ABSTRACT
Circular (circ) RNAs are differentially expressed in gastric cancer (GC) and participate in the biological growth of tumor cells. Given that, investigations were performed to unravel the function of circ_0000654 in GC. GC tissue and normal tissue specimens were obtained, in which circ_0000654, microRNA (miR)-149-5p, and inhibin-beta A (INHBA) levels were examined. GC cell line (BGC-823) was transfected to alter circ_0000654 and miR-149-5p expression, thereby observing cell malignancy. Stably-transfected BGC-823 cells were injected into nude mice to observe tumor growth in vivo. The interaction circ_0000654, miR-149-5p, and INHBA was validated. circ_0000654 and INHBA were up-regulated but miR-149-5p was down-regulated in GC. circ_0000654 absorbed miR-149-5p to target INHBA. Silencing circ_0000654inhibited the progress of GC cell biology. Oppositely, restoring circ_0000654 enhanced the growth of GC cells. Inhibiting miR-149-5p rescued down-regulated circ_0000654-induced anti-tumor effect on GC. circ_0000654 silence or miR-149-5p overexpression limited the growth of GC tumors in vivo. Obviously, circ_0000654 facilitates the growth of GC cells through absorbing miR-149-5p to up-regulate INHBA.
Introduction
Gastric cancer (GC) is a highly heterogeneous disease in terms of molecular phenotype. It is diagnosed through histology after endoscopic biopsy and is staged based on CT, endoscopic ultrasound, positive emission tomography, and laparoscopy [Citation1]. Helicobacter pylori infection is believed to be the main cause of GC and GC in the early stage is usually asymptomatic or accompanied by nonspecific symptoms [Citation2]. Complete surgical resection is still the only chance to cure GC, and the classic surgical methods include total gastrectomy and subtotal gastrectomy depending on tumor location [Citation3]. For advanced GC, the combination of immunotherapy, molecular-targeted therapy, and neoadjuvant chemoradiotherapy are mainly referred [Citation4]. Even though with advanced techniques, searching for novel biomarkers and improving the survival of cancer patients are considerably challenging.
Circular RNA (circRNA) is a type of endogenous RNA that is highly stable in mammalian cells, emerging as a therapeutic target for human cancers [Citation5,Citation6]. A large number of papers have clarified the pro-tumor properties of some circRNAs in GC, such as nuclear receptor-interacting protein 1 [Citation7], circ-0000670 [Citation8], Nance-Horan syndrome-like 1 [Citation9] and circRNA_102231 [Citation10]. Concerning circ_0000654, Zhiqiao Xu et al. have identified its pro-tumorigenic property in esophageal squamous cell carcinoma (ESCC) [Citation11], but its mechanism in GC was very little discussed. In the context of GC, miR-149 could regulate metastasis [Citation12], proliferation [Citation13], and drug resistance [Citation14]. circRNA can influence miRNA activity through miRNA absorption [Citation15]. circRNA-modulated miR-149-5p has been previously discussed in GC which blocks anti-tumorigenic activities of GC cells [Citation7], but little was known about the interaction between circ_0000654 and miR-149-5p in GC. Inhibin-beta A (INHBA), a ligand belonging to the transforming growth factor-beta superfamily, is highly expressed in GC, presenting a positive association with tumor size and invasion depth [Citation16]. Functionally, inhibition of INHBA could restrain the aggressiveness of GC cells [Citation17]. On the bioinformatics website, INHBA was found as a target of miR-149-5p. For this reason, our study was planned with the hypothesis that circ_0000654 acts as a sponge of miR-149-5p to target INHBA, thereby promoting the biological activities of GC cells. It is hoped that this novel molecular axis facilitates to develop a potential clinical treatment plan.
Methods and Materials
Ethics statement
The study was approved by the Ethics Committee of the First People’s Hospital of Shangqiu City. The written informed consent was collected from each participant. Animal treatments passed the review of the Animal Ethics Committee.
Clinical specimens
A total of 80 pairs of GC tissue and normal tissue specimens were obtained from patients undergoing radical gastrectomy without preoperative radiotherapy or chemotherapy in the First People’s Hospital of Shangqiu City. All fresh specimens were placed in liquid nitrogen and stored at −80°C.
Cell culture and transfection
Human gastric epithelial cell line (GES-1) and human GC cell lines (AGS, SGC-7901, BGC-823, MKN-45 and MGC-803) were obtained from ATCC (VA, USA) and grown in Dulbecco’s modified Eagle’s medium (Invitrogen, CA, USA). The medium contained 10% fetal bovine serum (FBS; Invitrogen) [Citation18].
The -untreated BGC-823 cells were set up as the blank group. BGC-823 were respectively transfected with si-NC, si-circ_0000654, oe-circ_0000654, oe-NC, NC-mimic, miR-149-5p-mimic, si-circ_0000654 + NC-inhibitor, and si-circ_0000654 + miR-149-5p-inhibitor using Lipofectamine 2000 (Invitrogen) [Citation19]. The plasmids or oligonucleotides were synthesized by GenePharma (Shanghai, China). si-RNA sequences: si-hsa_circ_0000654-1 (GGGAAAUACUGUUAUAUAUAAAG) and si-hsa_circ_0000654-2 (GGUGUAUAUCAAUAGGAUUAA). After reverse transcription quantitative polymerase chain reaction (RT-qPCR), si-hsa_circ_0000654-1 with better knockdown efficiency was selected for subsequent experiments.
RT-qPCR
Total RNA was extracted from cells and tissues with Trizol reagent (Takara, Otsu, Japan). PCR primers were listed in Supplementary . Reverse-transcription of RNA into cDNA was performed with script RT reagent kit or Mir-X miR First-Strand synthesis kit (Takara), followed by RT-qPCR with SYBR Premix Ex Taq II (Takara) in a real-time PCR system (Applied Biosystems, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used for internal control. Gene levels were analyzed through 2−ΔΔCt method [Citation19].
Table 1. The relation between circ_0000654 expression and the characteristics of patients with GC
Circular characteristic of circ_0000654
RNA (3 μ g) was digested with 9 U RNase R for 50 min. The resistance of circ_0000654 to RNase R was evaluated by RT-qPCR detection of RNA levels. All experiments were performed in triplicate [Citation20].
Cell counting kit (CCK)-8 assay
Cells (2 × 103 cells/well) were grown for 24, 48, 72, and 96 h, respectively and supplemented with 10 μL CCK-8 solution (Roche). Two hours later, optical density at 450 nm was determined on a microplate reader (Bio-Rad, CA, USA). All experiments were performed in triplicate [Citation18].
Flow cytometry
Cells obtained after detachment and centrifugation were appended with 75% ethanol, followed by dyeing with fluorescein isothiocyanate-AnnexinV and propidium iodide. Finally, the stained cells were placed in the FACSCalibur system (BD Pharmingen, NJ, USA) to measure cell apoptosis. All experiments were performed in triplicate [Citation21].
Scratch test
Cells (5 × 105 cells/well) at 90% confluence were scratched with a sterile pipette tip. After removal of the floating cells, the scratched cells were recovered in serum-free medium and photographed in 0–24 h. The cell migration distance was measured using Image-Pro Plus analysis software (Media Cybernetics, MD, USA). All experiments were performed in triplicate [Citation17].
Transwell assay
Boyden chamber (Corning, NY, USA) with 8-μm porous polycarbonate membrane was used to evaluate the invasion ability of GC cells. In the upper chamber pre-coated with Matrigel, 5 × 104 cells combined with 200 μL FBS-free medium were added; in the lower chamber, 20% FBS-DMEM was dispersed. After 48 h, the invaded cells were fixed with methanol for staining with 0.1% crystal violet. All experiments were performed in triplicate [Citation22].
Western blot assay
Total protein was collected using radio-immunoprecipitation assay lysis buffer (Beyotime, Shanghai, China) and protease inhibitors (Roche, Mannheim, Germany), and protein concentration was examined using BCATM protein assay kit (Pierce, IL, USA). The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene fluoride membrane, blocked with skim milk, and combined with the primary antibodies INHBA (1:3200, Millipore/Sigma, MA, USA) and GAPDH (1:1000, CST, MA, USA). Afterward, the membrane was incubated with horseradish peroxidase-labeled immunoglobulin G (1:1000, CST), immersed in enhanced chemiluminescence solution (Pierce) and detected in a chemiluminescence detection system. All experiments were performed in triplicate [Citation19].
Dual-luciferase reporter gene assay
The circ_0000654 or INHBA fragment containing the miR-149-5p target sequence was inserted into the pmirGLO (Promega, WI, USA) to form circ_0000654-wild type (WT) or INHBA-WT while circ_0000654-mutant (MUT) or INHBA-MUT contained mutated binding site. The reporter vector, along with miR-149-5p-mimic or NC-mimic was co-transfected into BGC-823 cells with Lipofectamine 2000 (Invitrogen), and firefly/Renilla activities were captured with the dual-luciferase reporter gene detection kit (Promega). The experiment was conducted 3 times [Citation18].
RNA pull-down analysis
Biotinylated miR-149-5p was constructed to generate biotin-miR-149-5p, and biotinylated miR-NC (biotin-NC) was used as a control. Cell extract (2 μg) was incubated with 100 pmol biotinylated RNA, added with agarose beads (Invitrogen) and detected by RT-qPCR. The experiment was conducted 3 times [Citation23].
Tumor xenografts in nude mice
Female BALB/c nude mice of specific pathogen-free grade were 4–6 weeks old with a body weight of 18–20 g. The mice were reared under sterilized conditions and subcutaneously injected with BGC-823 cells (1 × 107 cells/mL, 0.2 mL) in the back near the armpit. Each group contained 5 mice. The largest long axis (L) and short axis (W) perpendicular to the largest long axis were measured every day to calculate tumor volume (L/2 × W2). Nude mice were euthanized after 30 d and subcutaneous tumors were isolated, weighed and photographed.
Immunohistochemistry (IHC)
The tumor tissue sections were embedded in paraffin (Sigma-Aldrich, CA, USA), deparaffinized with xylene, and hydrated with gradient ethanol in turn. After treatment with 3% hydrogen peroxide, the sections were incubated with primary antibody INHBA (1:3200, MilliporeSigma) and homologous secondary antibody. After staining with 3,3ʹ-diaminobenzidine and hematoxylin, the sections were observed through a microscope (Nikon, Tokyo, Japan). The experiment was conducted 3 times [Citation18].
Statistical analysis
Statistical analysis was performed using SPSS 18.0 software (IBM, NY, USA). Values were represented as mean ± standard deviation. Data comparison was evaluated using t-test or one-way analysis of variance (ANOVA). P less than 0.05 was significant [Citation24].
Results
Circ_0000654 is up-regulated in GC
Circ_0000654 is highly expressed in prostate cancer [Citation25]. However, its function in GC is unclear. To determine circ_0000654 expression in GC, 80 pairs of GC tissues and normal tissues were harvested, and higher circ_0000654 was measured in GC tissues than normal tissues (). Next, through RT-qPCR analysis, the same expression trend of circ_0000654 was also found in GC cell lines (AGS, SGC-7901, BGC-823, MKN-45 and MGC-803) (). Circ_0000654 was derived from the ‘reverse splicing’ of the exon of its host gene CHD2 (), and circ_0000654 was resistant to RNase R (), indicating that circ_0000654 is indeed a cyclic transcript.
Figure 1. Circ_0000654 is up-regulated in GC. A. circ_0000654 expression inGC tissues and normal tissues (RT-qPCR); B. circ_0000654 expression in GES-1 and GC cell lines (RT-qPCR); C. Genomic location and structural characteristics of circ_0000654; D. Resistance of circ_0000654 to RNase R; The value was expressed as mean ± standard deviation. Experimental repetitions = 3. Data comparison used t test or one way AVONA, * P < 0.05 vs. GES1 cells.
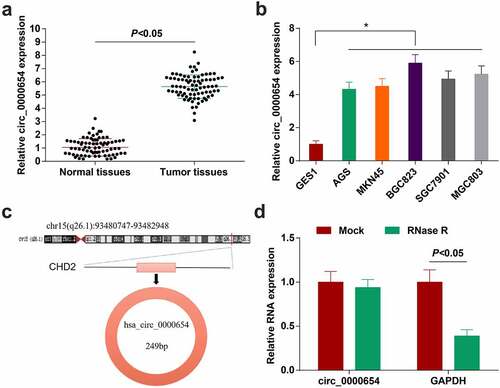
Taking the average relative expression of circ_0000654 as the boundary, GC samples were divided into circ_0000654 high expression group (54 cases) and low expression group (26 cases). It was confirmed that circ_0000654 expression was related to tumor differentiation, tumor node metastasis (TNM) staging and lymph node metastasis ().
Silencing circ_0000654 inhibits the progress of GC cell biology while overexpressing circ_0000654 acts oppositely
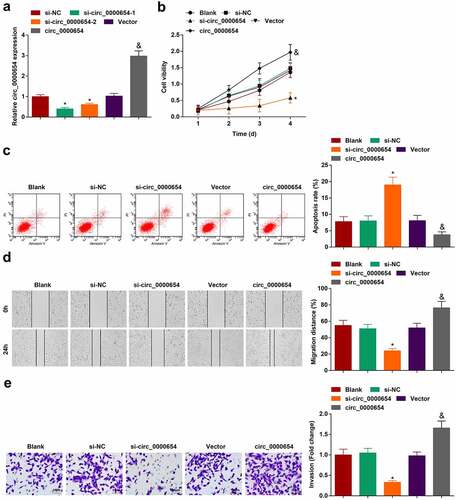
For exploring the biological functions of circ_0000654 in GC, BGC-823 cell line with the highest circ_0000654 expression was opted, and transfected with circ_0000654 siRNAs or overexpression plasmid, respectively. RT-qPCR found that si-circ_0000654-1 lowered circ_0000654 expression more effectively than si-circ_0000654-2 (). Therefore, follow-up experiments used si-circ_0000654-1 for loss-of-function analysis. Meanwhile, circ_0000654 overexpression plasmid raised circ_0000654 expression in BGC-823 cells (). Proliferation, apoptosis, migration and invasion were examined in circ_0000654-modified BGC-823 cells through CCK-8 (), flow cytometry (), scratch test () and Transwell assay (). It was manifested that silencing circ_0000654 depressed proliferation, migration and invasion but increased the apoptosis rate of BGC-823 cells. On the other hand, up-regulating circ_0000654 caused the opposite consequences. In summary, silent circ_0000654 could repress the progress of GC cell biology while restored circ_0000654 serves to accelerate GC development.
Figure 2. Silencing circ_0000654 inhibits the progress of GC cell biology while overexpressing circ_0000654 acts oppositely. A. circ_0000654 expression in BGC-823 cells transfected with circ_0000654 siRNA or overexpression plasmid (RT-qPCR); B. Proliferation of BGC-823 cells transfected with circ_0000654 siRNA or overexpression plasmid (CCK-8 assay); C. Apoptosis of BGC-823 cells transfected with circ_0000654 siRNA or overexpression plasmid (flow cytometry); D. Migration of BGC-823 cells transfected with circ_0000654 siRNA or overexpression plasmid (scratch test); E. Invasion of BGC-823 cells transfected with circ_0000654 siRNA or overexpression plasmid (Transwell assay); The value was expressed as mean ± standard deviation. Experimental repetitions = 3. Data comparison used t test or one way AVONA, * P < 0.05 vs. the si-NC group; & P < 0.05 vs. the vector.
miR-149-5p is down-regulated in GC; overexpression of miR-149-5p retards the progress of GC cells
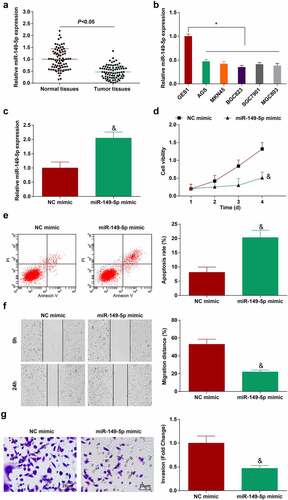
miR-149-5p is down-regulated in GC and overexpression of miR-149-5p inhibits GC growth [Citation26]. However, its mechanism has not yet been fully ascertained. In the present research, miR-149-5p expression was detected to be decreased in GC tissues and cell lines ()). Subsequently, miR-149-5p mimic was transfected into BGC-823 cells which successfully increased cellular miR-149-5p expression (). Then, the inhibitory effect of overexpressed miR-149-5p on the biological progress of GC cells was clarified, as represented by reduced proliferation (), migration () and invasion (), and augmented apoptosis rate (). In conclusion, overexpression of miR-149-5p can inhibit GC cell progression.
Figure 3. miR-149-5p is down-regulated in GC; overexpression of miR-149-5p retards the progress of GC cells. A. miR-149-5p expression in 80 pairs of GC tissues and normal tissues (RT-qPCR); B. miR-149-5p expression in GES-1 and GC cell lines (RT-qPCR); C. miR-149-5p expression in BGC-823 cells transfected with miR-149-5p mimic (RT-qPCR); D. Proliferation of BGC-823 cells transfected with miR-149-5p mimic (CCK-8 assay); E. Apoptosis of BGC-823 cells transfected with miR-149-5p mimic (flow cytometry); F. Migration of BGC-823 cells transfected with miR-149-5p mimic (scratch test); G. Invasion of BGC-823 cells transfected with miR-149-5p mimic (Transwell assay); The value was expressed as mean ± standard deviation. Experimental repetitions = 3. Data comparison used t test, * P < 0.05 vs. GES1 cells, & P < 0.05 vs. the NC mimic group.
Circ_0000654 absorbs miR-149-5p to target INHBA
Silencing circular RNA can regulate the expression of miRNA [Citation27]. miR-149-5p expression was increased in circ_0000654-silenced BGC-823 cells (). Therefore, a correlation was speculated between miR-149-5p and circ_0000654. Next, bioinformatics software http://starbase.sysu.edu.cn/ found that circ_0000654 and miR-149-5p had potential binding sites () and dual-luciferase report experiment showed that the luciferase activity of circ_0000654-WT was dropped by miR-149-5p-mimic (). In the RNA pull-down test, when biotin-miR-149-5p was used, circ_0000654 was enriched (). Taken together, miR-149-5p was a target of circ_0000654.
Figure 4. Circ_0000654 absorbs miR-149-5p to target INHBA. A. miR-149-5p in BGC-823 cells transfected with si-circ_0000654 (RT-qPCR); B. The presence of binding sites between miR-149-5p and circ_0000654 (http://starbase.sysu.edu.cn/); C. Interaction between miR-149-5p and circ_0000654 (dual luciferase reporter experiment); D. Interaction between miR-149-5p and circ_0000654 (RNA pull-down assay); E. INHBA expression in 80 pairs of GC tissues and normal tissues (RT-qPCR); F. INHBA expression in GES-1 and BGC-823 cells (RT-qPCR); G. The binding sites of miR-149-5p and INHBA (https://cm.jefferson.edu/); H. Interaction between miR-149-5p and INHBA (dual luciferase reporter experiment); I. Interaction between miR-149-5p and INHBA (RNA pull-down assay); J. INHBA expression in BGC-823 cells transfected with si-circ_0000654 (RT-qPCR and Western blot); K. INHBA expression in BGC-823 cells transfected with miR-149-5p mimic (RT-qPCR and Western blot). The value was expressed as mean ± standard deviation. Experimental repetitions = 3. Data comparison used t test, * P < 0.05 vs. the si-NC group, & P < 0.05 vs. the NC mimic group; ^ P < 0.05 vs. GES1 cells.
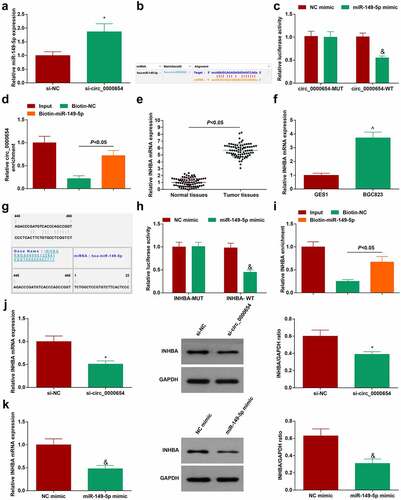
INHBA gene silencing inhibits the migration and invasion of GC cells [Citation17]. We further clarified whether INHBA is a downstream target gene of miR-149-5p. Highly expressed INHBA was tested in GC tissues and cells ()). To verify that INHBA was regulated by miR-149-5p, bioinformatics software https://cm.jefferson.edu/ was utilized to study the targeting relationship between miR-149-5p and INHBA (). In addition, the dual-luciferase report experiment showed that miR-149-5p-mimic reduced the luciferase activity of INHBA-WT (), and the RNA pull-down test also proved that miR-149-5p had a binding relationship with INHBA (). Next, whether the circ_0000654/miR-149-5p axis can regulate INHBA expression was examined and the finding implied that after silencing circ_0000654 or overexpressing miR-149-5p, INHBA expression in BGC-823 cells was reduced ()). Overall, INHBA was a downstream target of miR-149-5p and could be regulated by circ_0000654/miR-149-5p axis.
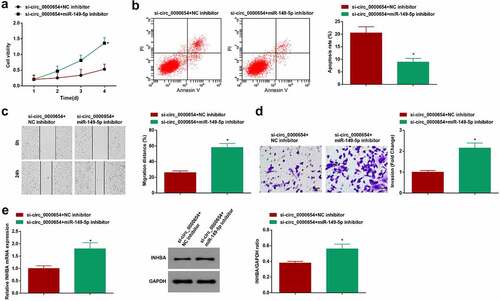
Inhibiting miR-149-5p rescues down-regulated circ_0000654-induced anti-tumor effect on GC
To clarify that circ_0000654/miR-149-5p/INHBA axis regulates the progress of GC cell biology, si-circ_0000654 and miR-149-5p-inhibitor were co-transfected into BGC-823 cells. Data analysis disclosed that the anti-GC role of si-circ_0000654 was rescued after miR-149-5p-inhibitor treatment ()). Moreover, miR-149-5p-inhibitor could increase INHBA expression regulated by si-circ_0000654 (). Obviously, circ_0000654/miR-149-5p/INHBA axis regulated the progress of GC cell biology.
Figure 5. Inhibiting miR-149-5p rescues down-regulated circ_0000654-induced anti-tumor effect on GC. A. Proliferation of BGC-823 cells co-transfected with si-circ_0000654 and miR-149-5p-inhibitor (CCK-8 assay); B. Apoptosis of BGC-823 cells co-transfected with si-circ_0000654 and miR-149-5p-inhibitor (flow cytometry); C. Migration of BGC-823 cells co-transfected with si-circ_0000654 and miR-149-5p-inhibitor (scratch test); D. Invasion of BGC-823 cells co-transfected with si-circ_0000654 and miR-149-5p-inhibitor (Transwell assay); E. INHBA expression in BGC-823 cells co-transfected with si-circ_0000654 and miR-149-5p-inhibitor (RT-qPCR and Western blot). The value was expressed as mean ± standard deviation. Experimental repetitions = 3. Data comparison used t test, * P < 0.05 vs. the si-circ_0000654 + NC inhibitor group.
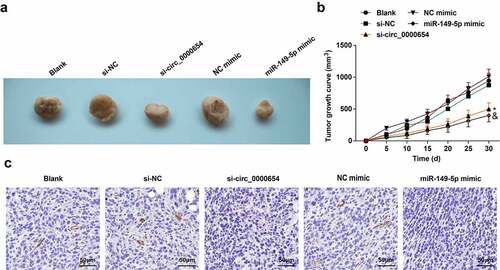
Circ_0000654 silence or miR-149-5p overexpression limits the growth of GC tumors
In attempting to understand how circ_0000654 and miR-149-5p affect the biological progress of GC tumors in vivo, BGC-823 cells under-expressing circ_0000654 or overexpressing miR-149-5p were injected into mice to form tumors. Reduced tumor volume and growth were noticed in mice injected with circ_0000654-deficient or miR-149-5p-sufficient BGC-823 cells ()). IHC analysis found reduced INHBA in tumors after silencing circ_0000654 or overexpressing miR-149-5p (). In a word, silencing circ_0000654 or overexpression of miR-149-5p limited the growth of GC tumors in vivo.
Figure 6. Circ_0000654 silence or miR-149-5p overexpression limits the growth of GC tumors. A. Tumors of mice after silencing circ_0000654 or overexpression of miR-149-5p (n = 5); B. Tumor growth of mice after silencing circ_0000654 or overexpression of miR-149-5p (n = 5); C. INHBA expression in tumors after silencing circ_0000654 or overexpression of miR-149 (IHC). The value was expressed as mean ± standard deviation. Experimental repetitions = 3. Data comparison used one-way ANOVA, * P < 0.05 vs. the si-NC group; & P < 0.05 vs. the NC mimic group.
Discussion
GC is the leading cause of cancer-related mortality across the world, with short overall survival for advanced disease [Citation28]. A great number of GC-related circRNAs are validated with potentials for tumor diagnosis and therapy [Citation29]. In the research, the exact role of circ_0000654 in GC was described and the main outcome depicted that silencing circ_0000654 positively regulated INHBA expression via targeting miR-149-5p, thereafter to retard the outgrowth of GC cells.
At first, measurement of circ_0000654 expression was performed to reveal that circ_0000654 was up-regulated in GC patients and was related to tumor differentiation, TNM staging and lymph node metastasis. For GC cells, the silent circ_0000654 could depress the proliferation, invasion, and migration but induce apoptosis in vitro, as well as impede tumor formation in vivo. In an experimental study concerning ESCC, the raised circ_0000654 expression is correlated with local lymph node metastasis; and ectopic expression of circ_0000654 activates tumor cell progression while the inhibited level of circ_0000654 imposes the opposite effect through mediating miR-149-5p [Citation11]. To our best knowledge, the very limited paper has described the accurate mechanism of circ_0000654 in GC.
Next, miR-149-5p was a proved target of circ_0000654 which was down-regulated in GC tissues and cell lines. The action of miR-149-5p in GC was explored with the findings explaining that overexpression of miR-149-5p impaired the malignant behaviors of GC cells in vitro and in vivo, whilst inhibition of miR-149-5p rescued the anti-tumor influence of silent circ_0000654 on GC in vitro. In line with our findings, Xing Zhang et al. have discovered the phenomenon that elevating miR-149-5p in GC cells is accountable to blunt the malignant activities in vitro [Citation7]. Mechanistically, miR-149-5p expression reduction is witnessed in GC samples, but enhancement of miR-149-5p hinders the migratory and invasive properties of tumor cells [Citation30]. An innovative article about GC also reports that miR-149-5p is abated in tumor samples, and further unravels that up-regulated miR-149-5p has a damaging effect on the biological activities of GC cells [Citation31]. Notably, Yang Yang et al. have brought forward an idea that circ_0044516-regulated miR-149-5p enhancement obstructs the development of GC cells in vitro and in vivo [Citation32]. The deregulation of miR-149-5p has also been widely probed including but not limited to GC. For instance, in the context of breast cancer, by targeting miR-149-5p, depletion of circ_0072995 could depress the malignant phenotypes and glycolysis of tumor cells [Citation33]. Also, the decreased miR-149-5p level is determined in medullary thyroid carcinoma, and overexpressed miR-149-5p accounts for proliferation and invasion inhibition of tumor cells [Citation34]. As commonly discussed before, aberrantly expressed miR-149-5p is related to tumorigenesis and modification of miR-149-5p could orchestrate the development of tumors.
Afterward, INHBA was identified as a downstream actor of miR-149-5p, which could be regulated by circ_0000654/miR-149-5p axis. As observed experimentally, GC clinical samples exhibit high INHBA expression [Citation17]. Moreover, promoted expression of INHBA detected in GC tissue has a correlation with poor survival [Citation16,Citation35]. Besides, the up-regulated INHBA tested in ovarian cancer [Citation36], ESCC [Citation37] and nasopharyngeal carcinoma [Citation38] are all validated to promote tumor development. Finitely, the relative mechanism of INHBA as the downstream actor of circ_0000654/miR-149-5p axis needs more investigations.
Conclusion
To sum up, our research exhibited the manifestation of circ_0000654, miR-149-5p and INHBA in GC, and captured the innovative conclusion that deficiency of circ_0000654 attenuated the malignancy of GC through enhancing miR-149-5p and reducing INHBA levels. This work has addressed a novel theoretical reference for the treatment options for GC, and the sample size shall be enriched to further validate the conclusion.
Supplemental Material
Download MS Word (13.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Smyth EC, Verheij M, Allum W, et al. Gastric cancer. Lancet. 2020;396(10251):635–648.
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211–217.
- Johnston FM, Beckman M. Updates on management of gastric cancer. Curr Oncol Rep. 2019;21(8):67.
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626.
- Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136.
- Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301.
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20.
- Liu P, Cai S, Li N., et al. Circular RNA-hsa-circ-0000670 promotes gastric cancer progression through the microRNA-384/SIX4 axis. Exp Cell Res. 2020;394(2):112141.
- Zhu Z, Rong Z, Luo Z, et al. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer. 2019;18(1):126.
- Yuan G, Ding W, Sun B, et al. Upregulated circRNA_102231 promotes gastric cancer progression and its clinical significance. Bioengineered. 2021;12(1):4936–4945.
- Xu Z, Tie X, Li N, et al. Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. IUBMB Life. 2020;72(3):426–439.
- Luo X, Wang GH, Bian ZL, et al. Long non-coding RNA CCAL/miR-149/FOXM1 axis promotes metastasis in gastric cancer. Cell Death Dis. 2018;9(10):993.
- Fang J, Chen W, Meng X., et al. Downregulating circRNA_0044516 inhibits cell proliferation in gastric cancer through miR-149/Wnt1/beta-catenin pathway. J Gastrointest Surg. 2020;(7). DOI: 10.1007/s11605-020-04834-w.
- Li X, Liang J, Liu YX, et al. miR-149 reverses cisplatin resistance of gastric cancer SGC7901/DDP cells by targeting FoxM1. Pharmazie. 2016;71(11):640–643.
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83.
- Chen ZL, Qin L, Peng XB, et al. INHBA gene silencing inhibits gastric cancer cell migration and invasion by impeding activation of the TGF-beta signaling pathway. J Cell Physiol. 2019;234(10):18065–18074.
- Wang D, Liu K, Chen E., et al. LINC00511 promotes proliferation and invasion by sponging miR-515-5p in gastric cancer. Cell Mol Biol Lett. 2020;25(1):4.
- Sun CB, Wang HY, Han XQ, et al. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J Gastrointest Oncol. 2020;12(4):394–404.
- Ren K, Li B, Jiang L, et al. circ_0023461 silencing protects cardiomyocytes from hypoxia-Induced dysfunction through targeting miR-370-3p/PDE4D signaling. Oxid Med Cell Longev. 2021;2021:8379962.
- Huang HG, Tang XL, Huang XS, et al. Long noncoding RNA LINC00511 promoted cell proliferation and invasion via regulating miR-124-3p/EZH2 pathway in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24(8):4232–4245.
- Qi H, Xiao Z, Wang Y., et al. Long non-coding RNA LINC00665 gastric cancer tumorigenesis by regulation miR-149-3p/RNF2 axis. Onco Targets Ther. 2019;12:6981–6990.
- Ma Y, Ren Y, Wen H, et al. circCOL1A1 promotes the progression of gastric cancer cells through sponging miR-145 to enhance RABL3 expression. J Immunol Res. 2021;2021:6724854.
- Liu X, Jiao T, Wang Y, et al. Long non-coding RNA GAS5 acts as a molecular sponge to regulate miR-23a in gastric cancer. Minerva Med. 2016; 11.
- Li T, Sun X, Chen L., et al. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121(3):2118–2126.
- Cao D, et al. 18beta-glycyrrhetinic acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1 signaling. Oncotarget. 2016;7(44):71960–71973.
- Hu J, Wang L, Chen J, et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505(1):222–228.
- Zhang XY, Zhang PY. Gastric cancer: somatic genetics as a guide to therapy. J Med Genet. 2017;54(5):305–312.
- Li R, Jiang JJ, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661–1680.
- Hui C, Tian L, He X., et al. Circular RNA circNHSL1 contributes to gastric cancer progression through the miR-149-5p/YWHAZ axis. Cancer Manag Res. 2020;12:7117–7130.
- Wang Z, Liu X, Liu X, et al. Long non-Coding RNA BLACAT1 promotes the tumorigenesis of gastric cancer by sponging microRNA-149-5p and targeting KIF2A. Cancer Manag Res. 2020;12:6629–6640.
- Yang Y, Cai B, Shi X, et al. circ_0044516 functions in the progression of gastric cancer by modulating MicroRNA-149-5p/HuR axis. Mol Cell Biochem. 2021. DOI:10.1007/s11010-020-04026-9.
- Qi C, Qi X, Zhou Z, et al. Circ_0072995 promotes cell carcinogenesis via up-Regulating miR-149-5p-Mediated SHMT2 in Breast Cancer. Cancer Manag Res. 2020;12:11169–11181.
- Ye X, Chen X. miR-149-5p inhibits cell proliferation and invasion through targeting GIT1 in medullary thyroid carcinoma. Oncol Lett. 2019;17(1):372–378.
- Oshima T, Yoshihara K, Aoyama T, et al. Relation of INHBA gene expression to outcomes in gastric cancer after curative surgery. Anticancer Res. 2014;34(5):2303–2309.
- Li X, Yang Z, Xu S, et al. Targeting INHBA in ovarian cancer cells suppresses cancer xenograft growth by attenuating stromal fibroblast activation. Dis Markers. 2019;2019:7275289.
- Lyu S, Chao, J, Xu, R, et al. INHBA upregulation correlates with poorer prognosis in patients with esophageal squamous cell carcinoma. Cancer Manag Res. 2018;10:1585–1596.
- Peng S, Wang J, Hu P, et al. INHBA knockdown inhibits proliferation and invasion of nasopharyngeal carcinoma SUNE1 cells in vitro. Int J Clin Exp Pathol. 2020;13(5):854–868.
