ABSTRACT
Circular RNAs (circRNAs) have shown pivotal regulatory roles in tumorigenesis and progression. Our purpose was to analyze the role of circRNA La ribonucleoprotein 1B (circ-LARP1B; hsa_circ_0070934) in cutaneous squamous cell carcinoma (CSCC) progression and its associated mechanism. Cell viability, colony formation ability, migration, and invasion were analyzed by 3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyltetrazolium bromide (MTT) assay, colony formation assay, wound healing assay, and transwell invasion assay. Flow cytometry was performed to analyze cell apoptosis and cell cycle progression. Cell glycolytic metabolism was analyzed using Glucose Uptake Colorimetric Assay kit, Lactate Assay Kit II, and ATP colorimetric Assay kit. Dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were performed to verify the interaction between microRNA-515-5p (miR-515-5p) and circ-LARP1B or TPX2 microtubule nucleation factor (TPX2). Circ-LARP1B expression was up-regulated in CSCC tissues and cell lines. Circ-LARP1B knockdown suppressed cell viability, colony formation ability, migration, invasion, cell cycle progression, and glycolysis and triggered cell apoptosis in CSCC cells. miR-515-5p was a direct target of circ-LARP1B in CSCC cells, and circ-LARP1B silencing-mediated anti-tumor effects were largely counteracted by miR-515-5p knockdown. miR-515-5p directly interacted with the 3ʹ untranslated region (3ʹUTR) of TPX2. TPX2 overexpression largely overturned miR-515-5p-mediated anti-tumor effects in CSCC cells. Circ-LARP1B could up-regulate TPX2 expression by sponging miR-515-5p in CSCC cells. Circ-LARP1B knockdown suppressed tumor growth in vivo. In conclusion, circ-LARP1B contributed to CSCC progression by targeting miR-515-5p/TPX2 axis. The circ-LARP1B/miR-515-5p/TPX2 axis might provide novel therapeutic targets for CSCC patients.
Introduction
Cutaneous squamous cell carcinoma (CSCC) is a common type of non-melanoma skin cancer (NMSC) [Citation1]. Epidemiological studies have found that the occurrence of CSCC is related to long-term sunlight exposure and ionizing radiation [Citation2,Citation3]. Although considerable improvement has been made in the therapeutic strategy of CSCC, the outcome of patients with metastatic CSCC is still dismal [Citation4]. Therefore, exploring the molecular mechanism of CSCC progression is essential to develop novel effective therapeutic methods for CSCC patients.
Circular RNAs (circRNAs) have attracted much attention in the field of human diseases since Andreeva et al. found in 2014 that circRNAs have many specific biological characteristics and are widely existed in plants and animals [Citation5,Citation6]. CircRNAs have shown important regulatory roles in cancers [Citation7] and cardiovascular diseases [Citation8]. A previous study reported that circRNA La ribonucleoprotein 1B (circ-LARP1B; hsa_circ_0070934) is up-regulated in CSCC, and it contributes to the growth and invasion of CSCC cells [Citation9]. Here, we further explored the molecular mechanism of circ-LARP1B in CSCC tumorigenesis.
It has been widely accepted that circRNAs can regulate cell biological phenotypes by sponging microRNAs (miRNAs) [Citation10,Citation11]. For example, circ-BPTF is reported to promote bladder cancer progression by sponging miR-31-5p [Citation12]. Previous articles have demonstrated that circ-LARP1B can contribute to CSCC development by sponging miR-1236-3p, miR-1238 or miR-1247-5p [Citation9,Citation13]. In this study, miR-515-5p was identified as a candidate target of circ-LARP1B by searching bioinformatics database. Yan et al. reported that circANKS1B contributes to the progression and cisplatin resistance of oral squamous cell carcinoma by sponging miR-515-5p to up-regulate TGF-β1 [Citation14], suggesting the tumor suppressor role of miR-515-5p in oral squamous cell carcinoma. Zhang et al. found that SP1-induced ZFPM2-AS1 facilitates glioma development by up-regulating SOD2 via sponging miR-515-5p [Citation15], indicating the anti-tumor role of miR-515-5p in glioma. However, the role of miR-515-5p in CSCC progression and its interaction with circ-LARP1B have never been reported.
Through bioinformatics analysis, TPX2 microtubule nucleation factor (TPX2) is predicted as a candidate target of miR-515-5p in this study. TPX2 is a cell cycle-associated molecule [Citation16]. TPX2 silencing is reported to suppress the proliferation of cervical cancer cells [Citation17]. Feng et al. found that miR-216b hampers the malignant phenotypes of CSCC cells by down-regulating TPX2 [Citation18]. These studies suggested that TPX2 might be a novel anti-tumor target. In this study, we tested the interaction and functional correlation between TPX2 and miR-515-5p in CSCC cells.
In this study, we first identified that circ-LARP1B was highly expressed in CSCC tissues and cell lines. Loss-of-function experiments were conducted to explore the role of circ-LARP1B in CSCC cells. We proposed the hypothesis that circ-LARP1B functioned in CSCC cells by targeting miRNA/messenger RNA (mRNA) axis, and rescue experiments were conducted to verify the hypothesis. Taking these findings together, circ-LARP1B may be a new effective target for CSCC therapy.
Materials and methods
Clinical tissue samples
CSCC tissues (n = 38) and matched non-tumor tissues (n = 38) were collected at General Hospital of Ningxia Medical University. The clinical study was approved by the ethics committee of General Hospital of Ningxia Medical University. CSCC patients who had received pre-operative anti-tumor therapy were excluded in this clinical study. All participates had signed the written informed consent before tissue collection. The association between circ-LARP1B expression and the clinical pathological features of CSCC patients is shown in .
Table 1. The association between circ-LARP1B expression and the clinical pathological features of CSCC patients
Cell lines
Two CSCC cell lines (A431 and HSC-1) and normal human epidermal keratinocytes (NHEK) were purchased from BeNa Culture Collection (Beijing, China) and were cultured with Dulbecco’s modified Eagle medium (DMEM, Gibco, Carlsbad, CA, USA) added with 10% fetal bovine serum (FBS, Gibco) or serum-free keratinocyte growth medium (Life Technologies, Gaithersburg, MD, USA) at 37°C with 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA samples were isolated from tissues and cells with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and were dissolved in 30 μL of enzyme-free water. Complementary DNA (cDNA) was synthesized using Script RT reagent kit (for circ-LARP1B and TPX2; Takara, Dalian, China) and Mir-X miR First-Strand Kit (for miR-515-5p; Takara). qPCR reaction was conducted using SYBR reagent (Takara) and specific primers (). The relative expression was calculated by the 2−ΔΔCt method [Citation19] and was normalized to the house-keeping gene, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6.
Table 2. Primers used in RT-qPCR
Subcellular fractionation assay
The subcellular localization of circ-LARP1B was analyzed using the PARISTM Kit Protein and RNA Isolation system (Thermo Fisher Scientific, Waltham, MA, USA) [Citation20].
Cell transfection
Small interfering (si)RNAs targeting circ-LARP1B (si-circ-LARP1B#1 and #2), siRNA negative control (si-NC), circ-LARP1B ectopic expression plasmid (circ-LARP1B), pLCDH-cir empty vector (pLCDH-cir), short hairpin (sh)RNA targeting circ-LARP1B (sh-circ-LARP1B), shRNA NC (sh-NC), miR-515-5p mimics (miR-515-5p), miR-NC, miR-515-5p inhibitor (anti-miR-515-5p), anti-miR-NC, TPX2 ectopic expression plasmid (TPX2), and pcDNA empty vector (pcDNA) were purchased from Ribobio (Shanghai, China) and GenePharma (Shanghai, China). CSCC cells were transfected with small RNAs and plasmids with Lipofectamine 3000 reagent (Invitrogen).
3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyltetrazolium bromide (MTT) assay
MTT assay was conducted as previously described [Citation21]. In brief, a total of 20 μL of 5% MTT reagent (SenBeJia Biological Technology, Jiangsu, China) was incubated with CSCC cells for 4 h. The formazan products were dissolved with 100 μL of dimethyl sulfoxide (DMSO; Sangon Biotech, Shanghai, China). The absorbance at the wavelength of 570 nm was examined.
Colony formation assay
Colony formation assay was performed as previously described [Citation22]. CSCC cells were seeded onto 12-well plates at low density and were cultured for 14 d. The culture medium was replenished every 3 d. The colonies were rinsed with phosphate-buffered saline (PBS) solution (Sangon Biotech), immobilized with 4% paraformaldehyde for 15 min at room temperature and stained with 0.5% crystal violet for 10 min at room temperature. Finally, the number of colonies was counted.
Cell apoptosis analysis
Flow cytometry was carried out to analyze cell apoptosis as previously described [Citation22]. CSCC cells were simultaneously stained with Annexin V-fluorescein isothiocyanate (FITC) (BioVision, Mountain View, CA, USA) and propidium iodide (PI; BioVision) for 15 min at room temperature in the dark. The fluorescence intensities of FITC and PI were measured with the excitation at 488 nm.
Wound healing assay
Wound healing assay was conducted as previously described [Citation23]. When cell confluence reached 90%-100%, the wound was created in cell monolayer using the 200 μL pipette tip. After washing three times with PBS buffer (Sangon Biotech), CSCC cells were cultured with serum-free medium for 24 h. Cell images were captured at 0 h and 24 h. The width of the scratch was analyzed at the magnification of 40 × .
Transwell invasion assay
Transwell invasion assay was conducted as previously described [Citation21]. The diluted Matrigel (50 μL/well; BD Biosciences, San Jose, CA, USA) was added to the upper compartments. CSCC cells in serum-free medium were seeded onto the upper compartments. The lower compartments were added with 600 μL culture medium plus 10% FBS. After incubation for 24 h, invaded CSCC cells were fixed, stained, and then photographed.
Cell cycle analysis
Flow cytometry was performed to analyze cell cycle progression as previously described [Citation24]. CSCC cells were immobilized with 80% ethanol (Sangon Biotech) at 4°C overnight. The next day, CSCC cells were incubated with RNase A for 30 min. A total of 400 μL of PI (Sigma, St. Louis, MO, USA) was added to incubate with CSCC cells. Cell cycle progression was assessed by the FACS CantoII flow cytometer (BD Biosciences).
Analysis of glycolytic metabolism
Cell glycolytic metabolism was analyzed using Glucose Uptake Colorimetric Assay kit (Biovision), Lactate Assay Kit II (Biovision), and ATP colorimetric Assay kit (Biovision) according to the manufacturer’s instructions.
Western blot assay
Protein samples (30 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were shifted onto the polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA). After blocking with 5% skimmed milk for 1 h at room temperature, the membrane was incubated with the primary antibodies at 4°C overnight. The primary antibodies are listed as below: anti-hexokinase 2 (anti-HK-2; 2867S; Cell Signaling Technology, Danvers, Massachusetts, USA), anti-cleaved caspase 3 (9661S; Cell Signaling Technology), anti-matrix metallopeptidase 2 (anti-MMP2; 40994S; Cell Signaling Technology), anti-cyclin D1 (55506S; Cell Signaling Technology), anti-TPX2 (12245S; Cell Signaling Technology), anti-phosphoinositide-3-kinase (anti-PI3K; 4257S; Cell Signaling Technology), anti-AKT (14982S; Cell Signaling Technology), anti-phosphorylated-AKT (anti-p-AKT; 4060S; Cell Signaling Technology), and anti-β-actin (ab8226; Abcam, Cambridge, MA, USA). Subsequently, the membrane was incubated with the secondary antibody for 1 h at room temperature. Protein bands were visualized using the imaging system (Thmoregan Biological Technology, Beijing, China). Western blot assay was carried out as previously described [Citation21].
Dual-luciferase reporter assay
The downstream targets of circ-LARP1B and miR-515-5p were searched by Circular RNA interactome (https://circinteractome.irp.nia.nih.gov) and Starbase (http://starbase.sysu.edu.cn).
Dual-luciferase reporter assay was conducted to verify the interaction between miR-515-5p and circ-LARP1B or TPX2 as previously described [Citation25]. The partial fragment of circ-LARP1B or TPX2 3ʹ untranslated region (3ʹUTR) was inserted to pmirGLO vector (Promega, Madison, WI, USA). These reporter plasmids were co-transfected with small RNAs into CSCC cells. Luciferase intensities were determined using a dual-luciferase reporter assay kit (Promega).
RNA immunoprecipitation (RIP) assay
CSCC cells were disrupted, and cell lysates were incubated with sepharose beads (Millipore, Billerica, MA, USA) coated with Argonaute-2 (Ago2) antibody (Bio-Rad) or Immunoglobulin G (IgG) antibody (Bio-Rad). RNA enrichment was analyzed by RT-qPCR. RIP assay was performed as previously described [Citation26].
Tumor xenograft assay
A total of 10 BALB/c male nude mice were purchased from Vital River Laboratory Animal Technology (Beijing, China). A431 cells were subcutaneously injected into the right flank of the nude mice. After inoculation for 7 d, tumor volume was measured every 5 d as width2 × length × 0.5. After inoculation for 32 d, all nude mice were killed, and the weight of xenograft tumors was recorded. All procedures were implemented with the approval of the Animal Care and Use Committee of General Hospital of Ningxia Medical University.
Statistical analysis
All data were analyzed by GraphPad Prism 7.0 software (GraphPad, La Jolla, CA, USA) and were represented as mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) were utilized to analyze the differences in two groups or multiple groups. The differences were considered statistically significant with P < 0.05.
Results
Previous studies demonstrated that circ-LARP1B is up-regulated in CSCC, and it plays an oncogenic role in CSCC progression [Citation9,Citation13]. However, the molecular mechanism underlying the oncogenic role of circ-LARP1B in CSCC remains largely unclear. Here, we first confirmed the expression pattern of circ-LARP1B in CSCC. Then, loss- and gain-of-function experiments were conducted to explore the role of circ-LARP1B in regulating the biological phenotypes of CSCC cells. After predicting and confirming the downstream miRNA/mRNA axis of circ-LARP1B, we successfully established circ-LARP1B/miR-515-5p/TPX2 axis. Then, a series of rescue experiments were conducted to verify whether the oncogenic role of circ-LARP1B was dependent on its regulation of miR-515-5p/TPX2 axis. Our findings provide a new insight into the molecular mechanism of circ-LARP1B and may offer potential targets for CSCC treatment.
Circ-LARP1B is highly expressed in CSCC tissues and cell lines
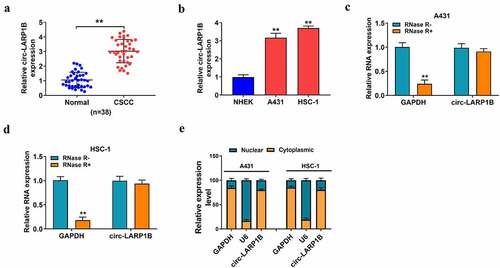
We collected 38 pairs of CSCC tissues and adjacent normal tissues to analyze the expression pattern of circ-LARP1B. Circ-LARP1B was highly expressed in CSCC tissues compared with that in adjacent normal tissues ()). The association between circ-LARP1B expression and the clinical pathological features of CSCC patients is shown in . Of note, CSCC patients with high expression of circ-LARP1B were closely associated with positive lymph node metastasis and poor histopathological grade (). The expression of circ-LARP1B was also analyzed in normal human epidermal keratinocytes (NHEK) and two CSCC cell lines (A431 and HSC-1). The data revealed that circ-LARP1B expression was significantly elevated in CSCC cell lines compared with that in NHEK cell line ()). CircRNAs are resistant to exonuclease due to the closed loop structure. Then, we used exonuclease RNase R to test whether circ_LARP1B was a circular transcript. Circ-LARP1B was to RNase R, while the expression of GAPDH was notably reduced by RNase R treatment (). The subcellular localization of circRNAs is closely associated with their biological functions. We found that circ-LARP1B was majorly distributed in the cytoplasmic fraction of CSCC cells ()), indicating that circ-LARP1B might function in the post-transcriptional level. These data suggested that circ-LARP1B might be implicated in the regulation of CSCC progression.
Figure 1. Circ-LARP1B is highly expressed in CSCC tissues and cell lines. (a) The expression of circ-LARP1B was determined in CSCC tissues (n = 38) and adjacent normal tissues (n = 38) by RT-qPCR. (b) The expression of circ-LARP1B in two CSCC cell lines (A431 and HSC-1) and normal human epidermal keratinocytes (NHEK) was measured by RT-qPCR. (c and d) The stability of circ-LARP1B was tested using exonuclease RNase R, and GAPDH was used as the control. The levels of circ-LARP1B and GAPDH were examined in RNase R digestion (RNase R+) group or RNase R- group by RT-qPCR. (e) The subcellular localization of circ-LARP1B was analyzed using the PARISTM Kit Protein and RNA Isolation system. GAPDH or U6 was served as the cytoplasmic or nuclear marker, respectively. **P < 0.01.
Circ-LARP1B silencing suppresses the malignant phenotypes of CSCC cells
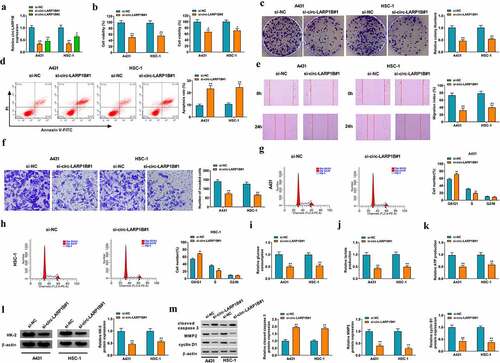
We designed two siRNAs targeting circ-LARP1B, named si-circ-LARP1B#1 and si-circ-LARP1B#2. RT-qPCR verified the transfection efficiencies of si-circ-LARP1B#1 and si-circ-LARP1B#2 in CSCC cells ()). Transfection with si-circ-LARP1B#1 or si-circ-LARP1B#2 reduced the viability of CSCC cells ()). Si-circ-LARP1B#1 was chosen for the following experiments due to its higher knockdown efficiency and stronger inhibitory effect on cell viability than si-circ-LARP1B#2. Then, we analyzed the roles of circ-LARP1B in regulating the colony formation ability, apoptosis, migration, invasion, cell cycle progression, and glycolytic metabolism of CSCC cells. As shown in ), circ-LARP1B silencing notably reduced the number of colonies, suggesting that circ-LARP1B knockdown suppressed the colony formation ability of CSCC cells. Cell apoptosis was triggered by circ-LARP1B silencing ()). Circ-LARP1B knockdown markedly reduced the distance of cell migration ()). The number of invaded cells was notably decreased by circ-LARP1B knockdown ()). These data demonstrated that circ-LARP1B silencing suppressed the migration and invasion abilities of CSCC cells. Circ-LARP1B knockdown increased the percentage of CSCC cells in G0/G1 phase and reduced the percentage of CSCC cells in S phase (), demonstrating that circ-LARP1B knockdown suppressed cell cycle progression in G1/S transition. It is widely accepted that the metabolism of cancer cells is markedly different from that of normal cells. Cancer cells exhibit a special metabolic phenotype known as the Warburg effect [Citation27], featured by the enhanced aerobic glycolysis and reduced oxidative phosphorylation. The Warburg effect promotes the survival and inhibits the apoptosis of cancer cells [Citation28]. We found that circ-LARP1B silencing restrained the consumption of glucose and the production of lactate and ATP in CSCC cells (), suggesting that circ-LARP1B knockdown suppressed the glycolytic metabolism of CSCC cells. Circ-LARP1B knockdown reduced the expression of a glycolysis-related rate-limiting enzyme HK-2 in CSCC cells ()). Meanwhile, we found that circ-LARP1B interference up-regulated the level of cleaved caspase 3 and reduced the expression of MMP2 and cyclin D1 ()), further demonstrating that circ-LARP1B knockdown induced the apoptosis and suppressed the metastasis and proliferation of CSCC cells. Taken together, circ-LARP1B silencing suppressed the malignant behaviors of CSCC cells. Si-circ-LARP1B#1 was written as si-circ-LARP1B in the following experiments.
Figure 2. Circ-LARP1B silencing suppresses the malignant phenotypes of CSCC cells. (a) RT-qPCR was conducted to analyze the knockdown efficiencies of two siRNAs of circ-LARP1B (si-circ-LARP1B#1 and si-circ-LARP1B#2). (b-m) CSCC cells were transfected with si-NC or si-circ-LARP1B#1. (b) Cell viability was analyzed by MTT assay. (c) The colony formation ability of CSCC cells was analyzed by colony formation assay. (d) Cell apoptosis was measured by flow cytometry. (e) Cell migration ability was assessed by wound healing assay. (f) Cell invasion capacity was detected by transwell invasion assay. (g and h) Cell cycle progression was analyzed by flow cytometry. (i-k) Cell glycolytic metabolism was analyzed using Glucose Uptake Colorimetric Assay kit, Lactate Assay Kit II, and ATP colorimetric Assay kit. (l) Western blot assay was conducted to determine the protein expression of HK-2 in CSCC cells. (m) The levels of apoptosis-, metastasis-, and proliferation-associated proteins (cleaved caspase 3, MMP2, and cyclin D1) were detected by Western blot assay. *P < 0.05, **P < 0.01.
To further confirm the role of circ-LARP1B in CSCC cells, overexpression experiments were conducted. We found that circ-LARP1B overexpression promoted the viability, invasion, and cell cycle progression and suppressed the apoptosis of CSCC cells (Supplementary Figure S1a-e). In addition, circ-LARP1B overexpression increased the protein expression of HK-2 (Supplementary figure S1f), suggesting that circ-LARP1B might promote cell glycolysis in CSCC cells. These data further demonstrated that circ-LARP1B played an oncogenic role in CSCC cells.
miR-515-5p is a direct target of circ-LARP1B in CSCC cells
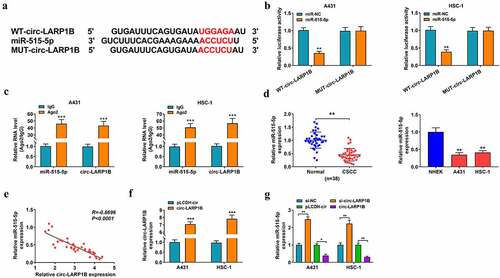
The potential binding sites between circ-LARP1B and miR-515-5p were predicted by Circular RNA interactome software ()). The luciferase activity of wild-type reporter plasmid (WT-circ-LARP1B) was notably decreased by miR-515-5p overexpression, and the luciferase activity of mutant reporter plasmid (MUT-circ-LARP1B) was unaffected by the transfection of miR-NC or miR-515-5p ()), suggesting that miR-515-5p directly interacted with circ-LARP1B in CSCC cells. RIP assay revealed that miR-515-5p and circ-LARP1B were simultaneously enriched in RNA induced silencing complex (RISC) when using Ago2 antibody ()), further suggesting the interaction between miR-515-5p and circ-LARP1B. As shown in ), miR-515-5p expression was down-regulated in CSCC tissues (n = 38) compared with adjacent normal tissues (n = 38). miR-515-5p expression was also down-regulated in A431 and HSC-1 cells compared with NHEK cells ()). There was a negative linear correlation between the expression of miR-515-5p and circ-LARP1B in CSCC tissues ()). The overexpression efficiency of circ-LARP1B plasmid was verified by RT-qPCR ()). Circ-LARP1B interference up-regulated miR-515-5p expression, while circ-LARP1B overexpression reduced miR-515-5p expression in CSCC cells ()). Overall, miR-515-5p was a direct target of circ-LARP1B in CSCC cells.
Figure 3. miR-515-5p is a direct target of circ-LARP1B in CSCC cells. (a) Circular RNA interactome database predicted the interaction between circ-LARP1B and miR-515-5p. (b) The interaction between circ-LARP1B and miR-515-5p was confirmed by dual-luciferase reporter assay. (c) The target relationship between circ-LARP1B and miR-515-5p was verified by RIP assay. (d) The expression of miR-515-5p in CSCC tissues and cell lines was detected by RT-qPCR. (e) The linear correlation between the expression of circ-LARP1B and miR-515-5p in CSCC tissues was analyzed by Pearson’s correlation coefficient. (f) The overexpression efficiency of circ-LARP1B ectopic expression plasmid in CSCC cells was analyzed by RT-qPCR. (g) The expression of miR-515-5p was detected in CSCC cells transfected with si-NC, si-circ-LARP1B, pLCDH-cir, or circ-LARP1B plasmid by RT-qPCR. *P < 0.05, **P < 0.01, ***P < 0.001.
miR-515-5p interference reverses circ-LARP1B absence-induced anti-tumor effects in CSCC cells
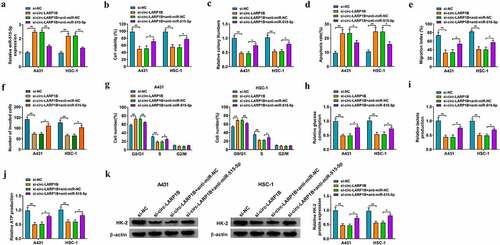
To explore whether circ-LARP1B silencing suppressed CSCC progression by up-regulating miR-515-5p in vitro, we performed rescue experiments. As shown in ), circ-LARP1B silencing elevated miR-515-5p expression, which was reversed by the addition of anti-miR-515-5p in CSCC cells. Circ-LARP1B interference-induced suppressive effects on the viability and colony formation ability of CSCC cells were overturned by miR-515-5p knockdown (). Circ-LARP1B interference-induced cell apoptosis was offset by the addition of anti-miR-515-5p ()). Circ-LARP1B knockdown suppressed the migration and invasion of CSCC cells, and the addition of anti-miR-515-5p rescued cell motility in CSCC cells (). Cell cycle progression was arrested in G0/G1 phase by circ-LARP1B knockdown, and the introduction of anti-miR-515-5p rescued cell cycle progression ()). Circ-LARP1B silencing restrained cell glycolysis, which was rescued in si-circ-LARP1B and anti-miR-515-5p co-transfected group (). Circ-LARP1B silencing reduced the expression of HK-2, which was recovered by the addition of anti-miR-515-5p in CSCC cells ()). Collectively, circ-LARP1B knockdown restrained CSCC progression partly by up-regulating miR-515-5p in CSCC cells.
Figure 4. miR-515-5p interference reverses circ-LARP1B absence-induced anti-tumor effects in CSCC cells. (a-k) A431 and HSC-1 cells were transfected with si-NC, si-circ-LARP1B, si-circ-LARP1B + anti-miR-NC or si-circ-LARP1B + anti-miR-515-5p. (a) RT-qPCR was conducted to detect the expression of miR-515-5p in transfected CSCC cells. (b) MTT assay was conducted to assess cell viability. (c) Colony formation assay was used to analyze the colony formation ability of transfected CSCC cells. (d) The apoptosis rate was measured by flow cytometry. (e) Wound healing assay was utilized to analyze the migration ability of CSCC cells. (f) Transwell invasion assay was performed to measure invasion ability of CSCC cells. (g) Cell cycle progression was evaluated by flow cytometry. (h-j) Cell glycolytic metabolism was analyzed using Glucose Uptake Colorimetric Assay kit, Lactate Assay Kit II, and ATP colorimetric Assay kit. (k) The protein expression of HK-2 was measured in CSCC cells by Western blot assay. *P < 0.05, **P < 0.01.
miR-515-5p directly interacts with TPX2 3ʹUTR in CSCC cells
TPX2 was predicted as a target of miR-515-5p by Starbase database ()). miR-515-5p overexpression significantly reduced the luciferase activity of wild-type reporter plasmid (WT-TPX2 3ʹUTR) but not that of mutant reporter plasmid (MUT-TPX2 3ʹUTR) ()), suggesting that miR-515-5p directly interacted with the 3ʹUTR of TPX2 in CSCC cells. RIP assay further demonstrated the interaction between miR-515-5p and TPX2 in CSCC cells ()). The mRNA and protein levels of TPX2 were up-regulated in CSCC tissues and cell lines (). TPX2 expression in CSCC tissues was negatively correlated with the expression of miR-515-5p ()). The overexpression efficiency of miR-515-5p mimics in CSCC cells was confirmed by RT-qPCR assay ()). TPX2 protein level was decreased by miR-515-5p overexpression, and miR-515-5p silencing up-regulated the protein expression of TPX2 in CSCC cells (). Overall, TPX2 was a direct target of miR-515-5p in CSCC cells.
Figure 5. miR-515-5p directly interacts with TPX2 3ʹUTR in CSCC cells. (a) The potential binding sites between miR-515-5p and TPX2 3ʹUTR were predicted by Starbase software. (b) Dual-luciferase reporter assay was performed to confirm the target interaction between miR-515-5p and TPX2 in CSCC cells. (c) RIP assay was conducted to verify the interaction between miR-515-5p and TPX2 in CSCC cells. (d) The expression of TPX2 mRNA in CSCC tissues (n = 38) and adjacent non-tumor tissues (n = 38) was determined by RT-qPCR. (e) The level of TPX2 mRNA in NHEK cells and two CSCC cell lines (A431 and HSC-1) was examined by RT-qPCR. (f) Western blot assay was conducted to analyze the protein expression of TPX2 in CSCC tissues and adjacent normal tissues. (g) The protein level of TPX2 in NHEK, A431 and HSC-1 cells was measured by Western blot assay. (h) The linear correlation between the expression of miR-515-5p and TPX2 in CSCC tissues was assessed by Pearson’s correlation coefficient. (i) The overexpression efficiency of miR-515-5p mimics in CSCC cells was analyzed by RT-qPCR. (j and k) Western blot assay was performed to measure the protein expression of TPX2 in A431 and HSC-1 cells transfected with miR-NC, miR-515-5p, anti-miR-NC or anti-miR-515-5p. *P < 0.05, **P < 0.01, ***P < 0.001.
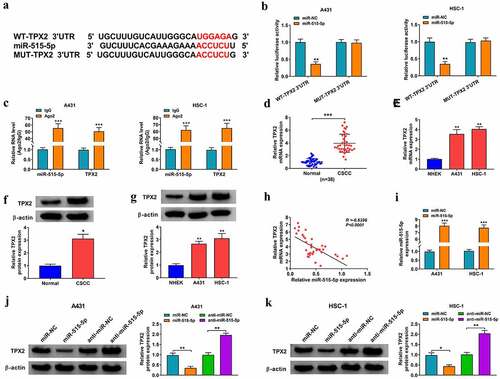
miR-515-5p overexpression-mediated anti-tumor effects in CSCC cells are overturned by the accumulation of TPX2
We performed overexpression experiments to analyze the role of TPX2 in CSCC cells. Western blot assay verified the overexpression efficiency of TPX2 plasmid in CSCC cells (Supplementary Figure S2a). TPX2 overexpression facilitated the viability, invasion, and cell cycle progression and inhibited the apoptosis of CSCC cells (Supplementary Figure S2b-2e). TPX2 overexpression also elevated HK-2 protein expression in CSCC cells (Supplementary figure S2f), indicating that TPX2 overexpression might contribute to cell glycolytic metabolism. Overall, TPX2 overexpression promoted the progression of CSCC in vitro.
To investigate whether miR-515-5p played a tumor suppressor role in CSCC cells by targeting TPX2, we performed rescue experiments. As shown in , miR-515-5p overexpression reduced the mRNA and protein expression of TPX2, which was recovered in miR-515-5p and TPX2 co-transfected group. miR-515-5p overexpression suppressed cell viability and colony formation ability, and these suppressive effects were attenuated by the addition of TPX2 plasmid in CSCC cells (). miR-515-5p overexpression-induced cell apoptosis was largely alleviated by the addition of TPX2 plasmid in CSCC cells ()). miR-515-5p overexpression suppressed the migration and invasion abilities of CSCC cells, which were largely recovered by the addition of TPX2 overexpression plasmid (). miR-515-5p overexpression-induced suppressive effect on cell cycle progression was partly attenuated by the addition of TPX2 plasmid in CSCC cells ()). miR-515-5p overexpression inhibited the uptake of glucose and the production of lactate and ATP, and these inhibitory effects were largely overturned by the introduction of TPX2 plasmid (). HK-2 expression was reduced by miR-515-5p overexpression, and the addition of TPX2 plasmid largely recovered the expression of HK-2 in CSCC cells ()). Taken together, miR-515-5p suppressed the malignant behaviors of CSCC cells partly by targeting TPX2.
Figure 6. miR-515-5p overexpression-mediated anti-tumor effects in CSCC cells are overturned by the accumulation of TPX2. (a-l) CSCC cells were transfected with miR-NC, miR-515-5p, miR-515-5p + pcDNA or miR-515-5p + TPX2. (a and b) The mRNA and protein expression of TPX2 was examined in CSCC cells by RT-qPCR and Western blot assay. (c) Cell viability was analyzed by MTT assay. (d) The colony formation ability was analyzed by colony formation assay. (e) Cell apoptosis rate was analyzed by flow cytometry. (f and g) Cell migration and invasion abilities were analyzed by wound healing assay and transwell invasion assay. (h) The percentages of CSCC cells in G0/G1 phase, S phase, and G2/M phase were analyzed by flow cytometry. (i-k) Cell glycolytic metabolism was analyzed using Glucose Uptake Colorimetric Assay kit, Lactate Assay Kit II, and ATP colorimetric Assay kit. (l) Western blot assay was conducted to analyze the protein level of HK-2 in CSCC cells. *P < 0.05, **P < 0.01.
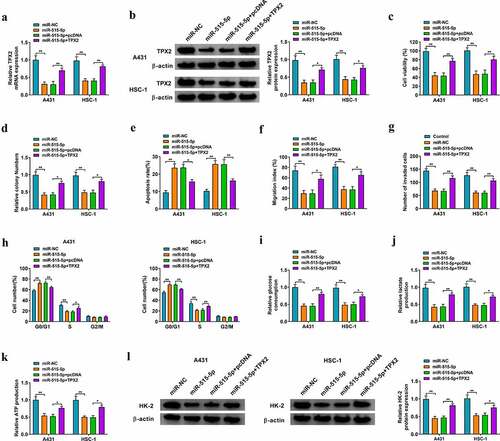
Circ-LARP1B can up-regulate TPX2 expression by sponging miR-515-5p in CSCC cells
Circ-LARP1B silencing reduced the protein expression of TPX2 in CSCC cells, which was recovered by the introduction of anti-miR-515-5p (), suggesting that circ-LARP1B could up-regulate the expression of TPX2 by sponging miR-515-5p in CSCC cells. Then, we analyzed the effect of circ-LARP1B/miR-515-5p/TPX2 axis on the activity of PI3K/AKT signaling pathway. Circ-LARP1B knockdown reduced PI3K expression and the phosphorylation level of AKT, which were offset by the addition of anti-miR-515-5p (), suggesting that circ-LARP1B could activate PI3K/AKT signaling by targeting miR-515-5p/TPX2 axis in CSCC cells.
Figure 7. Circ-LARP1B can up-regulate TPX2 expression by sponging miR-515-5p in CSCC cells. (a-d) CSCC cells were transfected with si-NC, si-circ-LARP1B, si-circ-LARP1B + anti-miR-NC, or si-circ-LARP1B + anti-miR-515-5p. (a and b) The protein level of TPX2 was analyzed by Western blot assay. (c and d) The activity of PI3K/AKT signaling was analyzed by Western blot assay. *P < 0.05, **P < 0.01.
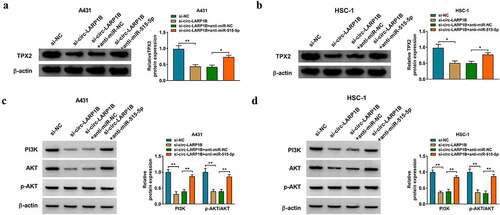
Circ-LARP1B silencing inhibits tumor growth in vivo
A431 cell line stably expressing sh-circ-LARP1B or sh-NC was established. A total of 2 × 106 A431 cells were injected into the right flank of the nude mice (n = 5/group). After injection for 7 d, tumor length and width were measured every 5 d, and tumor volume was analyzed as width2 × length × 0.5. After injection for 32 d, all nude mice were killed, and xenograft tumors were resected and weighed. As shown in ), circ-LARP1B silencing notably restrained tumor growth in vivo. IHC assay showed that tumors formed from circ-LARP1B-silenced A431 cells displayed lower-intensity staining of Ki-67, HK-2, MMP2, and cyclin D1 and higher-intensity staining of cleaved caspase 3 than those in sh-NC group ()). The expression of circ-LARP1B and TPX2 mRNA and protein was reduced in sh-circ-LARP1B group compared with sh-NC group, while miR-515-5p level was elevated in sh-circ-LARP1B group compared with sh-NC group (). Collectively, circ-LARP1B silencing suppressed tumor growth in vivo.
Figure 8. Circ-LARP1B silencing inhibits tumor growth in vivo. (a) After injection for 7 d, the tumor volume was measured every 5 d as (width2 × length)/2. (b) The tumor images in two groups were shown. (c) After injection for 32 d, all nude mice were euthanized, and tumor weight was recorded. (d) IHC assay was conducted to analyze the protein levels of proliferation-, glycolysis-, apoptosis-, and metastasis-associated markers in tumor tissues. (e) The expression of circ-LARP1B, miR-515-5p and TPX2 mRNA was measured by RT-qPCR. (f) Western blot assay was conducted to detect the protein expression of TPX2 in tumor tissues. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
CircRNAs are a unique class of non-coding RNAs, characterized by covalently linked ends, high stability, abundant expression, and tissue/cell-specific expression pattern, which are ideal bio-markers for human diseases [Citation29,Citation30]. Accumulating evidence have shown that genes have many transcriptional regulatory modes [Citation31,Citation32]. miRNAs are a class of small non-coding RNAs that can negatively regulate gene expression by inducing degradation or suppressing the translation of target mRNAs [Citation33]. Meanwhile, circRNAs share binding sites with miRNAs to sequester them, thus releasing downstream mRNAs from the inhibition of miRNAs and resulting in the up-regulation of gene expression, which is also known as the competitive endogenous RNA (ceRNA) mechanism [Citation10,Citation34]. Previous studies have shown that circRNAs can regulate the biological behaviors of cancer cells through the ceRNA mechanism. For instance, Niu et al. reported that circ_0001829 contributes to the development of gastric cancer by sponging miR-155-5p to induce the expression of SMAD2 [Citation35]. Sun et al. demonstrated that circ_0000105 facilitates the progression of liver cancer by binding to miR-498 to induce PIK3R1 expression [Citation36]. CircRNAs have also shown important regulatory role in CSCC progression [Citation37,Citation38]. An et al. demonstrated that circ-LARP1B aggravates CSCC progression by sequestering miR-1238 and miR-1247-5p [Citation9]. Zhang et al. found that circ-LARP1B contributes to CSCC progression by up-regulating HOXB7 expression via sponging miR-1236-3p [Citation13]. We found that circ-LARP1B expression was aberrantly up-regulated in CSCC tissues and cell lines, which was consistent with the previous studies [Citation9,Citation13]. The results of loss- and gain-of-function experiments together demonstrated that circ-LARP1B played an oncogenic role in CSCC by promoting cell viability, colony formation, migration, invasion, cell cycle progression, and glycolysis and inhibiting cell apoptosis.
The function of circRNAs is closely associated with their subcellular localization in the cells. We found that circ-LARP1B was majorly distributed in the cytoplasm of CSCC cells, indicating that circ-LARP1B might serve as a ceRNA. We identified miR-515-5p as a direct target of circ-LARP1B in CSCC cells. Previous articles have demonstrated that miR-515-5p played an anti-tumor role in a variety of human malignancies. For instance, miR-515-5p silencing is reported to promote the proliferation and motility of bladder cancer cells by targeting GINS2 [Citation39]. Liu et al. found that miR-515-5p restrains gastric cancer development by targeting HIF-1α [Citation40]. Huang et al. proved that circ_0008039 elevates the malignant potential of breast cancer cells by sponging miR-515-5p to up-regulate CBX4 [Citation41], suggesting the tumor suppressor role of miR-515-5p in breast cancer. However, the role of miR-515-5p in CSCC has never been reported. We found that miR-515-5p was down-regulated in CSCC tissues and cell lines, and it was negatively regulated by circ-LARP1B in CSCC cells. miR-515-5p overexpression suppressed the malignant phenotypes of CSCC cells. To explore whether circ-LARP1B knockdown-mediated anti-tumor effects in CSCC cells were dependent on the up-regulation of its target miR-515-5p, rescue experiments were conducted. The data showed that circ-LARP1B silencing suppressed the malignant behaviors of CSCC cells by up-regulating miR-515-5p.
We confirmed that miR-515-5p could directly bind to the 3ʹUTR of TPX2. TPX2 is a microtubule-associated protein that can regulate the formation of mitotic spindles, suggesting its important role in cell cycle progression [Citation16]. Increasing articles have reported that TPX2 is implicated in the tumorigenesis of multiple malignancies, including pancreatic cancer [Citation42], and colon cancer [Citation43]. Additionally, TPX2 is identified as a potential prognostic marker for gastric cancer [Citation44] and esophageal cancer [Citation45]. For instance, TPX2 is reported to be up-regulated in gastric cancer tissues, and high expression of TPX2 is related to dismal prognosis of gastric cancer patients [Citation44]. Sui et al. reported that TPX2 is an independent factor for the metastasis and prognosis of esophageal cancer patients [Citation45]. In CSCC, Feng et al. demonstrated that miR-216b suppresses CSCC progression by down-regulating TPX2 expression [Citation18], indicating the oncogenic role of TPX2. We found that TPX2 was up-regulated in CSCC tissues and cell lines. Besides, TPX2 was negatively regulated by miR-515-5p in CSCC cells. Rescue experiments uncovered that miR-515-5p overexpression hampered CSCC development by down-regulating TPX2. Circ-LARP1B could up-regulate the expression of TPX2 by sponging miR-515-5p in CSCC cells. Additionally, we found that circ-LARP1B silencing inactivated PI3K/AKT signaling by targeting miR-515-5p/TPX2 axis in CSCC cells.
Considering the oncogenic role of circ-LARP1B in regulating the biological behaviors of CSCC cells in vitro, we then assessed the role of circ-LARP1B in regulating tumor growth in vivo by xenograft tumor model. The data revealed that circ-LARP1B interference inhibited tumor growth in vivo, at least partly through the miR-515-5p/TPX2 axis.
Conclusions
In conclusion, circ-LARP1B played an oncogenic role in CSCC, and it contributed to CSCC progression through mediating miR-515-5p/TPX2/PI3K/AKT axis. Further studies are needed to explore the therapeutic potential of circ-LARP1B/miR-515-5p/TPX2 axis for CSCC patients.
Supplemental Material
Download Zip (14.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Ratushny V, Gober MD, Hick R, et al. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122(2):464–472.
- Liu T, Lei Z, Pan Z, et al. Genetic association between p53 codon 72 polymorphism and risk of cutaneous squamous cell carcinoma. Tumour Biol. 2014;35(4):3899–3903.
- Ishitsuka Y, Kawachi Y, Taguchi S, et al. Pituitary tumor-transforming gene 1 as a proliferation marker lacking prognostic value in cutaneous squamous cell carcinoma. Exp Dermatol. 2013;22(5):318–322.
- Xu N, Zhang L, Meisgen F, et al. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem. 2012;287(35):29899–29908.
- Andreeva K, Cooper NG. MicroRNAs in the neural retina. Int J Genomics. 2014;2014(165897):1–14.
- Zhu LP, He YJ, Hou JC, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37(5). DOI:10.1042/BSR20170750.
- Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48(121–127):121–127.
- Fan X, Weng X, Zhao Y, et al. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int. 2017;2017(5135781):1–9.
- An X, Liu X, Ma G, et al. Upregulated circular RNA circ_0070934 facilitates cutaneous squamous cell carcinoma cell growth and invasion by sponging miR-1238 and miR-1247-5p. Biochem Biophys Res Commun. 2019;513(2):380–385.
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238(42–51):42–51.
- Bi J, Liu H, Cai Z, et al. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging (Albany NY). 2018;10(8):1964–1976.
- Zhang DW, Wu HY, Zhu CR, et al. CircRNA hsa_circ_0070934 functions as a competitive endogenous RNA to regulate HOXB7 expression by sponging miR‑1236‑3p in cutaneous squamous cell carcinoma. Int J Oncol. 2020;57(2):478–487.
- Yan J, Xu H. Regulation of transforming growth factor-beta1 by circANKS1B/miR-515-5p affects the metastatic potential and cisplatin resistance in oral squamous cell carcinoma. Bioengineered. 2021;12(2):12420–12430.
- Zhang Y, Zhang Y, Wang S, et al. SP1-induced lncRNA ZFPM2 antisense RNA 1 (ZFPM2-AS1) aggravates glioma progression via the miR-515-5p/Superoxide dismutase 2 (SOD2) axis. Bioengineered. 2021;12(1):2299–2310.
- Li B, Qi XQ, Chen X, et al. Expression of targeting protein for xenopus kinesin-like protein 2 is associated with progression of human malignant astrocytoma. Brain Res. 2010;1352(200–207):200–207.
- Chang H, Wang J, Tian Y, et al. The TPX2 gene is a promising diagnostic and therapeutic target for cervical cancer. Oncol Rep. 2012;27(5):1353–1359.
- Feng C, Zhang HL, Zeng A, et al. Tumor-Suppressive Microrna-216b binds to TPX2, activating the p53 signaling in human cutaneous squamous cell carcinoma. Mol Ther Nucleic Acids. 2020;20(186–195). DOI:10.1016/j.omtn.2020.01.022.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Zheng S, Qian Z, Jiang F, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11(7):4126–4138.
- Wang X, Tao G, Huang D, et al. Circular RNA NOX4 promotes the development of colorectal cancer via the microRNA‑485‑5p/CKS1B axis. Oncol Rep. 2020;44(5):2009–2020.
- Wu D, Jia H, Zhang Z, et al. Circ_0000511 accelerates the proliferation, migration and invasion, and restrains the apoptosis of breast cancer cells through the miR326/TAZ axis. Int J Oncol. 2021;58(4): 1.
- Wang L, Hu J, Qiu D, et al. Dual-specificity phosphatase 5 suppresses ovarian cancer progression by inhibiting IL-33 signaling. Am J Transl Res. 2019;11(2):844–854.
- Xiao B, Tan L, He B, et al. MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J Transl Med. 2013;11(172). DOI:10.1186/1479-5876-11-172.
- Liu H, Liu N, Cheng Y, et al. Hexokinase 2 (HK2), the tumor promoter in glioma, is downregulated by miR-218/Bmi1 pathway. Plos One. 2017;12(12):e0189353.
- Zhu J, Bai J, Wang S, et al. Down-regulation of long non-coding RNA SNHG14 protects against acute lung injury induced by lipopolysaccharide through microRNA-34c-3p-dependent inhibition of WISP1. Respir Res. 2019;20(1):233.
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314.
- Zheng X, Boyer L, Jin M, et al. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5. DOI:10.7554/eLife.13374.
- Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
- Panni S, Lovering RC, Porras P, et al. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech. 2020;1863(6):194417.
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(351–379):351–379.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Niu Q, Dong Z, Liang M, et al. Circular RNA hsa_circ_0001829 promotes gastric cancer progression through miR-155-5p/SMAD2 axis. J Exp Clin Cancer Res. 2020;39(1):280.
- Sun Y, Sun X, Huang Q. Circ_0000105 promotes liver cancer by regulating miR-498/PIK3R1. J Gene Med. 2020;22(11):e3256.
- Chen S, Luo L, Chen H, et al. The current state of research regarding the role of non-coding RNAs in cutaneous squamous cell carcinoma. Onco Targets Ther. 2020;13(13151–13158). DOI:10.2147/OTT.S271346.
- Garofoli M, Volpicella M, Guida M, et al. The role of non-coding RNAs as prognostic factor, predictor of drug response or resistance and pharmacological targets, in the cutaneous squamous cell carcinoma. Cancers (Basel). 2020;12(9):2552.
- Dai G, Huang C, Yang J, et al. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell Mol Med. 2020;24(16):9231–9243.
- Liu J, Liu H, Zeng Q, et al. Circular RNA circ-MAT2B facilitates glycolysis and growth of gastric cancer through regulating the miR-515-5p/HIF-1α axis. Cancer Cell Int. 2020;20: 171.
- Huang FJ, Dang JQ, Zhang S, et al. Circular RNA hsa_circ_0008039 promotes proliferation, migration and invasion of breast cancer cells through upregulating CBX4 via sponging miR-515-5p. Eur Rev Med Pharmacol Sci. 2020;24(4):1887–1898.
- Ludwig R, Teran FJ, Teichgraeber U, et al. Nanoparticle-based hyperthermia distinctly impacts production of ROS, expression of Ki-67, TOP2A, and TPX2, and induction of apoptosis in pancreatic cancer. Int J Nanomedicine. 2017;12:1009–1018.
- Wei P, Zhang N, Xu Y, et al. TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J Transl Med. 2013;11(313). DOI:10.1186/1479-5876-11-313.
- Tomii C, Inokuchi M, Takagi Y, et al. TPX2 expression is associated with poor survival in gastric cancer. World J Surg Oncol. 2017;15(1):14.
- Sui C, Song Z, Yu H, et al. Prognostic significance of TPX2 and NIBP in esophageal cancer. Oncol Lett. 2019;18(4):4221–4229.
