blocked with a 5% BSA solution for 15 min at room temperature, and primary antibodies against E-cadherin (dilution 1:100; CST, 3195) and Twist (dilution 1:100; Proteintech, 25,465-1-AP) were incubated overnight at 4°C then washed with PBS and incubated with the secondary antibodies anti-rabbit IgG-HRP (boster, SV0002) and anti-mouse IgG-HRP (boster, SV0001). The DAB immunostaining kit (Boster Biological Technology) was subsequently used for color development, and the tissue sections were visualized using a DP-72 optical microscope (Olympus, Japan).
Construction of stable lentivirus cell line
The lentiviral expression vector was designed to constitutively downregulate hsa_circ_0030586 via the transfection of shRNA. A vector transfecting shNC was also generated to serve as a control. PC3 cells were infected with lentivirus according to the operation manual and plated on a 24-well plate for 24 h. After plating, the lentiviral suspension packaged by GemePharma Company with a green fluorescent protein reporter gene and puromycin resistance gene was added for infection according to the optimal multiplicity of infection of 100. Cells were then cultured with the infection-enhancing solution Polybrene to increase efficiency. After 48 h of infection, puromycin was added to screen for 3–5 days, and samples were collected for quantitative real-time PCR (qRT-PCR).
Tumor xenograft models
Animal studies were approved by the Laboratory Animal Ethics Committee of Guangzhou Forevergen Biotechnology Co., Ltd. Fourteen BALB/C mice (5–6 weeks old) were purchased from the Guangdong Medical Laboratory Animal Center and maintained in a specific pathogen-free facility. They were randomly divided into the shNC group (n = 7) and the shRNA group (n = 7). A 100 μL suspension containing 2 × 106 PC3-shNC or PC3-sh2 cells was injected into the back of nude mice. Tumor volume was monitored every 3 days. When the tumor size reached 800–1,000 mm3, the mice were sacrificed, followed by the measurement of tumor volumes and weights.
Statistical analysis
GraphPad_Prism_8 software was used for evaluating quantitative data in this study. Differences between groups were analyzed using the Student’s t-test. P values lower than 0.05 were considered to indicate statistically significant differences.
Results
In this study, we used the Gene Expression Omnibus (GEO) database (GSE77661), GEPIA database, and qRT-PCR to screen circRNAs that may be involved in the regulation of PCa progression. Hsa_circ_0030586 was found to be the most significantly upregulated circRNA. Then, we predicted its target genes involved in the PI3K-AKT signaling pathway. We found that hsa_circ_0030586 promotes EMT in PCa via PI3K-AKT signaling, likely by sponging hsa-miR-145-3p.
Expression of circRNAs in PCa cells
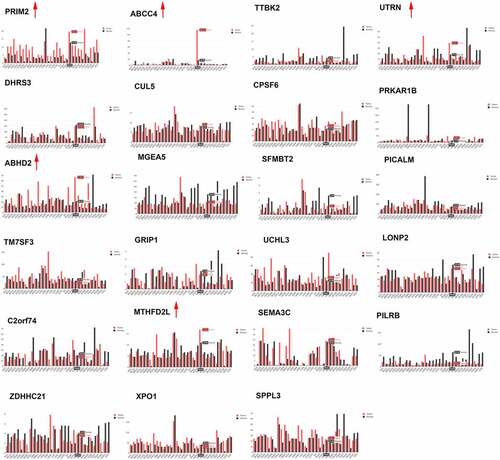
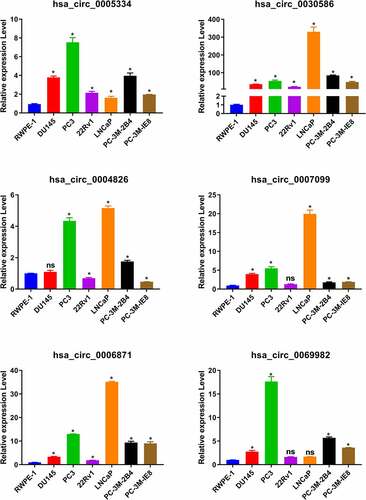
By the analysis of the dataset of GSE77661, we got 168 differentially expressed circRNAs between tumor and normal tissues (). Then we filter the differential expression circRNAs following the criterion: the length of circRNAs is between 300 and 3000 bp; the expression of circRNAs is upregulated in tumor than those in normal tissues. There 53 circRNA fit these criterion, and detail information was listed in . Furthermore, we searched the expression of parent gene of top 24 circRNAs from through GEPIA database. Results showed that the expression of five parent gene (PRIM2, ABCC4, UTRN, ABHD2, and MTHFD2L) was also significant increase in tumor compared to those in normal tissues (). The circRNAs corresponding to these parent genes include six circRNAs: hsa_circ_0005334, hsa_circ_0030586, hsa_circ_0004826, hsa_circ_0007099, hsa_circ_0006871, and hsa_circ_0069982). Afterward, qRT-PCR was used to detect these circRNAs expression in normal prostate epithelial cells RWPE-1 and PCa cells. Results indicated that the levels of hsa_circ_0005334, hsa_circ_0030586, and hsa_circ_0006871 were significantly upregulated in all PCa cells compared with those in normal prostate epithelial cells RWPE-1 (all p < 0.05; ). Among them, hsa_circ_0030586 had more greater difference between PCa cells and RWPE-1 than the others (). However, hsa_circ_004826, hsa_circ_0007099, and hsa_circ_0069982 expression in some PCa cells was not significant changes (). In addition, the function of hsa_circ_0030586 in PCa had no report, therefore hsa_circ_0030586 was selected for the following studies.
Table 2. The differentially expressed circular RNAs
Table 3. The upregulated differentially expressed circular RNAs (limite the length between 300 and 3000 bp)
Validation of the circular structure of hsa_circ_0030586
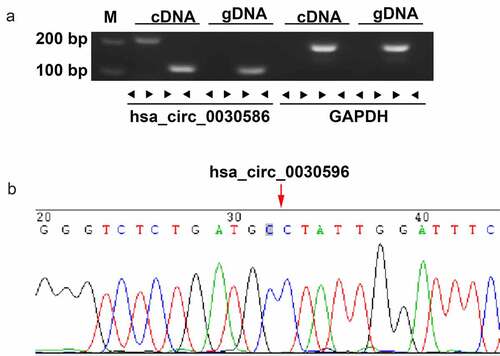
The circular structure of hsa_circ_0030586 was confirmed via agarose gel electrophoresis and Sanger sequencing. The results showed that hsa_circ_0030586 can only be amplified in cDNA by adopting divergent primers but not genomic DNA (). The sequence of the splice junction was further confirmed via Sanger sequencing (). These results convincingly showed that hsa_circ_0030586 can correctly form a ring structure.
Figure 3. Validation of the circular structure of hsa_circ_0030586. (a) Confirmation of the circular structure of hsa_circ_0030586 in PCa cells. RT-PCR was used to detect circular and linear cDNA using divergent and convergent primers, respectively. Primers targeting GAPDH and genomic DNA were considered as negative controls. (b) The junction site of hsa_circ_0030586 was confirmed via Sanger sequencing
Interfering of hsa_circ_0030586 suppressed the proliferation, migration, and invasion of PCa cells
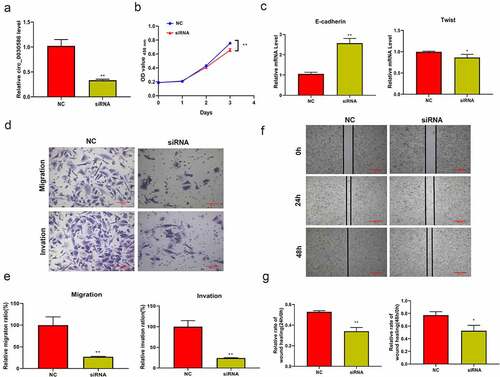
qRT-PCR results showed that transient interference fragments designed for hsa_circ_0030586 can significantly reduce its expression (p < 0.01; ). Furthermore, the CCK8 assay found that the inhibition of hsa_circ_0030586 expression can significantly inhibit cell viability at 72 h (p < 0.01; ). Moreover, qRT-PCR results showed that the EMT-related gene E-cadherin was significantly upregulated in the cells transfected with hsa_circ_0030586 siRNA compared to those transfected with siRNA NC (p < 0.01; ), whereas Twist was significantly downregulated (p < 0.05; ). The transwell assay and the wound healing assay showed that transfection with hsa_circ_0030586 siRNA can effectively inhibit migration (p < 0.01), invasion (p < 0.01), and the wound healing capacity (p < 0.05) of PC3 cells ()
Figure 4. Interfering with hsa_circ_0030586 suppressed the proliferation, migration, and invasion of PCa cells. (a) Transient interference fragments significantly reduced the expression of hsa_circ_0030586. (b) Transfection with hsa_circ_0030586 siRNA significantly inhibits cell viability at 72 h. (c) The EMT-related gene E-cadherin was significantly upregulated, and Twist was significantly downregulated in cells transfected with hsa_circ_0030586 siRNA. (d–e) Transfection with hsa_circ_0030586 siRNA significantly inhibited the migration and invasion of PC3 cells (scare bar: 100 μm). (f–g) Transfection with hsa_circ_0030586 siRNA significantly inhibited the wound healing capacity of PC3 cells (scare bar: 500 μm). *P < 0.05; ** P < 0.01
mRNA sequencing analysis
The scatter plot () showed 1,202 differentially expressed mRNAs between the NC and siRNA groups, among which 792 were upregulated and 410 were downregulated. KEGG analysis found that these differentially expressed mRNAs were mainly enriched in PI3K-AKT, cell cycle, p53 signaling, and other signaling pathways associated with tumor cell proliferation, apoptosis, and metastasis (). A heat map was drawn with 114 differentially expressed mRNAs involved in the 15 signaling pathways identified via KEGG analysis (). GO analysis showed that differentially expressed mRNAs were significantly enriched in various biological processes, including cellular processes, biological regulation, and binding (). Furthermore, based on the competing endogenous RNA (ceRNA) mechanism, the interactive networks were drawn with the miRNAs and target genes involved in the PI3K-AKT signaling pathway and their expression was consistent with that of hsa_circ_0030586 ().
Figure 5. mRNA sequencing analysis. (a) The scatter plot showed 1,202 differentially expressed mRNAs between the NC and siRNA groups, 792 were upregulated, and 410 were downregulated. (b) KEGG analysis showed that differentially expressed mRNAs are mainly enriched in PI3K-AKT, cell cycle, p53, and other signaling pathways associated with tumor cell proliferation, apoptosis, and metastasis. (c) Heat map showing the differentially expressed mRNAs between the NC and siRNA groups. (d) GO analysis showed the distribution of these differentially expressed genes on the BP (biological process), CC (cellular component), and MF (molecular function) rich GO Terms. (e) The circRNAs–miRNAs–mRNAs competing endogenous RNA (ceRNA) networks involved in the PI3K-AKT signaling pathway
Hsa_circ_0030586 activates the PI3K-AKT signaling pathway and promotes EMT
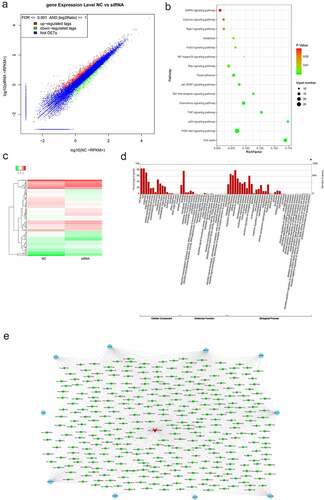
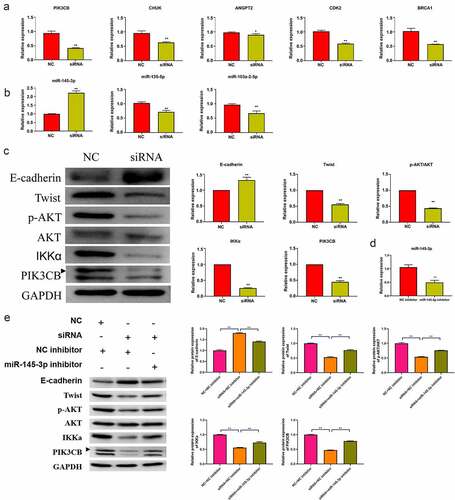
Five key molecules involved in the PI3K-AKT signaling pathway in the network were selected for qRT-PCR verification. The relative expressions of ANGPT2, CHUK, PIK3CB, CDK2, and BRCA1 were significantly downregulated in the hsa_circ_0030586 siRNA group compared to siRNA NC group (all p < 0.05; ). Among them, PIK3CB showed the highest downregulation folds, and it was an important upstream regulatory molecule of the PI3K-AKT signaling pathway (p < 0.01; ). Furthermore, the qRT-PCR verification of miRNA targeting PIK3CB was conducted. The miRNAs that were the binding sites of circRNA and miRNA > 1 that had been reported as having tumor suppressor effects in PCa [Citation17,Citation19–22] and were significantly negatively correlated with PIK3CB predicted by ENCORI (http://starbase.sysu.edu.cn/panMirCoExp.php#) were selected. Hsa-miR-145-3p, hsa-miR-135a-5p, and hsa-miR-103a-2-5p were screened out and verified. The results showed that the relative expression of hsa-miR-145-3p was significantly upregulated in the hsa_circ_0030586 siRNA group compared to siRNA NC group (p < 0.01), which was in line with our expectations (). Contrary to expectations, the relative expressions of hsa-miR-135a-5p and hsa-miR-103a-2-5p were significantly downregulated (p < 0.01; ). Meanwhile, a bioinformatics analysis conducted using miRanda version 3.3a also found that hsa-miR-145-3p can target and regulate BRCA1 and CDK2, which demonstrated that hsa-miR-145-3p played an important role in the process of hsa_circ_0030586, activating the PI3K-AKT signaling pathway. After interfering with hsa_circ_0030586, the expression of p-AKT, AKT, IKKα, and PIK3CB in the PI3K-AKT signaling pathway and the expression of the EMT markers E-cadherin and Twist were measured by WB. The relative protein expression of p-AKT/AKT, IKKα, PIK3CB, and Twist was significantly downregulated, whereas that of E-cadherin was significantly upregulated in siRNA group compared to those in siRNA NC group (all p < 0.01; ). The results indicated that interference with hsa_circ_0030586 can inhibit the PI3K-AKT signaling pathway and EMT in PCa cells. Furthermore, PC3 cells were transferred with an NC inhibitor and miR-145-3p inhibitor fragments for 48 h. The qRT-PCR results showed that miR-145-3p was significantly downregulated after transfection with miR-145-3p inhibitor than those in NC inhibitor group (p < 0.01; ). Moreover, the hsa_circ_0030586 interference fragment was transfected alone or combined with a miR-145-3p inhibitor to transfect into PC3 cells for 48 h. Transfecting an hsa_circ_0030586 interference fragment alone could downregulate the expression of p-AKT/AKT, IKKα, PIK3CB, and Twist and upregulate the expression of E-cadherin compared to the NC + NC inhibitor group (p < 0.01; ). It was found that the hsa_circ_0030586 interference fragment combined with miR-145-3p inhibitor transfection could reverse the effects caused by hsa_circ_003058 interfering (p < 0.01; ).
Figure 6. Hsa_circ_0030586 activates the PI3K-AKT signaling and promotes EMT. (a) The relative expression of five key molecules involved in the PI3K-AKT signaling pathway was detected by qRT-PCR. (b) The relative expression of three miRNAs targeting PIK3CB was detected by qRT-PCR. (c) The expression of E-cadherin, p-AKT, AKT, IKKα, PIK3CB, and Twist was tested via Western blot. GAPDH was used as an internal standard. (d) miR-145-3p expression was dectected in NC inhibitor and miR-145-3p inhibitor group; (e) The protein expression of E-cadherin, Twist, p-AKT, AKT, IKKα, and PIK3CB was tested via Western blot. GAPDH was used as an internal standard. *P < 0.05; ** P < 0.01
The tumor formation test in nude mice confirmed that hsa_circ_0030586 activates the PI3K-AKT signaling pathway and promotes EMT
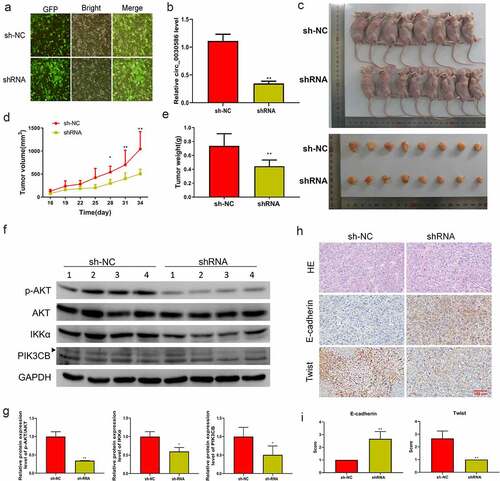
Male BALB/c nude mice were injected with PC3-shNC (shNC group) or PC3-sh (shRNA group) to induce tumorigenesis. A stable lentiviral cell line that interfered with hsa_circ_0030586 was constructed, and immunofluorescence staining showed that the fluorescence rate of the cells reached over 90% (). Furthermore, qRT-PCR results confirmed that the relative expression of hsa_circ_0030586 in the shRNA group was significantly downregulated compared with that in the shNC group (p < 0.01; ). The tumor volume and the body weight of mice in the shRNA group were significantly reduced compared with those in the shNC group (all p < 0.05; ). WB analysis showed that, after interfering with hsa_circ_0030586, the protein expression levels of p-AKT/AKT, IKKα, and PIK3CB were significantly decreased (all p < 0.05; ), indicating that the PI3K-AKT signaling pathway was significantly inhibited. The morphology of the tumor in two groups was observed via hematoxylin and eosin staining, which showed that the cancer cells of each group were of different sizes and shapes and arranged in nests, the nuclei were mostly oval, and there were necrotic areas of different sizes in the center of the cancer nests (). Moreover, IHC showed that E-cadherin expression was significantly upregulated and the expression of Twist was significantly downregulated after interfering with hsa_circ_0030586 (), further indicating that interference with hsa_circ_0030586 inhibited tumor EMT.
Figure 7. Interference with hsa_circ_0030586 inhibited tumorigenesis in nude mice. (a) Immunofluorescence staining shows the fluorescence rate of the cells. (b) Hsa_circ_0030586 expression in the shRNA and shNC groups was detected by qRT-PCR. (c) Nude mice and tumors injected with PC3-shRNA cells or PC3-shNC cells. (d, e) Tumor volumes (d) and weights (e) were calculated after nude mice were injected with PC3-shRNA cells or PC3-shNC cells. (f, g) p-AKT, AKT, IKKα, and PIK3CB protein expression was detected via Western blot. (h) HE and immunohistochemistry were used to detect the tumor morphology and expression of E-cadherin and Twist (scare bar: 100 μm). *p < 0.05; **p < 0.01
Discussion
Despite improvements in diagnostic and therapeutic treatments, several patients still developed advanced PCa at the time of diagnosis and missed the chance for in-time treatment because of a lack of effective early diagnostic markers [Citation23,Citation24]. Therefore, it is necessary to explore new early diagnostic molecular markers for PCa [Citation25,Citation26].
CircRNAs are potent regulators of various diseases. Recently, several circRNAs have been reported to regulate PCa development, mainly by acting as miRNA sponges and regulating target gene expression. Chen et al. identified 1,233 unique fusion genes and 76,311 distinct circRNAs from 144 localized prostate tumors via ultra-deep non-poly-A RNA-seq. In the differentially expressed circRNAs, the global burden of circRNAs was related to tumor aggressivity and 171 circRNAs were essential to PCa cell proliferation [Citation27]. Circ_0044516 was reported to play an important role in PCa cell survival and metastasis and may become a potential biomarker of PCa [Citation28]. It was reported that hsa_circ_0004870 is involved in the progression of enzalutamide resistance in CRPC [Citation29]. Feng et al. verified that circ0005276 interacted with FUS binding protein to activate the transcription of XIAP and promote proliferation and migration in PCa cells [Citation30]. circ_KATNAL1 was reported to play an anticancer role by regulating the miR-145-3p/WISP1 axis in PCa cells and may be the target for PCa diagnosis and treatment [Citation31]. These studies suggested that circRNAs play an important role in PCa development. Although plenty of circRNAs have been identified in PCa, some of them have not been thoroughly investigated yet, and their mechanism remains to be further studied.
In this study, hsa_circ_0030586 was significantly upregulated in PCa cells, and interfering with it suppressed the proliferation, migration, and invasion of PC3 cells. The in vivo study also proved that interfering with hsa_circ_0030586 significantly suppressed PCa cell growth. Our study suggested that hsa_circ_0030586 could promote cancer cell progression in PCa, which is consistent with previous studies arguing that circRNAs promote PCa progression [Citation32–35]. By contrast, circCRKL was lowly expressed in PCa tissues, and there was an upregulated expression of KLF5 by sponging miR-141 to inhibit PCa progression [Citation36]. Another study reported that circUCK2 expression was downregulated in enzalutamide-resistant (EnzR) cells, and circUCK2 overexpression inhibited the EnzR cell growth [Citation37]. This evidence suggested that circRNAs are abnormally expressed in PCa, functioning through various mechanisms.
CircRNAs act as miRNA sponges to control their expression and affect downstream target gene function [Citation38,Citation39]. For example, CircHIPK3 acts as a miR-338-3p sponge and promotes the expression of Cdc25B and Cdc2 to increase the proliferation ability of PCa [Citation40]. CircFMN2 acts as a miR-1238 sponge to promote LIM-homeobox Gene 2 expression, thereby regulating PCa progression [Citation41]. In this study, through a ceRNA mechanism, five key molecules involved in the PI3K-AKT signaling pathway were selected for qRT-PCR validation. We found that the relative expressions of ANGPT2, CHUK, PIK3CB, CDK2, and BRCA1 were significantly downregulated after interfering with hsa_circ_0030586. Among them, PIK3CB showed the highest downregulation, and it is an important upstream regulatory molecule of the PI3K-AKT signaling pathway. The PIK3CB gene was found to be involved in the progress of PCa in cell culture and nude mouse xenograft models [Citation42]. Additionally, elevated PI3K activity is one of the major mechanisms for many types of human cancers, including PCa [Citation43,Citation44]. It has been reported that the long noncoding DANCR promotes PCa via the FAK/PI3K/AKT/GSK3β/snail axis by targeting miR-185-5p [Citation45]. Another study revealed that lncRNA-SNHG1 regulates Wnt/β-catenin and the PI3K/AKT/mTOR signaling axis to affect proliferation, apoptosis, and autophagy in PCa cells [Citation46]. These studies supported our speculation that hsa_circ_0030586 may be regulating PCa progression via PI3K/AKT signaling. Furthermore, the qRT-PCR verification of miRNA targeting PIK3CB was conducted, and the relative expression of hsa-miR-145-3p was significantly upregulated after interfering with hsa_circ_0030586, which was in agreement with a ceRNA mechanism. Furthermore, the hsa_circ_003058 interference fragment combined with miR-145-3p. The inhibition of transfection could reverse the effects caused by hsa_circ_003058 interference, which demonstrated that the effects of hsa_circ_003058 on the PI3K-AKT pathway and EMT are dependent on miR-145-3p. The role of miR-145-3p in PCa has been largely reported. For example, miR-145-3p was found to be closely related to PCa and to be a target of metadherin (MTDH) [Citation19], miR-145-3p may regulate the expression of hsa_circRNA_008068 and hsa_circRNA_406557 in LNCaP cells [Citation17], and miR-145-5p and miR-145-3p were downregulated in CRPC [Citation20]. For further validation, the expression of key proteins p-AKT, AKT, IKKα, and PIK3CB in the PI3K-AKT signaling pathway and the expression of EMT markers E-cadherin and Twist were detected by WB. It was found that the relative protein expression of p-AKT/AKT, IKKα, PIK3CB, and Twist was significantly downregulated, whereas E-cadherin was significantly upregulated after interfering with hsa_circ_0030586. The above results were verified by both in vitro and in vivo studies. These results indicated that interfering with hsa_circ_0030586 may inhibit EMT via PI3K-AKT signaling in PCa cells.
However, studies on clinical tissue specimens were not reported in this article. In further studies, we will conduct an in-depth investigation of hsa_circ_0030586 using clinical specimens. Additionally, more studies are needed to explore the function and potential mechanism of downregulated circRNAs in GSE77661.
Conclusions
In conclusion, hsa_circ_0030586 is highly expressed in PCa cells and may sponge miR-145-3p to promote EMT via PI3K-AKT signaling. It is suggested that hsa_circ_0030586 can be used as a molecular marker for PCa, helping its early diagnosis and treatment.
Authors’ contributions
Z-J Y conceived and designed the study and critically revised the manuscript. G-C L conducted the experiments, analyzed the data, and drafted the manuscript. LC and JF participated in study design, study implementation, and manuscript revision. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kimura T, Egawa S. Epidemiology of prostate cancer in Asian countries. Int J Urol. 2018;25(6):524–531.
- Pernar CH, Ebot EM, Wilson KM, et al. The epidemiology of prostate cancer. Cold Spring Harb Perspect Med. 2018;8(12):a030361.
- Sartor O, de Bono JS, Longo DL. Metastatic prostate cancer. N Engl J Med. 2018;378(7):645–657.
- May EJ, Viers LD, Viers BR, et al. Prostate cancer post-treatment follow-up and recurrence evaluation. Abdom Radiol (NY). 2016;41(5):862–876.
- Evans AJ. Treatment effects in prostate cancer. Mod Pathol. 2018;31(S1):S110–121.
- Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005.
- Liu L, Wang J, Khanabdali R, et al. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14(12):1715–1721.
- Shabaninejad Z, Vafadar A, Movahedpour A, et al. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12(1):84.
- Cai F, Li J, Zhang J, et al. Knockdown of circ_CCNB2 sensitizes prostate cancer to radiation through repressing autophagy by the miR-30b-5p/KIF18A axis. Cancer Biother Radiopharm. 2020. DOI:10.1089/cbr.2019.3538
- Gao Y, Liu J, Huan J, et al. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020;20:334.
- Weng XD, Yan T, Liu CL. Circular RNA_LARP4 inhibits cell migration and invasion of prostate cancer by targeting FOXO3A. Eur Rev Med Pharmacol Sci. 2020;24(10):5303–5309.
- Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881.e813.
- Kong Z, Wan X, Lu Y, et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J Cell Mol Med. 2020;24(1):799–813.
- Shi J, Liu C, Chen C, et al. Circular RNA circMBOAT2 promotes prostate cancer progression via a miR-1271-5p/mTOR axis. Aging (Albany NY). 2020;12(13):13255–13280.
- Matin F, Jeet V, Moya L, et al. A plasma biomarker panel of four microRNAs for the diagnosis of prostate cancer. Sci Rep. 2018;8(1):6653.
- He JH, Zhang JZ, Han ZP, et al. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J Exp Clin Cancer Res. 2014;33(1):72.
- He JH, Han ZP, Zhou JB, et al. MiR-145 affected the circular RNA expression in prostate cancer LNCaP cells. J Cell Biochem. 2018;119(11):9168–9177.
- Ma Y, Ren Y, and Wen H, et al. CircCOL1A1 promotes the progression of gastric cancer cells through sponging miR-145 to enhance RABL3 expression. J ImmunolRes. 2021;2021:6724854.
- Pan D, Jia Z, Li W, et al. The targeting of MTDH by miR‑145‑5p or miR‑145‑3p is associated with prognosis and regulates the growth and metastasis of prostate cancer cells. Int J Oncol. 2019;54(6):1955–1968.
- Goto Y, Kurozumi A, Arai T, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117(3):409–420.
- Coarfa C, Fiskus W, Eedunuri VK, et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene. 2016;35(18):2345–2356.
- Chen W, Yao G, and Zhou K. MiR-103a-2-5p/miR-30c-1-3p inhibits the progression of prostate cancer resistance to androgen ablation therapy via targeting androgen receptor variant 7. J CellBiochem. 2019;120(8):14055–14064.
- Lu YT, Delijani K, Mecum A, et al. Current status of liquid biopsies for the detection and management of prostate cancer. Cancer Manag Res. 2019;11:5271–5291.
- Bozkurt S, Paul R, Coquet J, et al. Phenotyping severity of patient-centered outcomes using clinical notes: a prostate cancer use case. Learn Health Syst. 2020;4(4):e10237.
- Lachance J. Beyond stamp collecting: evolutionary and functional genomics advance our understanding of cancer biology. Cancer Res. 2021;81(7):1637–1638.
- Mokou M, Frantzi M, Mischak H, et al. Developing novel drug candidates and repurposed drugs for prostate cancer based on molecular profiles. Curr Med Chem. 2021;28. DOI:10.2174/0929867328666210525162730
- Chen S, Huang V, Xu X, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4):831–843.e822.
- Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121(3):2118–2126.
- Greene J, Baird AM, Casey O, et al. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep. 2019;9(1):10739.
- Feng Y, Yang Y, Zhao X, et al. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10(11):792.
- Zheng Y, Chen CJ, Lin ZY, et al. Circ_KATNAL1 regulates prostate cancer cell growth and invasiveness through the miR-145-3p/WISP1 pathway. Biochem Cell Biol. 2020;98(3):396–404.
- Wang P, Zhang L, Yin S, et al. Hsa_circ_0062019 promotes the proliferation, migration, and invasion of prostate cancer cells via the miR-195-5p/HMGA2 axis. Acta Biochim Biophys Sin (Shanghai). 2021;53:815–822.
- Li X, Azhati B, Wang W, et al. Circular RNA UBAP2 promotes the proliferation of prostate cancer cells via the miR-1244/MAP3K2 axis. Oncol Lett. 2021;21(6):486.
- Chen W, Cen S, Zhou X, et al. Circular RNA circNOLC1, upregulated by NF-KappaB, promotes the progression of prostate cancer via miR-647/PAQR4 axis. Front Cell Dev Biol. 2020;8:624764.
- Sha J, Xia L, Han Q, et al. Downregulation of circ-TRPS1 suppressed prostatic cancer prognoses by regulating miR-124-3p/EZH2 axis-mediated stemness. Am J Cancer Res. 2020;10(12):4372–4385.
- Nan C, Wang Y, Yang S, et al. CircCRKL suppresses the progression of prostate cancer cells by regulating the miR-141/KLF5 axis. Pathol Res Pract. 2020;216(11):153182.
- Xiang Z, Xu C, Wu G, et al. CircRNA-UCK2 increased TET1 inhibits proliferation and invasion of prostate cancer cells via sponge MiRNA-767-5p. Open Med (Wars). 2019;14:833–842.
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134.
- Gao Y, Wang J, Zheng Y, et al. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. 2016;7:12060.
- Liu F, Fan Y, Ou L, et al. CircHIPK3 facilitates the G2/M transition in prostate cancer cells by sponging miR-338-3p. Onco Targets Ther. 2020;13:4545–4558.
- Shan G, Shao B, Liu Q, et al. CircFMN2 sponges miR-1238 to promote the expression of LIM-Homeobox Gene 2 in prostate cancer cells. Mol Ther Nucleic Acids. 2020;21:133–146.
- Zhu Q, Youn H, Tang J, et al. Phosphoinositide 3-OH kinase p85alpha and p110beta are essential for androgen receptor transactivation and tumor progression in prostate cancers. Oncogene. 2008;27(33):4569–4579.
- Berenjeno IM, Guillermet-Guibert J, Pearce W, et al. Both p110α and p110β isoforms of PI3K can modulate the impact of loss-of-function of the PTEN tumour suppressor. Biochem J. 2012;442(1):151–159.
- He C, Duan S, Dong L, et al. Characterization of a novel p110β-specific inhibitor BL140 that overcomes MDV3100-resistance in castration-resistant prostate cancer cells. Prostate. 2017;77(11):1187–1198.
- Sun W, Zu S, Shao G, et al. Long non-coding DANCR targets miR-185-5p to upregulate LIM and SH3 protein 1 promoting prostate cancer via the FAK/PI3K/AKT/GSK3β/snail pathway. J Gene Med. 2021;23(7):e3344.
- Chen J, Wang F, Xu H, et al. Long non-coding RNA SNHG1 regulates the Wnt/β-Catenin and PI3K/AKT/mTOR signaling pathways via EZH2 to affect the proliferation, apoptosis, and autophagy of prostate cancer cell. Front Oncol. 2020;10:552907.