ABSTRACT
Long noncoding RNAs (lncRNAs) play critical roles in tumor progression regulation, including osteosarcoma. Evidence indicates that N6-methyladenosine (m6A) modification modulates mRNA stability to regulate osteosarcoma tumorigenesis. Here, present research aims to detect the roles of m6A-modified lncRNA FOXD2-AS1 in the osteosarcoma pathophysiological process. Clinical data unveiled that osteosarcoma patients with higher FOXD2-AS1 expression had a poorer overall survival rate compared to those with lower FOXD2-AS1 expression. Functional research illuminated that FOXD2-AS1 accelerated the migration, proliferation and tumor growth in vitro and in vivo. Mechanistically, a remarkable m6A-modified site was found on the 3ʹ-UTR of FOXD2-AS1, and m6A methyltransferase WTAP (Wilms’ tumor 1 associated protein) promoted the methylation modification, thus enhancing the stability of FOXD2-AS1 transcripts. Furthermore, FOXD2-AS1 interacted with downstream target FOXM1 mRNA through m6A sites, forming a FOXD2-AS1/m6A/FOXM1 complex to heighten FOXM1 mRNA stability. In conclusion, these findings propose a novel regulatory mechanism in which m6A-modified FOXD2-AS1 accelerates the osteosarcoma progression through m6A manner, which may provide new concepts for osteosarcoma tumorigenesis.
1. Introduction
Osteosarcoma is a primary malignancy of bone tissue, acting as one of the most common bone tumors [Citation1–3]. Given its high incidence, osteosarcoma is currently considered to be a substantial public health challenge across the globe in the last decades [Citation4]. The morbidity and mortality of osteosarcoma have brought about a huge social burden [Citation5]. Large epidemiological survey illustrates that the number of osteosarcoma patients are increasing in developed or developing countries [Citation6]. Although series of approaches have been used to treat osteosarcoma, the treatments are less effective as resistance developing. Therefore, it is critical to further explore the molecular mechanisms underlying the progression of osteosarcoma.
Long noncoding RNA (lncRNA) is a group of special RNA molecule longer than 200 nucleotides and without protein-coding potential [Citation7]. As an essential epigenetic regulator, lncRNA is involved in series of biological processes with diverse functions, such as binding with proteins, adsorbing microRNAs, recombining DNA. For example,
lncRNA SNHG10 up-regulates the FZD3 through sponging miR-182-5p and the SNHG10/miR-182-5p/FZD3 axis promotes the β-catenin transfer to nuclear accumulation to activate Wnt singling pathway [Citation8]. Therefore, these data illuminate that lncRNA significantly regulates osteosarcoma progression.
N [Citation6]-methyladenosine (m6A) has been identified to regulate the progression of various cancers via RNAs’ m6A modification [Citation9]. In osteosarcoma, m6A has been identified to regulate the biological characteristics through diverse mechanism [Citation10]. For example, m6A methyltransferase WTAP is involved in the metastasis and proliferation of osteosarcoma in vitro and vivo, and WTAP targets the 3ʹ-UTR of HMBOX1 mRNA to regulate the HMBOX1 stability depending on m6A modification [Citation11]. METTL3 knockdown decreases the m6A level of Drg1 mRNA levels, thereby reducing both the mRNA and protein levels of DRG1 in an m6A-dependent manner [Citation12]. METTL14 level is reduced in osteosarcoma cells compared with normal tissues and METTL14 overexpression significantly inhibited the proliferation, migration of osteosarcoma cells by activating caspase-3 [Citation13]. Moreover, m6A modification also induces the mRNA degradation in cancer. For instance, m6A-binding proteins YTHDF2 suppressed cell proliferation, tumor growth of hepatocellular carcinoma cells through directly bound the m6A modification site of EGFR 3ʹ-UTR to induce the degradation of EGFR mRNA [Citation14]. Another evidence is that, in prostate cancer, YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent manner to trigger AKT tumor progression pathway [Citation15]. These findings evince that m [Citation6]A modification exerts critical roles in the osteosarcoma.
Overall, our team tries to investigate the functions and its deep mechanism by which m [Citation6]A-modified lncRNA regulates the progression of osteosarcoma. Mechanistically, a remarkable m [Citation6]A-modified site was found on the 3ʹ-UTR of FOXD2-AS1, and m [Citation6]A methyltransferase WTAP (Wilms’ tumor 1 associated protein) installed the methylation modification, thus enhancing the stability of FOXD2-AS1 transcripts. Here, present research demonstrates a novel regulation by which WTAP-mediated lncRNA FOXD2-AS1 accelerates the osteosarcoma tumorigenesis via FOXD2-AS1/m [Citation6]A/FOXM1 complex.
2. Materials and Methods
2.1. Clinical samples collection
The clinical osteosarcoma tissues and matched corresponding adjacent normal tissue were collected during the surgery at Tianjin Hospital then immediately frozen in −80°C for RNA extraction. Survival times were recorded based on the date of surgery to the date of death or to the last follow-up. The collection of specimens and clinical research have been approved by the Ethics Committee of the Tianjin Hospital.
2.2. Cell lines and transfection
Osteosarcoma cell lines (MG63, Saos-2) and normal osteoblastic cell line (hFOB) were provided from Type Culture Collection Centre (Shanghai, China). Cells were cultured in RPMI-1640 medium containing 10% FBS, 1% streptomycin-penicillin antibiotic solution. For the overexpression of FOXD2-AS1, cDNAs were cloned into pCDH puro lentiviral vector and controls. For the silencing of FOXD2-AS1, the specific shRNA sequences were cloned into PLKO.1 vector and then transfected with packing and into 293TF cells with PAX2 plasmids. 48 h later, the lentivirus was collected and infected Saos-2 cells for 24 h. For the overexpression of WTAP, human WTAP cDNA (NM_001270531.2) were cloned into pCDH puro lentiviral vector (CD510B-1, System Biosciences). To screen the stable transfection, 1 μg/ml puromycin was used to treat the cells.
2.3. RNA extraction and qRT-PCR
RNA was extracted from the tissue and cells using TRIzol (Invitrogen). cDNA was synthesized with HiScript 1st Strand cDNA Synthesis Kit (Vazyme Biotech, China). For the quantification of mRNA and lncRNA, the cDNA was obtained by using PrimeScript RT reagent kit (Takara, Dalian, China). The real‐time PCR was performed by 7900 Real‐time PCR System using SYBR Premix Ex Taq (Takara). The primers using for quantitative RT-PCR analysis were listed in Table S1.
2.4. Western blot and antibodies
After transfection, cells were harvested and the total protein was extracted from cells using RIPA lysis buffer. The quality of protein was qualified using BCA detecting kit (Keygen, Nanjing, China). Sample was subjected SDS-PAGE (10%) and then transferred to PVDF microporous membrane (Millipore, Boston, MA, USA). Blots were probed by rabbit polyclonal antibodies anti-WTAP (1:1000, 195,380) at 4°C overnight. After that, membranes were incubated with secondary antibodies HRP-goat anti-rabbit (ABclonal) at room temperature. Protein bands were, respectively, detected on chemiluminescence imaging analysis system using Enhanced chemiluminescence (ECL, Millipore, Bredford, USA).
2.5. m6A quantitative analysis
Quantification of the m6A modification was performed using EpiQuik m6A RNA Methylation Quantification Kit (Epigentek, P-9005-48, colorimetric) following the manufacturer’s protocol [Citation16]. Total RNA was isolated from cells using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA (200 ng) was used to determine the percentage of m6A.The m6A level was colorimetrically quantified by reading the absorbance at a wavelength of 450 nm.
2.6. RNA stability
The stability of RNA isolated from osteosarcoma was detected using actinomycin D (1 μg/ml). In brief, RNA was extracted using Trizol reagent (Invitrogen) and then reversely transcripted using oligo (dT) primers. Finally, RNA level was measured using qRT-PCR.
2.7. Transwell migration assay
After transfection, cells (1 × 105) were suspended in serum-free medium and seeded into the upper chambers of transwell (8 mm pore size, Costar). In lower chamber, full-growth medium with FBS was added. At 37 C in 5% CO2, cells were incubated for 48 h and cells were removed with cotton swabs. The migrated cells were fixed with methanol with 0.1% crystal violet and counted in five randomly selected fields for each well using a light microscope (Olympus, Tokyo, Japan).
2.8. CCK-8 proliferation assay
The transfected cells were washed twice with PBS and subsequently seeded into a 96-well plate (1 × 103 cells in each well). After 24 h, cell counting kit-8 (CCK-8) reagent (10 mL/well) (Dojindo Laboratories, Kumamoto, Japan) was adminstered to the plate. After the mixed well, the cells, the optical density (OD) value of wells was detected at 450 nm using an automatic enzyme-mark reader (Multiskan, Thermo Fisher Scientific, Waltham, MA, USA).
2.9. m6A RNA immunoprecipitation PCR (MeRIP-PCR) assay
MeRIP-PCR was performed as previously described [Citation17]. Total RNA was extracted from cells with Trizol (Invitrogen). Anti-m [Citation6]A antibody (ab208577) was used to pull down the m6A-modified lncRNA or mRNA. Then, RNA (100 μg) was added to MeRIP buffer (150 mM NaCl, pH 7.5, 0.1% NP-40, 10 mM Tris-HCl) and then incubated with rabbit IgG (1 μl). Beads were then treated with Proteinase K (20 mg/ml) for 1 h at 42°C. After elution, m6A-combined RNA was extracted using Trizol and the RNA level was measured by qRT-PCR.
2.10. RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation (RIP) assay was performed using MagnaRIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions [Citation18]. RIP assay was performed using an anti-m [Citation6]A antibody (Abcam, ab208577, 1:1000) previously bound to magnetic Dynabeads in RIP Immunoprecipitation Buffer (Magna RIP Kit). Then, precipitated RNA was retrieved and then detected by qRT-PCR analysis.
2.11. In vivo
The in vivo mice assay had been approved by the Ethics Committee of the Tianjin Hospital. Approximately 5 × 106 MG63 cells (FOXD2-AS1 overexpression group, control group) were suspended in 100 μl PBS and then injected in the flank of BALB/c nude mice (4–5 weeks, male). During the3 weeks observation, the width and length were, respectively, recorded. The tumor size was calculated using formula = (width [Citation2] × length × 0.52). Finally, the mice were anesthetized and sacrificed for the neoplasm weighting.
2.12. Bioinformatics analysis
The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov) and GEPIA (http://gepia.cancer-pku.cn/) databases were used to compare the expression of WTAP and FOXM1, as well as the correlation within FOXM1 and FOXD2-AS1. Kaplan-Meier Plotter (http://kmplot.com/analysis/) was performed to calculate the survival.
2.13. Statistical analysis
Statistical analysis was carried out using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7.0 software. The interaction within FOXD2-AS1 and the osteosarcoma clinicopathologic feature of osteosarcoma patients was calculated using Chi-square test. Osteosarcoma survival was calculated using Kaplan–Meier method and log-rank test. Data was presented as mean ± SD. p less than 0.05 was considered as statistical significance.
3. Results
Clinically, high-expression of FOXD2-AS1 correlated with poorer overall survival rate. Functional research illuminated that FOXD2-AS1 accelerated the migration, proliferation and tumor growth in vitro and in vivo. Mechanistically, a remarkable m6A-modified site was found on the 3ʹ-UTR of FOXD2-AS1, and m6A methyltransferase WTAP promoted the methylation modification, thus enhancing the stability of FOXD2-AS1 transcripts. Furthermore, FOXD2-AS1 interacted with downstream target FOXM1 mRNA through m6A sites, forming a FOXD2-AS1/m [Citation6]A/FOXM1 complex to heighten FOXM1 mRNA stability. Overall, m [Citation6]A-modified FOXD2-AS1 accelerates the osteosarcoma progression through m [Citation6]A manner, which may provide new concepts for osteosarcoma tumorigenesis.
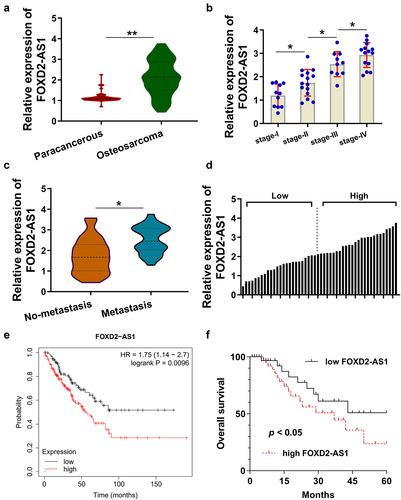
3.1. LncRNA FOXD2-AS1 was a poor prognosis-related RNA in osteosarcoma
In the enrolled osteosarcoma patients’ specimens (), RT-PCR analysis was performed and results indicated that FOXD2-AS1 expression up-regulated in the osteosarcoma tissues (). In other words, higher level of FOXD2-AS1 was observed in osteosarcoma tissues than those in paracancerous tissues. Moreover, FOXD2-AS1 expression was analyzed using PCR according to the tumor stage (I~ IV). The advanced stage osteosarcoma had higher FOXD2-AS1 expression (). Besides, FOXD2-AS1 expression up-regulated in the metastasis tissue specimens as compared to the non- metastasis (). The enrolled osteosarcoma specimens were divided into higher and lower cohorts based on the median value (). According to the TCGA dataset (http://gepia.cancer-pku.cn/), the large sample size analysis indicated that higher FOXD2-AS1 expression indicated the lower survival rate (). In the osteosarcoma cohort, the patients with higher FOXD2-AS1 expression illuminated the lower survival rate (). Collectively, these findings strongly suggested that lncRNA FOXD2-AS1 was a poor prognosis-related RNA in osteosarcoma.
Table 1. The relationship within FOXD2-AS1 and osteosarcoma patients’ clinicopathological characteristics
Figure 1. LncRNA FOXD2-AS1 was a poor prognosis-related RNA in osteosarcoma. (a) RT-PCR analysis illuminated the FOXD2-AS1 expression in the enrolled osteosarcoma patients’ specimens. (b) The FOXD2-AS1 expression was identified in the osteosarcoma tissues with osteosarcoma tissues tumor stage (I~ IV). (c) FOXD2-AS1 expression was analyzed in the metastasis tissue specimens and non-metastasis. (d) The enrolled osteosarcoma specimens were divided into higher and lower cohorts based on the median value. (e) TCGA dataset (http://gepia.cancer-pku.cn/) indicated the survival rate in patients with higher or lower FOXD2-AS1 expression. (f) Kaplan-Meier analysis of overall survival time of osteosarcoma patients according to the expression of FOXD2-AS1. **p < 0.01; *p < 0.05.
3.2. m6A methyltransferase WTAP enhanced the stability of FOXD2-AS1
In the osteosarcoma specimens, landscape data analysis (http://gepia.cancer-pku.cn/index) revealed that the level of WTAP increased in the osteosarcoma (). Moreover, the interaction analysis revealed that FOXD2-AS1 was positively correlated to the expression of WTAP (). In the osteosarcoma cell lines, m6A colorimetric quantitative analysis illuminated that m6A enrichment overtly up-regulated in the cells (). To investigate the role of WTAP on the FOXD2-AS1 transcript expression, the overexpression of WTAP was constructed in MG63 cell line (). RNA stability assay using Act D (5 μg/ml) discovered that WTAP overexpression enhanced the stability of FOXD2-AS1 transcript (). Bioinformatics online tools (http:// rna.sysu.edu.cn/rmbase/, http://rmvar.renlab.org/) inspired that there was a potential m6A modification site on the 3ʹ-UTR of FOXD2-AS1 (). The m6A motif of WTAP is GGACU, which was matched with the candidate m6A modification site of FOXD2-AS1 (). Once again, m6A colorimetric quantitative analysis revealed that WTAP promoted the m6A modification enrichment in MG63 cell (). MeRIP-qPCR demonstrated that WTAP overexpression promoted the binding within FOXD2-AS1 and m6A antibody (anti-m [Citation6]A) (). Taken together, these findings concluded that m6A methyltransferase WTAP enhanced the stability of FOXD2-AS1.
Figure 2. m6A methyltransferase WTAP enhanced the stability of FOXD2-AS1. (a) Online public dataset analysis (http://gepia.cancer-pku.cn/index) revealed the level of WTAP in osteosarcoma. (b) The interaction within FOXD2-AS1 expression and WTAP was analyzed using public dataset analysis (http://gepia.cancer-pku.cn/index). (c) m6A colorimetric quantitative analysis illuminated the m6A enrichment in the osteosarcoma cells. (d) Western blot analysis detected the WTAP protein level in MG63 cell transfected with WTAP overexpression. (e) RNA stability assay was performed in MG63 cell treated with Act D (5 μg/ml). The relative level of lncRNA FOXD2-AS1 was detected using RT-PCR. (f) Bioinformatics online tools (http:// rna.sysu.edu.cn/rmbase/, http://rmvar.renlab.org/) inspired the potential m6A modification site on the 3ʹ-UTR of FOXD2-AS1. (g) The candidate m6A modification site motif of FOXD2-AS1 is GGACU. (h) m6A colorimetric quantitative analysis revealed the m6A modification enrichment in MG63 cell with WTAP overexpression transfection. (i) MeRIP-qPCR demonstrated the FOXD2-AS1 level precipitated by m6A antibody (anti-m [Citation6]A). **p < 0.01; *p < 0.05.
![Figure 2. m6A methyltransferase WTAP enhanced the stability of FOXD2-AS1. (a) Online public dataset analysis (http://gepia.cancer-pku.cn/index) revealed the level of WTAP in osteosarcoma. (b) The interaction within FOXD2-AS1 expression and WTAP was analyzed using public dataset analysis (http://gepia.cancer-pku.cn/index). (c) m6A colorimetric quantitative analysis illuminated the m6A enrichment in the osteosarcoma cells. (d) Western blot analysis detected the WTAP protein level in MG63 cell transfected with WTAP overexpression. (e) RNA stability assay was performed in MG63 cell treated with Act D (5 μg/ml). The relative level of lncRNA FOXD2-AS1 was detected using RT-PCR. (f) Bioinformatics online tools (http:// rna.sysu.edu.cn/rmbase/, http://rmvar.renlab.org/) inspired the potential m6A modification site on the 3ʹ-UTR of FOXD2-AS1. (g) The candidate m6A modification site motif of FOXD2-AS1 is GGACU. (h) m6A colorimetric quantitative analysis revealed the m6A modification enrichment in MG63 cell with WTAP overexpression transfection. (i) MeRIP-qPCR demonstrated the FOXD2-AS1 level precipitated by m6A antibody (anti-m [Citation6]A). **p < 0.01; *p < 0.05.](/cms/asset/f90e2015-c275-43ed-a5b6-7190c104afe1/kbie_a_2008218_f0002_oc.jpg)
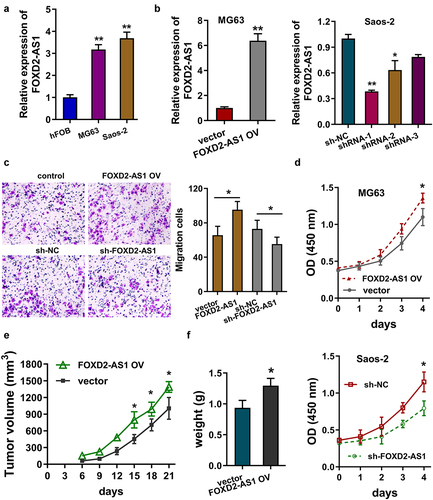
3.3. FOXD2-AS1 promoted the progression of osteosarcoma in vivo and vitro
In the osteosarcoma cells (MG63, Saos-2), FOXD2-AS1 expression up-regulated as compared to the normal osteoblastic cell line (hFOB) (). To explore the function of FOXD2-AS1 on osteosarcoma cells, the FOXD2-AS1 overexpression (FOXD2-AS1 OV) was transfected in MG63 cell and FOXD2-AS1 knockdown was transfected in Saos-2 cells (). Transwell migration assay illuminated that FOXD2-AS1 overexpression up-regulated the migrative cells for MG63 cells and FOXD2-AS1 knockdown reduced the migrative cells for Saos-2 cells (). Proliferative CCK-8 assay indicated that FOXD2-AS1 overexpression up-regulated the proliferative ability of MG63 cells and FOXD2-AS1 knockdown reduced the proliferative ability of Saos-2 cells (). In vivo animal assay, FOXD2-AS1 overexpression promoted the tumor growth in mice subcutaneous injection (). Collectively, these results indicated that FOXD2-AS1 promoted the progression of osteosarcoma in vivo and vitro.
Figure 3. FOXD2-AS1 promoted the progression of osteosarcoma in vivo and vitro. (a) RT-PCR indicated the FOXD2-AS1 expression in the normal osteoblastic cell line (hFOB) and osteosarcoma cells (MG63, Saos-2). (b) RT-qPCR analysis detected the FOXD2-AS1 level in the MG63 cells transfected with FOXD2-AS1 overexpression (FOXD2-AS1 OV) and in Saos-2 cells transfected with FOXD2-AS1 knockdown (sh-FOXD2-AS1). (c) Transwell migration assay illuminated the migrative cell number in MG63 cells with FOXD2-AS1 overexpression, and Saos-2 cells with FOXD2-AS1 knockdown. (d) Proliferative CCK-8 assay indicated the proliferative ability of MG63 cells and Saos-2 cells. (e, f) In vivo animal assay illustrated the tumor growth in mice subcutaneous injection using MG63 cells with FOXD2-AS1 overexpression. **p < 0.01; *p < 0.05.
3.4. FOXD2-AS1 enhanced the FOXM1 mRNA stability via m6A modification manner
Previous studies have reported finding that FOXM1 functions as an oncogene in the osteosarcoma [Citation19]. Thus, in order to investigate the potential regulation mediated by FOXD2-AS1, we tried to explore the interaction within FOXD2-AS1 and FOXM1 mRNA. Public database (http://gepia.cancer-pku.cn/index.html) indicated that FOXD2-AS1 level was positively correlated with FOXM1 in osteosarcoma specimens (). Besides, the level of FOXM1 was up-regulated in osteosarcoma specimens as compared to normal controls (). Kaplan Meier plotter (http://kmplot.com/analysis/) indicated that the osteosarcoma patients with higher FOXM1 level showed a lower survival rate (). RNA stability analysis indicated that FOXD2-AS1 overexpression enhanced the FOXM1 mRNA stability and FOXD2-AS1 knockdown repressed it (). The m6A binding site motif is GGACU, which was matched with the candidate m [Citation6]A modification site of FOXM1 mRNA (). Schematic diagram displayed the interaction within FOXD2-AS1 and FOXM1 mRNA (). RNA immunoprecipitation (RIP)-qPCR assay exhibited the interaction within FOXD2-AS1 and FOXM1 mRNA using anti-m [Citation6]A antibody upon FOXD2-AS1 overexpression or knockdown (). Overall, these findings showed that FOXD2-AS1 enhanced the FOXM1 mRNA stability via m [Citation6]A modification manner.
Figure 4. FOXD2-AS1 enhanced the FOXM1 mRNA stability via m [Citation6]A modification manner. (a) Public database (http://gepia.cancer-pku.cn/index.html) indicated the correlation within with FOXD2-AS1 level and FOXM1 in osteosarcoma specimens. (b) The level of FOXM1 in osteosarcoma specimens as compared to normal controls. (c) Kaplan Meier plotter (http://kmplot.com/analysis/) indicated the survival rate of osteosarcoma patients with higher or lower FOXM1 level. (d) RNA stability analysis using RT-qPCR indicated the FOXM1 mRNA remaining upon FOXD2-AS1 overexpression or FOXD2-AS1 knockdown. (e) The m [Citation6]A binding site motif of FOXM1 mRNA is GGACU. (f) Schematic diagram displayed the interaction within FOXD2-AS1 and FOXM1 mRNA. (g) RNA immunoprecipitation (RIP)-qPCR assay exhibited the FOXM1 mRNA level precipitated by anti-m [Citation6]A antibody upon FOXD2-AS1 overexpression or knockdown. **p < 0.01; *p < 0.05.
![Figure 4. FOXD2-AS1 enhanced the FOXM1 mRNA stability via m [Citation6]A modification manner. (a) Public database (http://gepia.cancer-pku.cn/index.html) indicated the correlation within with FOXD2-AS1 level and FOXM1 in osteosarcoma specimens. (b) The level of FOXM1 in osteosarcoma specimens as compared to normal controls. (c) Kaplan Meier plotter (http://kmplot.com/analysis/) indicated the survival rate of osteosarcoma patients with higher or lower FOXM1 level. (d) RNA stability analysis using RT-qPCR indicated the FOXM1 mRNA remaining upon FOXD2-AS1 overexpression or FOXD2-AS1 knockdown. (e) The m [Citation6]A binding site motif of FOXM1 mRNA is GGACU. (f) Schematic diagram displayed the interaction within FOXD2-AS1 and FOXM1 mRNA. (g) RNA immunoprecipitation (RIP)-qPCR assay exhibited the FOXM1 mRNA level precipitated by anti-m [Citation6]A antibody upon FOXD2-AS1 overexpression or knockdown. **p < 0.01; *p < 0.05.](/cms/asset/866d3073-2d2e-40ef-b78b-d9a0a4139cd6/kbie_a_2008218_f0004_oc.jpg)
4. Discussion
N [Citation6]-methyladenosine (m6A) and lncRNA have become critical regulators in human cancers, referring in particular to internal RNA modification on lncRNAs [Citation20–22]. LncRNA FOXD2-AS1 has been reported to accelerate the osteosarcoma [Citation23,Citation24]. Emerging evidence illuminates that m6A-modified lncRNAs play important roles in the development and progression of cancer [Citation25,Citation26]. Although series of therapeutic strategies have been applied to osteosarcoma, the treatment status is complex and untoward. Of which, the recrudesce and metastasis are the most important risk factors. Therefore, it’s crucial to explore the mechanism underling the occurrence and development of osteosarcoma.
The m6A-lncRNAs cooperation has been identified to regulate the human cancer progression. For example, LINRIS blocks the ubiquitination of IGF2BP2 to maintain the stability of m6A reader insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2). LINRIS knockdown attenuates MYC-mediated glycolysis in colorectal cancer cells [Citation27]. On the genomic location of lncRNA THOR, there are several m6A sites, including GG (m6A) CU, GA (m6A) CA and UG (m6A) CU sequences. RNA pull-down and RIP-qPCR assay reveal that m6A readers YTHDF1 and YTHDF2 recognize the m6A motifs and enhance lncRNA THOR stability [Citation28]. METTL3-mediated m6A modification led to the upregulation of LINC00958 by stabilizing its transcript. The up-regulation of LINC00958 aggravates the malignant phenotypes of hepatocellular carcinoma through sponging miR-3619-5p to upregulate HDGF expression [Citation29]. Taken together, these findings suggest that m6A-lncRNA play critical roles in the cancers’ progression.
Here, present research shows that m6A methyltransferase WTAP installs the m6A modification on the lncRNA FOXD2-AS1 (). The m6A-assembled FOXD2-AS1 display higher transcript stability when treated with Act D. Besides, the high-expression of FOXD2-AS1 could promote the migration and proliferation in vitro and accelerate tumor growth in vivo. Thus, WTAP exerts the oncogenic roles on osteosarcoma progression through installing the m6A modification on lncRNA FOXD2-AS1, thereby increasing the expression of oncogene FOXD2-AS1. Thus, our findings also prove the m6A-lncRNAs cooperation, which expands the understanding of oncogenic mechanism in osteosarcoma.
Figure 5. WTAP/FOXD2-AS1/m [Citation6]A/FOXM1 axis promotes the osteosarcoma progression.
![Figure 5. WTAP/FOXD2-AS1/m [Citation6]A/FOXM1 axis promotes the osteosarcoma progression.](/cms/asset/59371798-3724-44d1-8a19-d269174fcc76/kbie_a_2008218_f0005_oc.jpg)
In osteosarcoma, m6A modification has been gradually identified to participate the pathophysiological process. For instance, METTL3 is up-regulated in human osteosarcoma tissues and cell lines. The silencing of METTL3 decreases the m [Citation6]A methylation level of lymphoid enhancer-binding factor 1 (LEF1) to inhibit the activity of Wnt/β-catenin signaling pathway [Citation30]. Moreover, other research indicates that high-expression of METTL3, HNRNPA2B1 and KIAA1429 is significantly associated with the poor prognosis [Citation31]. M6A ‘writer’ METTL3 and m6A ‘reader’ YTHDF2 cooperatively regulate the aberrant m6A modification of TRIM7, thus to up-regulate TRIM7 expression in osteosarcoma, which promotes the migration and invasion through BRMS1 ubiquitination [Citation32]. Overall, these evidences indicate that m6A modification could regulate the progression of osteosarcoma.
6. Conclusion
Taken together, we identified that a novel interaction within WTAP and FOXD2-AS1 in osteosarcoma. WTAP enhances the stability of FOXD2-AS1 to enrich its abundance, thereby interacting with FOXM1 via m6A binding to increase FOXM1 expression. The findings further elucidate the function of WTAP-FOXD2-AS1 in osteosarcoma genesis, which may help researchers developing effective therapeutic strategies.
Supplemental Material
Download MS Word (17.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Garcia MB, Ness KK, Schadler KL. Exercise and Physical Activity in Patients with Osteosarcoma and Survivors. Adv Exp Med Biol. 2020;1257:193–207.
- Lilienthal I, Herold N. Targeting Molecular Mechanisms Underlying Treatment Efficacy and Resistance in Osteosarcoma: a Review of Current and Future Strategies. Int J Mol Sci. 2020;21(18):6885.
- Smrke A, Anderson PM, Gulia A, et al. Future Directions in the Treatment of Osteosarcoma. In: Cells. 2021;10(1):172.
- Limaiem F, Byerly DW, Singh R. Osteoblastoma. StatPearls. 2021;1:1–11.
- Sadykova LR, Ntekim AI, Muyangwa-Semenova M, et al. Epidemiology and Risk Factors of Osteosarcoma. Cancer Invest. 2020;38(5):259–269.
- Yang C, Tian Y, Zhao F, et al. Bone Microenvironment and Osteosarcoma Metastasis. Int J Mol Sci. 2020;21(19):6985.
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36.
- Zhu S, Liu Y, Wang X, et al. lncRNA SNHG10 Promotes the Proliferation and Invasion of Osteosarcoma via Wnt/β-Catenin Signaling. Mol Ther Nucleic Acids. 2020;22:957–970.
- Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20(1):18.
- Chen J, Tian Y, Zhang Q, et al. Novel Insights Into the Role of N6-Methyladenosine RNA Modification in Bone Pathophysiology. Stem Cells Dev. 2021;30(1):17–28.
- Chen S, Li Y, Zhi S, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6) A-dependentmanner. Cell Death Dis. 2020;11(8):659.
- Ling Z, Chen L, Zhao J. m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci Rep. 2020;40(4):BSR20200282.
- Liu Z, Liu N, Huang Z, et al. METTL14 Overexpression Promotes Osteosarcoma Cell Apoptosis and Slows Tumor Progression via Caspase 3 Activation. Cancer Manag Res. 2020;12:12759–12767.
- Zhong L, Liao D, Zhang M, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261.
- Li J, Xie H, Ying Y, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19(1):152.
- Wu Y, Xie L, Wang M, et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9(1):4772.
- Zhang S, Zhao BS, Zhou A, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31(591–606.e6):591–606.e6.
- Wu P, Fang X, Liu Y, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12(4):298.
- Zhu X, Lu K, Cao L, et al. FoxM1 is Upregulated in Osteosarcoma and Inhibition of FoxM1 Decreases Osteosarcoma Cell Proliferation, Migration, and Invasion. Cancer Manag Res. 2020;12:9857–9867.
- Ma Z, Ji J. N6-methyladenosine (m6A) RNA modification in cancer stem cells. Stem Cells. 2020;38(12):1511–1519.
- Zhou Z, Lv J, Yu H, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020;19(1):104.
- Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int J Biol Sci. 2020;16(11):1929–1940.
- Zhang QQ, Sl X, Ding C, et al. LncRNA FOXD2-AS1 knockdown inhibits the resistance of human osteosarcoma cells to cisplatin by inhibiting miR-143 expression. Eur Rev Med Pharmacol Sci. 2021;25:678–686.
- Zhang H, Lu Y, Wang J, et al. Downregulation of the long non‑coding RNA FOXD2‑AS1 inhibits cell proliferation, migration and invasion in osteosarcoma. Mol Med Rep. 2019;20(1):292–302.
- Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):310–318.
- Yi YC, Chen XY, Zhang J, et al. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):121.
- Wang Y, Lu J-H, Wu Q-N, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18(1):174.
- Liu H, Xu Y, Yao B, et al. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11:613.
- Zuo X, Chen Z, Gao W, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13(1):5.
- Miao W, Chen J, Jia L, et al. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516(3):719–725.
- Li J, Rao B, Yang J, et al. Dysregulated m6A-Related Regulators Are Associated With Tumor Metastasis and Poor Prognosis in Osteosarcoma. Front Oncol. 2020;10:769.
- Zhou C, Zhang Z, Zhu X, et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine. 2020;59:102955.
