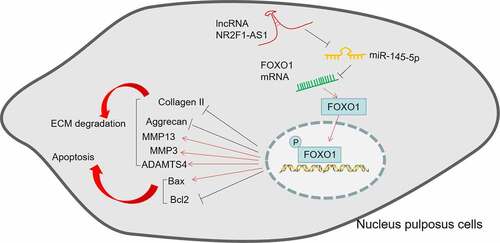
ABSTRACT
Emerging reports uncover that long noncoding RNAs (lncRNAs) help regulate intervertebral disc degeneration (IVDD). Here, we probe the function of lncRNA nuclear receptor subfamily 2 group F member 1 antisense RNA 1 (NR2F1-AS1) in IVDD. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was applied to verify the expression of NR2F1-AS1 and miR-145-5p in nucleus pulposus (NP) tissues from IVDD patients or NP cells dealt with IL-1β or TNF-α. Flow cytometry or the TdT-mediated dUTP nick end labeling (TUNEL) assay was performed to validate the apoptosis of NP cells with selective regulation of NR2F1-AS1 and miR-145-5p. ECM-related genes, FOXO1, Bax, and Bcl2 were evaluated by qRT-PCR or Western blot (WB). The targeted relationships between NR2F1-AS1 and miR-145-5p, miR-145-5p and FOXO1 were testified by the dual-luciferase reporter assay and the RNA immunoprecipitation (RIP) assay. Our outcomes substantiated that NR2F1-AS1 was up-regulated, while miR-145-5p was down-regulated in intervertebral disc tissues of IVDD patients or NP cells treated with IL-1β or TNF-α. Besides, overexpressing NR2F1-AS1 intensified ECM degradation and NP cell apoptosis induced by IL-1β, while knocking down NR2F1-AS1 or up-regulating miR-145-5p reversed IL-1β-mediated effects in NP cells. Meanwhile, NR2F1-AS1 choked miR-145-5p and abated its effects in NP cells. This study confirms that NR2F1-AS1 modulates IVDD progression by up-regulating the FOXO1 pathway through the sponge of miR-145-5p as a competitive endogenous RNA (ceRNA).
1. Introduction
Intervertebral disc degeneration (IVDD) is caused by repetitive mechanical loads or wear, leading to backache[Citation1]. IVDD is characterized by increased levels of the proinflammatory cytokines secreted by intervertebral disc cells[Citation2]. Current treatments for IVDD and backache include analgesics or surgery, which aim to reduce symptoms rather than eradicate the latent pathology [Citation3]. However, the progression mechanism of IVDD has not been well clarified [Citation4]. Recent reports have testified that long noncoding RNAs (lncRNAs) are powerful regulators of gene expression in IVDD [Citation5]. Hence, studying the molecular mechanism of IVDD is expected to provide a novel option for its treatment.
LncRNAs have become vital modulators of multiple biological processes, including cell proliferation, apoptosis, inflammation, metabolic modulation, et al [Citation6]. LncRNAs are abnormally expressed in tumors[Citation7], inflammation[Citation8], and cardiovascular diseases [Citation9] and exert a unique role in different diseases by regulating various downstream targets (such as chromatin, RNA, and proteins). The latest research indicated that lncRNAs are implicated in the pathological processes of various bone diseases, including extracellular matrix (ECM) degradation, inflammation, apoptosis, and angiogenesis [Citation10–12]. Thus, lncRNAs contribute to IVDD. As a vital member of lncRNAs, lncRNA nuclear receptor subfamily 2 group F member 1 antisense RNA 1 (NR2F1-AS1) is located at 5q15, with a length of 2814 bp. It is reported that NR2F1-AS1 affects tumor progression as an oncogene [Citation13–15]. However, the function of NR2F1-AS1 in IVDD is scarcely researched. Therefore, it is critical to make certain the expression and effect of NR2F1-AS1 in IVDD.
Evidence is mounting that microRNAs (miRNAs) subserve the development of IVDD. By reviewing previous studies, we concluded that miRNAs’ expression is closely related to nucleus pulposus (NP) cell apoptosis and proliferation [Citation16–20]. As a miRNA, miR-145-5p is confirmed to contribute to the progression of inflammatory diseases such as myocardial infarction[Citation21], spinal cord injuries[Citation22], and chronic glomerulonephritis[Citation23]. Additionally, miR-145-5p aggravates rheumatoid arthritis (RA) by activating the NF-κB pathway and enhances the secretion of matrix metalloproteinase-9 (MMP-9)[Citation24]. Disappointingly, the mechanism of miR-145-5p in IVDD remains largely unknown.
The forkhead box protein O (FOXO) family, also known as forkhead proteins, has four subtypes in mammals, namely FOXO1 (FKHR), FOXO3 (fkhrl1), FOXO4 (AFX) and FOXO6. FOXO1 plays a vital role in cell cycle control, apoptosis, metabolism and adipocyte differentiation[Citation25]. Also, the FOXO1 transcription network is critical in regulating homeostasis and ECM, and it mediates the abnormal expression of these pathways observed in the pathogenesis of osteoarthritis (OA)[Citation26]. Wang A et al. stated that MEG3 facilitates OA chondrocytes’ proliferation and hampers their apoptosis and ECM degradation through the miR-361-5p/FOXO1 pathway[Citation27]. Nevertheless, the mechanism of FOXO1 in IVDD remains elusive.
This study seeks to characterize the function of NR2F1-AS1 in IVDD and to reveal its underlying mechanisms. Our findings imply that NR2F1-AS1 was upregulated in the intervertebral disc tissues from IVDD patients, whereas miR-145-5p was downregulated. NRF2F1-AS1 overexpression promoted IL-1β-mediated NP cell apoptosis, up-regulates ECM-related genes, and inhibited miR-145-5p level. The bioinformatic analysis showed that NR2F1-AS1 acts as a potential ceRNA on miR-145-5p, which targets FOXO1. Thus, we guessed NR2F1-AS1 influences IVDD progression via the miR-145-5p/FOXO1 axis. We hope this study provides novel mechanisms and potential options for IVDD treatment.
2. Methods and materials
2.1 Patients and sample collection
This research was granted by the Research Ethics Committee of the Zhongnan Hospital of Wuhan University Wuhan University (Approved number: WUZN-2019-0344). Human NP specimens were harvested from patients with idiopathic scoliosis (non-IVDD, n = 21) and patients with IVDD (n = 35) (see for specific information). The patients were diagnosed as lumbar disc herniation or lumbar disc herniation combined with spinal stenosis and were treated by discectomy via a posterior. Those patients were excluded if they have bone metabolic disease, congenital bone malformation, gout, renal dysfunction, or hypercalcemia. Intervertebral disc tissues were collected during the surgery and immediately frozen in liquid nitrogen at −80°C. Immunohistochemistry (IHC) was performed for detecting Caspase3 (1:100, ab32351, Abcam, USA) in the intervertebral disc tissues [Citation28]. IVDD patients with degenerative spinal stenosis, tumor, infection, or a prior lumbar disc surgery were excluded. All patients underwent routine preoperative lumbar MRI, and the degree of IVDD was analyzed with the modified Pfirrmann grade based on magnetic resonance imaging (MRI) T2 weighted images and the severity of degeneration. Pfirrmann grade I indicates a normal, healthy disc as only found in children, whereas Pfirrmann grade V indicates the most severe degree of degeneration. When it is above Pfirrmann grade III, the pore structure of bone endplate changes significantly and the number of pores decreases gradually [Citation29–31].
Table 1. The primer sequences
Table 2. The clinical characteristics of IVDD patients and non-IVDD patients
2.2 Cell culture and treatment
Human denatured NP cells were extracted from NP tissues of IVDD patients. They were then resuspended in the RPMI-1640 complete culture medium comprising 10% fetal bovine serum and 1% penicillin/streptomycin and incubated at 37°C with saturated humidity and 5% CO2. The medium was substituted once every 2 to 3 days. When the cells were about to confluence, 0.25% trypsin was used for trypsinization and sub-culture. To establish an in-vitro cell model of IVDD, normal NP cells were processed with 20 ng/mL of interleukin (IL)-1β for 48 hours. Untreated NP cells were used as control cells [Citation32].
2.3 Cell transfection
pcDNA empty vector (NC), pcDNA-lncRNA NR2F1-AS1 (lncRNA NR2F1-AS1), lncRNA NR2F1-AS1’s short hairpin RNA negative control (sh-NC), lncRNA NR2F1-AS1’s short hairpin RNA (sh-NR2F1-AS1), miRNA control (miR-NC), and miR-145-5p mimics were acquired from GenePharma Co., Ltd. (Shanghai, China). Human NP cells were seeded into 24-well plates (3 × 105 cells/well) and cultured at 37°C with 5% CO2 for 24 hours before the transfection. The above vectors were transfected into NP cells with Lipofectamine®3000 (Invitrogen; Thermo Fisher Scientific, Inc.) as per the supplier’s specifications. The transfection validity was determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Finally, the cells were maintained at 37°C with 5% CO2 for 24 hours for later use [Citation33].
2.4 Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
The total RNAs cells or tissues were separated with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the concentration was examined, the miRNA and mRNA were subjected to reverse transcription into cDNA using the One Step PrimeScript miRNA cDNA synthesis kit (Bao Biological Engineering Co., Ltd., Dalian, China) and PrimeScript RT kit (Invitrogen, Shanghai, China), respectively. We then implemented qRT-PCR by utilizing SYBR®Premix-Ex-Taq™ (Takara, TX, USA) and the ABI7300 system. The expression profiles of NR2F1-AS1, miR-145-5p and FOXO1 were assessed with the 2−ΔΔCt method (U6 served as a housekeeping gene for miR-145-5p, and GAPDH was a housekeeping gene for NR2F1-AS1, FOXO1, MMP3, MMP13, ADAMTS4, aggrecan, and Collagen II). The primers were synthesized by Shanghai Sangon Biotech Co., Ltd. The primer sequences are exhibited in. qRT-PCR was conducted with 40 cycles of pre-denaturation at 95°C for 30 s, denaturation at 95°C for 5 s, and annealing/extension at 60°C for 30 s. The relative expression of the target gene was analyzed by the 2−∆∆CT method. ∆CT = target gene – GAPDH, while ∆∆ = ∆CT experiment group -∆CT control group [Citation34].
2.5 TdT-mediated dUTP nick end labeling (TUNEL)
Each group of cells in the plates were treated as described above. Then, the culture medium was discarded, and the cells were rinsed with PBS. Afterward, the cells were immobilized with immunostaining fixative solution for 30–60 minutes and flushed with PBS. Then, the immunostaining washing solution was added for ice incubation for 2 minutes. Next, 50 μL TUNEL detection solution was added to the sample and maintained for 60 minutes at 37°C in the dark. Subsequently, the cells were flushed 3 times with PBS. After mounting with the antifade mounting medium, the cells were reviewed under a fluorescence microscope. The excitation light was 450–500 nm, and the emission light was 515–565 nm (green fluorescence). Five fields of view were randomly chosen for each sample, and the apoptotic rate = apoptotic cells/total cells×100%[Citation35].
2.6 Cellular immunofluorescence
miR-NC and miR-145-5p were transfected into NP cells according to the instructions. Then, the IL-1β-treated cells were seeded on 24-well plates, and the coverslip was prepared. After 48 hours, the cells growing on the coverslip were cleaned 3 times with PBS, secured with 4% paraformaldehyde for 30 minutes, and permeated with 0.1% Triton X-100 for 10 minutes. After the cells were rinsed 3 times with PBS, they were kept with 3% H2O2 for 10 minutes and blocked with 10% goat serum + 3% bovine serum albumin for 30 minutes. Afterward, they were maintained with the primary antibody of p-FOXO1 (Proteintech, USA; 1:100) at 4°C overnight and goat anti-rabbit IgG (H + L) (1:500) for 1 hour at room temperature (RT). The nucleus was dyed with DAPI, and the cells were viewed after mounting and photographing [Citation36].
2.7 Annexin V-FITC-PI apoptosis detection assay
Annexin V-FITC-PI apoptosis detection kit (Cat:40,302, Yeasen, Shanghai, China) was used for evaluating apoptosis. NP cells were trypsinized with 0.25% trypsin, then collected by centrifugation (1000 rpm, 5 min). PBS was used for washing the cells three times, and the cells were incubated with containing 200 μL Annexin V-FITC and maintained in the dark for 10 minutes. They were then flushed with 200 μL PBS, and 10 μL PI was added. Cell apoptosis was examined by flow cytometry (FCM) (Beckman Coulter) [Citation37].
2.8 RNA fluorescence in situ hybridization (FISH)
NP cells were grown on 4-chamber glass slides for 48 hours. After rinsing with PBS, the cells were immobilized with 3.7% paraformaldehyde, permeated with 70% ethanol, and then rehydrated in 2× SSC and 50% formamide for 5 minutes. The cells were hybridized with biotin-labeled NR2F1-AS1 and miR-145-5p probe mixture overnight at 42°C. The mixture contained 10% dextran sulfate, 5× Denhardt reagent, 2× SSC, 50% formamide and 100 μg/mL denatured and fragmented salmon sperm DNA. Nonspecific probes were removed by 0.5× SSC comprising 50% formamide at 37°C. The anti-biotin monoclonal antibody and the secondary antibody conjugated to AlexaFluor®647 were utilized to detect biotin-labeled NR2F1-AS1 and miR-145-5p. The cells were cleaned with PBS and then put on the coverslip with a reagent containing DAPI (Cell Signaling Technology, Boston, MA, USA) [Citation38].
2.9 Protein isolation and Western blot (WB)
After the cells were processed with varying factors, the primary culture medium was discarded. The RIPA (containing 1% PMSF) lysis buffer was employed to lyse the cells, which were collected through low-speed centrifugation to isolate the total protein. Then, the protein quantification was made with the Bradford method, and the samples were boiled for 5 minutes and centrifuged for 30 s after ice-cooling. Afterward, the supernatant was taken for polyacrylamide gel electrophoresis, and 30 μg of total protein was loaded onto a 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore, USA). After being sealed with 10% skim milk powder solution for two hours, the membranes were maintained with the primary antibodies of FOXO1 (Abcam, 1:1000, ab52857, MA, USA), p-FOXO1 (1:1000, ab259337), Bax (1:1000, ab32503), Bcl2 (1:1000, ab182858), and GAPDH (ab9485, 1:1000) overnight. After that, the membranes were flushed with TBST twice and kept with the fluorescein-labeled secondary antibody at RT for 1 hour. Finally, the membranes were flushed 3 times, exposed with the ECL chromogenic agent, and imaged with the membrane scanner [Citation39].
2.10 Dual-luciferase reporter assay
TargetScan software indicated that FOXO1 was an underlying target of miR-145-5p, while miR-145-5p was that of NR2F1-AS1. The reporter plasmids of wild-type and mutant NR2F1-AS1 and SAMD3-3’UTR were constructed, and miR-145-5p mimics and their negative controls were transfected into NP cells. The experiment was carried out 48 hours later as per the dual-luciferase reporter assay instructions (Promega, Madison, WI, USA). The relative fluorescence intensity of different treatment groups was estimated following the ratio of firefly fluorescence intensity/renilla fluorescence intensity detected by the microplate reader [Citation40].
2.11 RNA immunoprecipitation (RIP)
RIP was conducted with the Magna RIP Kit (EMD Millipore, Billerica, MA, United States) as per the manufacturer’s instructions. Following cell lysis with the RIP lysis buffer, the human anti-Ago-2 antibody (microporous) or the control antibody (normal mouse immunoglobulin, micropores) was added and maintained overnight at 4°C. The expression of NR2F1-AS1 and FOXO1 was assessed by qRT-PCR [Citation41].
2.12 Statistical analysis
The SPSS17.0 statistical software (SPSS Inc., Chicago, IL, USA) was adopted for analysis. Measurements were presented as mean ± standard deviation (x ± s). Pearson analysis was adopted to determine the correlation between NR2F1-AS1 and miR-145-5p in NP tissues. StarBase (https://starbase.sysu.edu.cn/) was utilized to predict the miRNA targets of lncRNAs [Citation42]. The multi-factor comparison was made by one-way analysis of variance, and t test was utilized for comparison between the two groups. P< 0.05 signified statistical significance.
3. Results
3.1 LncRNA NR2F1-AS1 was up-regulated, and miR-145-5p was down-regulated in NP tissues of IVDD patients
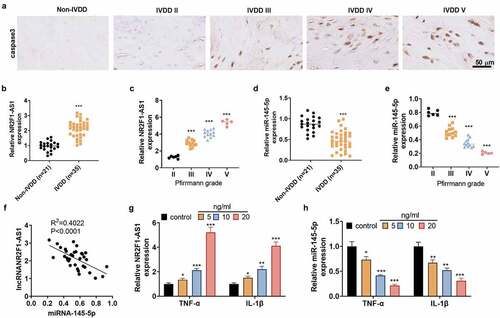
To figure out the expression of LncRNA NR2F1-AS1 in NP cells, we harvested human intervertebral disc tissues from IVDD patients and non-IVDD patients. IHC was performed for detecting Caspase3 in the tissues. We found that intervertebral disc tissues from non-IVDD patients had low expression of Caspase3, whereas Caspase3 was gradually increased with the increasing of Pfirrmann grades (). The NR2F1-AS1 expression was detected by RT-PCR. NR2F1-AS1 was confirmed to be up-regulated in intervertebral disc tissues from IVDD patients compared with that in the non-IVDD patients (). The expression level of NR2F1-AS1 was strengthened with the deterioration of IVDD (). The miR-145-5p expression was examined by qRT-PCR, and it was confirmed to be down-regulated in intervertebral disc tissues from IVDD patients compared with that in the non-IVDD patients () and gradually decreased with the deterioration of IVDD (). Linear regression analysis showed that NR2F1-AS1 was reversely related to miR-145-5p (). To probe the correlation between inflammation and the expression of NR2F1-AS1 and miR-145-5p, we treated normal NP cells with TNF-α (5, 10, 20 ng/mL) and IL-1β (5,10, 20 ng/mL) for 24 hours. As a result, TNF-α or IL-1β dose-dependently induced up-regulation of NR2F1-AS1 and down-regulation of miR-145-5p in NP cells versus the Con group (). These outcomes disclosed that NR2F1-AS1 and miR-145-5p were abnormally expressed in IVDD and contributed to its pathogenesis.
Figure 1. NR2F1-AS1 was up-regulated, and miR-145-5p was down-regulated in NP tissues of IVDD patients.
3.2 NR2F1-AS1 facilitated extracellular matrix denaturation and apoptosis in human NP cells
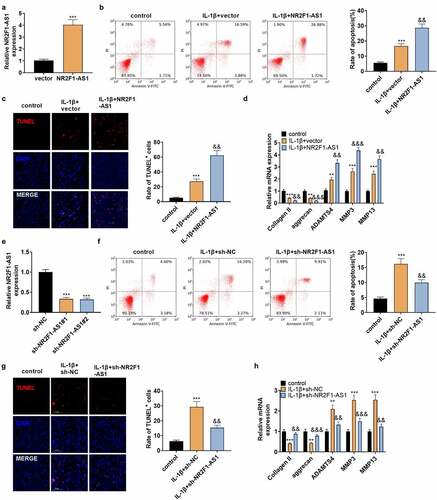
To make certain the impact of NR2F1-AS1 on NP cells, NP cells were separated from NP tissues of IVDD patients and transfected with NR2F1-AS1 overexpression plasmids. qPT-PCR verified that NR2F1-AS1 was up-regulated in NP cells (). IL-1β was adopted to treat the transfected NP cells, and NP cell apoptosis was verified by FCM and TUNEL staining. The outcomes illustrated that by contrast with the control group, the apoptotic rate and TUNEL-positive cell number in IL-1β-treated NP cells increased significantly, and the increase was strengthened after NR2F1-AS1 overexpression (). The profiles of Collagen II, aggrecan, ADAMTS4, MMP3, and MMP13 were examined by qRT-PCR. Notably, compared with the control group, ADAMTS4, MMP3, and MMP13 were highly expressed, while collagen II and aggrecan were down-regulated in IL-1β-treated NP cells. After NR2F1-AS1 overexpression, Collagen II, and aggrecan was downregulated, and ADAMTS4, MMP3, and MMP13 were further upregulated (compared with IL-1β+vector group, ). Additionally, we transfected IL-1β-treated NP cells with sh-NR2F1-AS1 to reversely verify the influence of NR2F1-AS1 on NP cells. As testified by qPT-PCR, NR2F1-AS1 was down-regulated in NP cells (vs. IL-1β+sh-NC group, ). NP cell apoptosis was lower in the IL-1β+NR2F1-AS1 group than that of the IL-1β+sh-NC group (). Followed by NR2F1-AS1 downregulation, the expression of ADAMTS4, MMP3, and MMP13 was decreased, while collagen II and aggrecan were up-regulated (p < 0.05 vs.the IL-1β+sh-NC group, ). Thus, NR2F1-AS1 intensifixed apoptosis and ECM degeneration in NP cells, while inhibiting NR2F1-AS1 had the reverse effect.
Figure 2. NR2F1-AS1 enhanced ECM degeneration and human NP cell apoptosis.
3.3 miR-145-5p hindered extracellular matrix degeneration and apoptosis in human NP cells
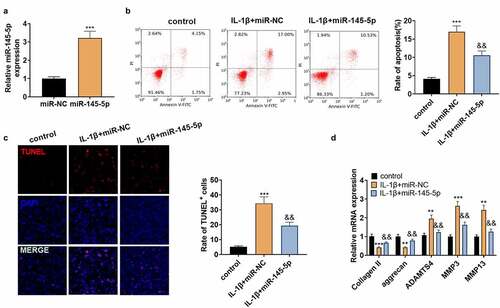
The above study revealed that miR-145-5p was down-regulated in NP tissues of IVDD patients, but it is not clear about the role of miR-145-5p in IVDD. Hence, NP cells were transfected with miR-145-5p mimics, and then qRT-PCR revealed that 145–5p was up-regulated in NP cells (vs.IL-1β+miR-NC group, ). Meanwhile, in comparison to the IL-1β+miR-NC group, up-regulating miR-145-5p reduced the apoptotic rate of NP cells and the TUNEL-positive cell number (). Subsequently, qRT-PCR manifested that ADAMTS4, MMP3, and MMP13 were significantly down-regulated following the transfection of miR-145-5p mimics, but the expression of collagen II and aggrecan were increased (). These results manifested that up-regulating miR-145-5p repressed NP cell apoptosis and ECM degradation.
Figure 3. miR-145-5p dampened ECM degeneration and NP cell apoptosis.
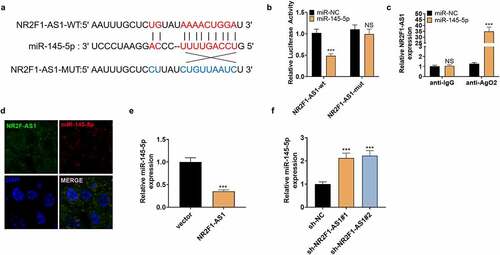
3.4 NR2F1-AS1 targeted miR-145-5p
We searched the target genes of NR2F1-AS1 on StarBase (http://starbase.sysu.edu.cn/index.php), which revealed that miR-145-5p was targeted by NR2F1-AS1 ( A). The dual-luciferase reporter assay was performed to affirm the association between the two. Notably, miR-145-5p overexpression weakened the luciferase activity of NP cells transfected with NR2F1-AS1-WT, while it had little impact on that of NP cells transfected with the NR2F1-AS1-mut vector (). Furthermore, the RIP experiment was implemented to clarify the association between the two. It turned out that after the miR-145-5p transfection, the amount of NR2F1-AS1 precipitated in the Ago2 antibody group was more than that in the IgG group, hinting that NR2F1-AS1 bound to Ago2 via miR-145-5p (). The FISH experiment exhibited that NR2F1-AS1 and miR-145-5p were expressed in the cytoplasm (). Finally, we observed that overexpressing NR2F1-AS1 reduced the miR-145-5p expression, while knocking down NR2F1-AS1 exerted opposite effects (). The above results verified that NR2F1-AS1 targeted miR-145-5p.
Figure 4. NR2F1-AS1 targeted miR-145-5p.
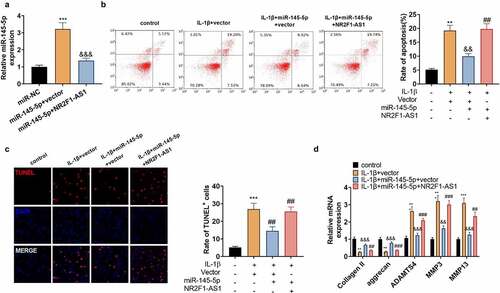
3.5 NR2F1-AS1 strengthened extracellular matrix degradation and apoptosis in human NP cells through miR-145-5p/FOXO1
The above data corroborated that NR2F1-AS1 targeted miR-145-5p. Next, we probed whether NR2F1-AS1 exerted a pro-apoptotic effect on NP cells via miR-145-5p. We transfected NP cells with miR-145-5p mimics with, or without NR2F1-AS1 overexpression plasmids to determine whether NR2F1-AS1 enhanced NP cell apoptosis and ECM degradation by targeting miR-145-5p. qRT-PCR data illustrated that compared with the miR-145-5p+vector group, miR-145-5p was down-regulated after overexpressing NR2F1-AS1 (). Besides, NR2F1-AS1 overexpression led to strengthened NP cell apoptosis versus the IL+1β+miR-145-5p+vector group (). qRT-PCR confirmed that ADAMTS4, MMP3, and MMP13 were up-regulated, while collagen II and aggrecan were down-regulated following NR2F1-AS1 overexpression versus the IL+1β+miR-145-5p+vector group (). These outcomes uncovered that NR2F1-AS1 increased ECM degradation and apoptosis in human NP cells by down-regulating miR-145-5p.
Figure 5. NR2F1-AS1 strengthened the ECM denaturation and apoptosis in human NP cells via miR-145-5p/FOXO1.
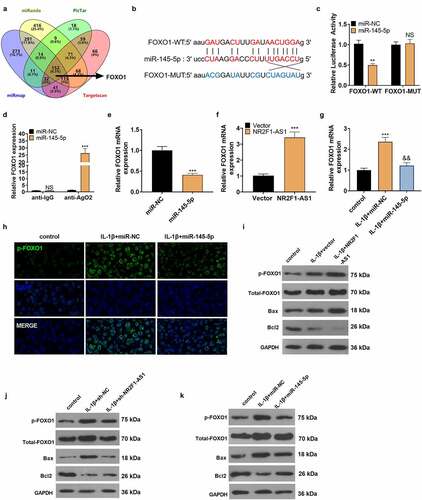
3.6 miR-145-5p targeted FOXO1
By searching the miRanda, PicTar, miRmap, and Targetscan websites, we discovered that miR-145-5p has a total of 152 potential mRNA targets, including FOXO1 (). Starbase software revealed that miR-145-5p had binding sites with FOXO1 (). To make certain whether miR-145-5p is bound to the predicted target site in FOXO1, we constructed wild-type and mutant (it was assumed that the binding site of miR-145-5p was mutated) FOXO1 luciferase reporter vectors. As expected, miR-145-5p overexpression distinctly abated the luciferase activity of NP cells transfected with FOXO1-WT, but it had no influence on the that of FOXO1-MUT (). Additionally, RIP analysis signified that FOXO1 and miR-145-5p were rich in Ago2 microribonucleoprotein complexes, indicating that Ago2 is directly bound to FOXO1 and miR-145-5p in NP cells (). miR-145-5p mimics were transfected into NP cells, and qRT-PCR confirmed that the FOXO1 mRNA expression was suppressed versus the control group (). In addition, we conducted qRT-PCR, which displayed that up-regulation of NR2F1-AS1 uplifted the mRNA expression of FOXO1 versus the Vector group (). Then miR-145-5p mimics were transfected into IL-1β-processed NP cells. As testified by qRT-PCR, FOXO1 was notably up-regulated in IL-1β-induced NP cells versus the Con group, and up-regulation of miR-145-5p reduced FOXO1 expression in NP cells (). Cellular immunofluorescence showed that the fluorescence intensity of p-FOXO1 in NP cells transfected with miR-145-5p was weaker than that of the IL-1β+miR-NC group (). IL-1β-treated NP cells were transfected with NR2F1-AS1, sh-NR2F1-AS1, and miR-145-5p. WB results illustrated that by contrast with the IL-1β+vector group, NR2F1-AS1 increased p-FOXO1, total FOXO1 and the expression of the pro-apoptotic protein Bax, and repressed the expression of the anti-apoptotic protein Bcl2. On the contrary, down-regulating NR2F1-AS1 or up-regulating miR-145-5p exerted the opposite effects (). The above results verified that FOXO1 was the target of miR-145-5p. miR-145-5p was negatively correlated with FOXO1, and NR2F1-AS1 was positively correlated with FOXO1 ().
Figure 6. miR-145-5p targeted FOXO1.
4. Discussion
IVDD is one primary reason for backache, which is a serious socio-economic burden. IVDD is usually defined by changes in tissue function and structure, including excessive ECM degradation and increased intervertebral disc cell senescence and death [Citation43]. Inflammatory processes exacerbated by TNF-α, IL-1, and IL-6 are key mediators of IVDD and backache [Citation44]. Therefore, the association between lncRNAs, ECM degradation and inflammation was studied in this paper. The possible mechanism was that NR2F1-AS1 increases the FOXO1 expression by down-regulating miR-145-5p, thus facilitating inflammation-mediated NP cell apoptosis and ECM degradation ().
Emerging evidence shows that noncoding RNAs exert an essential role in the biological process of IVDD [Citation5,Citation6]. For example, the ectopic expression of LINC00958 promotes NP cell proliferation, dampens the expression of collagen II and aggrecan, and facilitates the expression of MMP-2 and MMP-13[Citation45]. Overexpressing lncRNA RMRP enhances NP cell growth, elevates the expression of collagen II and aggrecan, and abates the expression of MMP13 and ADAMTS4[Citation46]. Besides, MALAT1[Citation47], lncRNA TRPC7-AS1[Citation48], and lncRNA-RP11-296A18.3 [Citation49] all contribute to IVDD. The role of lncRNA NR2F1-AS1 in tumors has been extensively studied [Citation50,Citation51]. Here, we confirmed that NR2F1-AS1 was up-regulated during IVDD progression. NR2F1-AS1 overexpression increased IL-1β-mediated NP cell apoptosis and ECM degradation, suggesting NR2F1-AS1 is a potential biomarker in IVDD evolvement and treatment.
miRNAs have been confirmed to contribute to diversified pathological processes of IVDD, such as apoptosis, ECM degradation, cell proliferation and inflammation[Citation52]. For instance, Zhao K et al. found that miRNA-143 enhances NP cell apoptosis by directly targeting BCL2, providing an underlying treatment option for IVDD[Citation53]. Wang J et al. stated that the miR-154 level is elevated in NP cells in IVDD patients. Besides, the inhibition of miR-154 strengthens the protein profile of collagen II and aggrecan and reduces the mRNA expression of MMP13 and ADAMTS4, while miR-154 overexpression reverses the effect[Citation54]. Also, Zhang et al. confirmed that miR-222 is uplifted in IVDD tissues and LPS-treated nucleus pulposus cells, and miR-222 significantly promotes the generation of TNF-α, IL-1β, and IL-6[Citation55]. Moreover, miR-98, miR-149, miR-27b, and miR-133a are all related to the degree of IVDD [Citation56–59]. Nevertheless, the function of miR-145-5p in IVDD remains elusive. Here, we testified that miR-145-5p was downregulated in IVDD patients and overexpressing miR-145-5p relieved IL-1β induced NP cell apoptosis and ECM degradation. As a downstream target of NR2F1-AS1, miR-145-5p was inhibited by the latter. The rescue experiments indicated that the NR2F1-AS1/ miR-145-5p has a potential role during IVDD progression.
A previous report indicates that FOXOs are key regulators of IVDD homeostasis during aging. Maintaining or restoring FOXO expression can be used as a therapeutic strategy to delay the onset of IVDD [Citation60,Citation61]. FOXO3, one member of FOXO families, has potent effects in protecting nucleus pulposus cells against apoptosis by repressing inflammation, ECM degradation, oxidative stress. [Citation62–64] In addition, studies by Chai X et al. showed that FOXO1 is up-regulated in LPS-treated NP cells, and overexpressing FOXO1 aggravates LPS-induced NP cell damages [Citation65]. Previous studies have found that FOXO1, a vital transcription factor in cells, promotes inflammatory reactions and oxidative stress [Citation66,Citation67]. FOXO1a also gets involved in IVDD progression by driving annulus fibrosus (AF) cells apoptosis through mitochondrial-related pathway[Citation68]. which is consistent with our study. Here, we substantiated that FOXO1 was up-regulated in degenerated human NP cells, and miR-145-5p mimics could down-regulate FOXO1. At the same time, the expression of NR2F1-AS1 and FOXO1 was positively correlated in NP cells, suggesting that NR2F1-AS1 and miRNA-145-5p targeted and regulated FOXO1 to affect IVDD progression.
5. Conclusion
Collectively, this study demonstrated that NR2F1-AS1 modulates the FOXO1 axis by sponging miR-145-5p as a ceRNA, thus regulating NP cell apoptosis and ECM degradation. This research reveals the mechanism of the lncRNA NR2F1-AS1/miR-145-5p/FOXO1 axis in NP cell damage in in-vitro experiments, providing potential therapeutic targets for IVDD. However, further experiment should be conducted for confirming the role of lncRNA NR2F1-AS1/miR-145-5p/FOXO1 axis in IVDD animal model.
Authors’ contribution
Conceived and designed the experiments: Longlong Du, Yu Deng, Baohui Wang
Performed the experiments: Longlong Du, Xuefeng Li, Qimeng Gao, Puwei Yuan, Yindi Sun, Yingpu Chen, Bo Huang.
Statistical analysis: Longlong Du, Xuefeng Li, Baohui Wang
Wrote the paper: Longlong Du, Xuefeng Li, Bo Huang, Baohui Wang
All authors read and approved the final manuscript.
Ethics Approval
Our study was approved by the Research Ethics Committee of Zhongnan Hospital of Wuhan University (Approved number: WUZN-2019-0344).
Consent for Publication
The written informed consent from 35 IVDD patients and 21 healthy donors of this study were obtained.
Data Availability
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
ceRNA: competitive endogenous RNA; ECM: extracellular matrix; FCM: Flow cytometry; FISH: fluorescence in situ hybridization; FOXO1: forkhead box protein O 1; IHC: immunohistochemistry; IL-1β: interleukin–1β; IVDD: Intervertebral disc degeneration; lncRNAs: long noncoding RNAs; LPS: lipopolysaccharide; MMP9: matrix metalloproteinase-9; MUT: mutant; NC: negative control; NP: nucleus pulposus; NR2F1-AS1: nuclear receptor subfamily 2 group F member 1 antisense RNA 1; NS: no significance; OA: osteoarthritis; OS: osteosarcoma; PVDF: polyvinylidene fluoride; RA rheumatoid arthritis; RIP: RNA immunoprecipitation; RT-PCR: Reverse transcription-polymerase chain reaction; shRNA: short hairpin RNA; TUNEL: TdT-mediated dUTP nick end labeling; UTR: untranslated region; WT: wild type.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Oichi T, Taniguchi Y, Oshima Y, et al. Pathomechanism of intervertebral disc degeneration. JOR Spine. 2020;3(1):e1076.
- Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1):44‐56.
- Reid PC, Morr S, Kaiser MG. State of the union: a review of lumbar fusion indications and techniques for degenerative spine disease. J Neurosurg Spine. 2019;31(1):1–14.
- Kadow T, Sowa G, Vo N, et al. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthop Relat Res. 2015;473(6):1903–1912.
- Guo HY, Guo MK, Wan ZY, et al. Emerging evidence on noncoding-RNA regulatory machinery in intervertebral disc degeneration: a narrative review. Arthritis Res Ther. 2020;22(1):270.
- Simion V, Zhou H, Haemmig S, et al. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat Commun. 2020;11(1):6135.
- Jiang N, Meng X, Mi H, et al. Circulating LncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin Chim Acta. 2018;486:26‐33.
- Liao K, Xu J, Yang W, et al. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol Immunol. 2018;101:182‐188.
- Sallam T, Sandhu J, Tontonoz P. Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ Res. 2018;122(1):155‐166.
- Chen WK, Yu XH, Yang W, et al. LncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(1):e12313.
- Xi Y, Jiang T, Wang W, et al. Long noncoding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep. 2017;7(1):13234.
- Zhan S, Wang K, Xiang Q, et al. LncRNA HOTAIR up-regulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J Cell Physiol. 2020;235(3):2195‐2208.
- Zhang Y, Zheng A, Xu R, et al. NR2F1-induced NR2F1-AS1 promotes esophageal squamous cell carcinoma progression via activating Hedgehog signaling pathway. Biochem Biophys Res Commun. 2019;519(3):497‐504.
- Ren P, Zhang H, Chang L, et al. LncRNA NR2F1-AS1 promotes proliferation and metastasis of ESCC cells via regulating EMT. Eur Rev Med Pharmacol Sci. 2020;24(7):3686‐3693.
- Li S, Zheng K, Pei Y, et al. Long noncoding RNA NR2F1-AS1 enhances the malignant properties of osteosarcoma by increasing forkhead box A1 expression via sponging of microRNA-483-3p. Aging (Albany NY). 2019;11(23):11609–11623.
- Li Z, Yu X, Shen J, et al. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48(3):278‐283.
- Chai X, Si H, Song J, et al. miR-486-5p inhibits inflammatory response, matrix degradation and apoptosis of nucleus pulposus cells through directly targeting FOXO1 in intervertebral disc degeneration. Cell Physiol Biochem. 2019;52(1):109‐118.
- Sun JC, Zheng B, Sun RX, et al. MiR-499a-5p suppresses apoptosis of human nucleus pulposus cells and degradation of their extracellular matrix by targeting SOX4. Biomed Pharmacother. 2019;113:108652.
- Yang Q, Guo XP, Cheng YL, et al. MicroRNA-143-5p targeting eEF2 gene mediates intervertebral disc degeneration through the AMPK signaling pathway. Arthritis Res Ther. 2019;21(1):97.
- Hua WB, Wu XH, Zhang YK, et al. Dysregulated miR-127-5p contributes to type II collagen degradation by targeting matrix metalloproteinase-13 in human intervertebral disc degeneration. Biochimie. 2017;139:74‐80.
- Yuan M, Zhang L, You F, et al. MiR-145-5p regulates hypoxia-induced inflammatory response and apoptosis in cardiomyocytes by targeting CD40. Mol Cell Biochem. 2017;431(1–2):123–131.
- Ma S, Zhang C, Zhang Z, et al. Geniposide protects PC12 cells from lipopolysaccharide-evoked inflammatory injury via up-regulation of miR-145-5p. Artif Cells Nanomed Biotechnol. 2019;47(1):2875–2881.
- Wu J, He Y, Luo Y, et al. MiR-145-5p inhibits proliferation and inflammatory responses of RMC through regulating AKT/GSK pathway by targeting CXCL16. J Cell Physiol. 2018;233(4):3648–3659.
- Wang X, Tang K, Wang Y, et al. Elevated microRNA-145-5p increases matrix metalloproteinase-9 by activating the nuclear factor-κB pathway in rheumatoid arthritis. Mol Med Rep. 2019;20(3):2703–2711.
- Xing YQ, Li A, Yang Y, et al. The regulation of FOXO1 and its role in disease progression. Life Sci. 2018;193:124–131.
- Duffy T, Bekki H, Lotz MK. Genome-wide occupancy profiling reveals critical roles of FoxO1 in regulating extracellular matrix and circadian rhythm genes in human chondrocytes. Arthritis Rheumatol. 2020;72(9):1514–1523.
- Wang A, Hu N, Zhang Y, et al. MEG3 promotes proliferation and inhibits apoptosis in osteoarthritis chondrocytes by miR-361-5p/FOXO1 axis. BMC Med Genomics. 2019;12(1):201.
- Liu S, Yang SD, Huo XW, et al. 17β-Estradiol inhibits intervertebral disc degeneration by down-regulating MMP-3 and MMP-13 and up-regulating type II collagen in a rat model. Artif Cells Nanomed Biotechnol. 2018;46(sup2):182–191.
- Che YJ, Guo JB, Liang T, et al. Assessment of changes in the micro-nano environment of intervertebral disc degeneration based on Pfirrmann grade. Spine J. 2019 Jul;19(7):1242–1253. Epub 2019 Jan 30.
- Shao J, Yu M, Jiang L, et al. Differences in calcification and osteogenic potential of herniated discs according to the severity of degeneration based on Pfirrmann grade: a cross-sectional study. BMC Musculoskelet Disord. 2016;17(1):191.
- Pfirrmann CW, Metzdorf A, Zanetti M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976). 2001;26(17):1873–1878.
- Tang N, Dong Y, Chen C, et al. Anisodamine maintains the stability of intervertebral disc tissue by inhibiting the senescence of nucleus pulposus cells and degradation of extracellular matrix via Interleukin-6/janus kinases/signal transducer and activator of transcription 3 pathway. Front Pharmacol. 2020 Published 2020 Dec 15;11:519172.
- Shi J, Cao F, Chang Y, et al. Long non-coding RNA MCM3AP-AS1 protects chondrocytes ATDC5 and CHON-001 from IL-1β-induced inflammation via regulating miR-138-5p/SIRT1. Bioengineered. 2021;12(1):1445–1456.
- Liang Z, Xu J, Ma Z, et al. MiR-187 suppresses non-small-cell lung cancer cell proliferation by targeting FGF9. Bioengineered. 2020 Dec;11(1):70–80.
- Tong L, Tang C, Cai C, et al. Upregulation of the microRNA rno-miR-146b-5p may be involved in the development of intestinal injury through inhibition of Kruppel-like factor 4 in intestinal sepsis. Bioengineered. 2020 Dec;11(1):1334–1349.
- Han B, Ge Y, Cui J, et al. Down-regulation of lncRNA DNAJC3-AS1 inhibits colon cancer via regulating miR-214-3p/LIVIN axis. Bioengineered. 2020 Dec;11(1):524–535.
- Zhong X, Xiu H, Bi Y, et al. Targeting eIF5A2 inhibits prostate carcinogenesis, migration, invasion and metastasis in vitro and in vivo. Bioengineered. 2020 Jan 1;11(1):619–627.
- Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020 Jun 3;19(1):101.
- Zhang N, Liu JF. MicroRNA (MiR)-301a-3p regulates the proliferation of esophageal squamous cells via targeting PTEN. Bioengineered. 2020 Dec;11(1):972–983.
- Jiao H, Chen R, Jiang Z, et al. miR-22 protect PC12 from ischemia/reperfusion-induced injury by targeting p53 up-regulated modulator of apoptosis (PUMA). Bioengineered. 2020 Dec;11(1):209–218.
- Sun Z, Zhang A, Hou M, et al. Circular RNA hsa_circ_0000034 promotes the progression of retinoblastoma via sponging microRNA-361-3p. Bioengineered. 2020 Dec;11(1):949–957.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014 Jan;42(D1):D92–7. Epub 2013 Dec 1.
- Wang F, Cai F, Shi R, et al. Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthritis Cartilage. 2016;24(3):398–408.
- Johnson ZI, Schoepflin ZR, Choi H, et al. Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur Cell Mater. 2015;30:104–117.
- Zhao K, Zhang Y, Yuan H, et al. Long noncoding RNA LINC00958 accelerates the proliferation and matrix degradation of the nucleus pulposus by regulating miR-203/FOXO1. Aging (Albany NY). 2019;11(23):10814–10825.
- Wang X, Peng L, Gong X, et al. LncRNA-RMRP promotes nucleus pulposus cell proliferation through regulating miR-206 expression. J Cell Mol Med. 2018;22(11):5468–5476.
- Zheng H, Wang T, Li X, et al. LncRNA MALAT1 exhibits positive effects on nucleus pulposus cell biology in vivo and in vitro by sponging miR-503. BMC Mol Cell Biol. 2020;21(1):23.
- Wang X, Li D, Wu H, et al. LncRNA TRPC7-AS1 regulates nucleus pulposus cellular senescence and ECM synthesis via competing with HPN for miR-4769-5p binding. Mech Ageing Dev. 2020;190:111293.
- Wang X, Lv G, Li J, et al. LncRNA-RP11-296A18.3/miR-138/HIF1A pathway regulates the proliferation ECM synthesis of human nucleus pulposus cells (HNPCs). J Cell Biochem. 2017;118(12):4862–4871.
- Wang L, Zhao S, Mingxin YU. LncRNA NR2F1-AS1 is involved in the progression of endometrial cancer by sponging miR-363 to target SOX4. Pharmazie. 2019;74(5):295–300.
- Peng J, Hou F, Zhu W, et al. LncRNA NR2F1-AS1 regulates miR-17/SIK1 axis to suppress the invasion and migration of cervical squamous cell carcinoma cells. Reprod Sci. 2020;27(7):1534–1539.
- Wang C, Wang WJ, Yan YG, et al. MicroRNAs: new players in intervertebral disc degeneration. Clin Chim Acta. 2015;450:333–341.
- Zhao K, Zhang Y, Kang L, et al. Epigenetic silencing of miRNA-143 regulates apoptosis by targeting BCL2 in human intervertebral disc degeneration. Gene. 2017;628:259–266.
- Wang J, Liu X, Sun B, et al. up-regulated miR-154 promotes ECM degradation in intervertebral disc degeneration. J Cell Biochem. 2019. DOI:10.1002/jcb.28471.10.1002/jcb.28471
- Zhang Y, Yang J, Zhou X, et al. Knockdown of miR-222 inhibits inflammation and the apoptosis of LPS-stimulated human intervertebral disc nucleus pulposus cells. Int J Mol Med. 2019;44(4):1357–1365.
- Ji ML, Lu J, Shi PL, et al. Dysregulated miR-98 contributes to extracellular matrix degradation by targeting IL-6/STAT3 signaling pathway in human intervertebral disc degeneration. J Bone Miner Res. 2016;31(4):900–909.
- Qin C, Lv Y, Zhao H, et al. MicroRNA-149 suppresses inflammation in nucleus pulposus cells of intervertebral discs by regulating MyD88. Med Sci Monit. 2019;25:4892–4900.
- Li HR, Cui Q, Dong ZY, et al. Downregulation of miR-27b is involved in loss of Type II collagen by directly targeting matrix metalloproteinase 13 (MMP13) in human intervertebral disc degeneration. Spine(Phila Pa 1976). 2016;41(3):E116–E123.
- Xu YQ, Zhang ZH, Zheng YF, et al. Dysregulated miR-133a mediates loss of Type II collagen by directly targeting matrix metalloproteinase 9 (MMP9) in human intervertebral disc degeneration. Spine (Phila Pa 1976). 2016;41(12):E717–E724. Phila Pa 1976.
- Alvarez-Garcia O, Matsuzaki T, Olmer M, et al. Age-related reduction in the expression of FOXO transcription factors and correlations with intervertebral disc degeneration. J Orthop Res. 2017;35(12):2682–2691.
- Alvarez-Garcia O, Matsuzaki T, Olmer M, et al. FOXO are required for intervertebral disk homeostasis during aging and their deficiency promotes disk degeneration. Aging Cell. 2018;17(5):e12800.
- Liu Q, Tan Z, Xie C, et al. Oxidative stress as a critical factor might involve in intervertebral disc degeneration via regulating NOXs/FOXOs [published online ahead of print, 2021 Nov 9]. J Orthop Sci. 2021;S0949-2658(21):338–339.
- Xia P, Gao X, Li F, et al. Down-Regulation of microRNA-30d alleviates intervertebral disc degeneration through the promotion of FOXO3 and suppression of CXCL10. Calcif Tissue Int. 2021;108(2):252–264.
- Wang Y, Yang Y, Zuo R, et al. FOXO3 protects nucleus pulposus cells against apoptosis under nutrient deficiency via autophagy. Biochem Biophys Res Commun. 2020;524(3):756–763.
- Chai X, Si H, Song J, et al. miR-486-5p inhibits inflammatory response, matrix degradation and apoptosis of nucleus pulposus cells through directly targeting FOXO1 in intervertebral disc degeneration. Cell Physiol Biochem. 2019;52(1):109–118.
- Li Y, Yao N, Gao Y, et al. MiR-1224-5p attenuates polycystic ovary syndrome through inhibiting NOD-like receptor protein 3 inflammasome activation via targeting Forkhead box O 1. Bioengineered. 2021;12(1):8555–8569.
- Mai C, Qiu L, Zeng Y, et al. Lactobacillus casei strain shirota enhances the ability of geniposide to activate SIRT1 and decrease inflammation and oxidative stress in septic mice. Front Physiol. 2021 Published 2021 Sep 20;12:678838.
- Jing D, Wu W, Deng X, et al. FoxO1a mediated cadmium-induced annulus fibrosus cells apoptosis contributes to intervertebral disc degeneration in smoking. J Cell Physiol. 2021;236(1):677–687.