ABSTRACT
The involvement of certain circular RNAs (circRNAs) in the development of hepatocellular carcinoma (HCC) has been reported. Herein, this study aimed to investigate the function and mechanism of circ_0001955 in HCC tumorigenesis. Expression of circ_0001955, miR-655-3p, and alkaline ceramidase 3 (ACER3) was evaluated by quantitative real-time PCR and Western blot. Cell counting kit-8, colony formation, transwell, tube formation, flow cytometry and tumor xenograft assays were adopted to perform in vitro and in vivo experiments. The direct interaction between miR-655-3p and circ_0001955 or ACER3 was verified using dual-luciferase reporter and RNA immunoprecipitation assays. Circ_0001955 was highly expression in HCC tissues and cells. Functionally, circ_0001955 deletion suppressed HCC tumorigenesis in vitro by suppressing cell growth, metastasis and angiogenesis. Mechanistically, circ_0001955 could competitively sponge miR-655-3p, which targeted ACER3. Besides that, miR-655-3p silencing abolished the anticancer action of circ_0001955 silencing on HCC cells. Moreover, miR-655-3p overexpression inhibited HCC cell oncogenic phenotypes mentioned above, which were attenuated by ACER3 up-regulation. Additionally, circ_0001955 knockdown also impeded HCC growth in a mouse model. In all, this study suggested a novel circ_0001955/miR-655-3p/ACER3 pathway in HCC progression.
Graphical abstract

Introduction
Hepatocellular carcinoma (HCC) is the second most common cancer-related death, accounting for approximately 80% of the total liver cancer all over the world [Citation1,Citation2]. HCC patients usually have poor clinical outcomes due to the high frequencies of recurrence and metastasis [Citation3], besides that, patients with advanced HCC have few treatment options. Thus, further clarifications on the mechanism of HCC progression are necessary.
Growing evidence has supported that circular RNA (circRNA) deregulation is closely associated with HCC progression [Citation4,Citation5]. Unlike traditional linear RNAs, circRNAs have closed ring structures that lack 5’ and 3’ ends, which allow them away from the impaction of RNA exonuclease, thus, circRNAs are more stably expressed and difficult to degrade [Citation6,Citation7]. Functionally, circRNAs can participate in modulating cell biological processes, including cell angiogenesis, proliferation, metastasis, and metabolism [Citation8,Citation9]. Currently, multiple lines of evidence have shown the abnormal expression of circRNAs in HCC, and proposed the involvement of circRNAs in HCC tumorigenesis. For example, Su et al. revealed that CircRNA Cdr1as facilitated HCC cells to proliferate and migrate by up-regulating alpha fetoprotein level through absorbing miR-1270 [Citation10]. CircRNA-104718 accelerated HCC cell proliferative and migratory abilities in vitro and in vivo via miR-218-5p/Thioredoxin Domain Containing 5 pathway [Citation11]. Besides, Gao et al. showed that circRNA circ_0008274 up-regulated granulin by sequestering miR-140-3p to promote HCC cell proliferation, invasion and migration in vitro [Citation12]. Circ_0001955 (ID: has_circ_0001955) is a circRNA derived from exons 5 to 10 of the Casein Kinase 1 Gamma 1 (CSNK1G1) gene and locates at chr15:64,495,280–64,508,912. In HCC, circ_0001955 expression was found to be higher, and knockdown of circ_0001955 attenuated HCC growth and metastasis [Citation13,Citation14]. However, large-scale identifications of the function and mechanism of circ_0001955 HCC tumorigenesis were not yet reported.
Hence, this study aimed to probe the function of circ_0001955 in HCC progression. Moreover, the potential molecular mechanisms of circ_0001955 that was related to HCC tumorigenesis were also explored, which may provide a novel insight into the development of efficient therapeutics for HCC patients.
Materials and methods
Tissue specimens
A total of 35 cases of HCC patients from the First Affiliated Hospital of Zhengzhou University were included in this study. They were diagnosed as HCC by pathological examination and did not undergo preoperative chemotherapy or radiotherapy. The tumor tissues and adjacent non-cancer tissues were obtained and then immediately stored at −80°C until RNA extraction. Written informed consent was collected from all participants before specimen collection and the Ethics Committee of the First Affiliated Hospital of Zhengzhou University allowed this study.
Cell culture
Human HCC cell lines (HCCLM3, SNU-387, MHCC97H, and Huh-7), and normal THLE-2 were obtained from Chinese Academy of Medical Sciences (Beijing, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) adding with 100 U/mL of penicillin and streptomycin and 10% fetal bovine serum (FBS; Hyclone, Logan, Utah, USA). HUVECs were purchased the same institution and then grown in endothelial cell medium (ECM; Gibco) supplemented with 10% FBS (Hyclone), 1% endothelial cell growth supplement (Gibco), and 1% penicillin and streptomycin. Then, all cells were maintained with 5% CO2 at 37°C.
Quantitative real-time PCR (qRT-PCR)
Nuclear and cytoplasmic separation was performed referring to the instruction of RNA Subcellular Isolation Kit (Life Technologies, Waltham, MA, USA). Isolation of total RNA was accomplished using an RNeasy mini kit (Qiagen), and the purity of total RNA was detected by a spectrophotometer (NanoDrop 2000 C, Thermo Fisher Scientific, Rochester, NY, USA). For circRNA detection, RNase R (3 U/μg) was used to interact with 2 μg of RNA extractions for 1 h at 37°C. Total cDNA was synthesized using the SuperScriptIII reverse transcriptase (Takara Biotech, Otsu, Japan). Then, the expression of circ_0001955, linear CSNK1G1 mRNA, alkaline ceramidase 3 (ACER3) mRNA and miR-655-3p, was determined using SYBR Green PCR Master Mix (Qiagen) with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 as the loading control. The relative fold changes were shown as a cycle threshold (Ct) value. The primer sequences for qRT-PCR are shown in .
Table 1. Primers sequences used for qRT-PCR
Transient transfection of cells
Transient transfection with plasmids, small interfering RNA (siRNA), miRNA mimic, miRNA inhibitor or respective controls was implemented with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) when HCCLM3 and Huh-7 cells reached 80% confluency of dishes. The circ_0001955-specific siRNA (si-circ_0001955) and negative control (NC) (si-NC), pcDNA-ACER3 overexpression plasmid (ACER3) and plasmid containing scrambled sequences (vector) were synthesized by Geneseed (Shanghai, China). The miR-655-3p inhibitors (anti-miR-655-3p), mimic (miR-655-3p), and NC (miR-NC or anti-miR-NC) were provided by GenePharma (Shanghai, China).
Cell counting kit-8 (CCK-8) assay
Transfected HCCLM3 and Huh-7 cells were plated in 96-well plates, then cell proliferation was assayed using the CCK-8 solution (Beyotime, Shanghai, China), and then the absorbance at 450 nm was read using a microplate reader at indicated times.
Colony formation assay
Transfected HCCLM3 and Huh-7 cells were plated in a 6 cm plate with medium at 1000 cells per well. Cells were grown at 37°C with 5% CO2 for 14 days. Cell colonies (≥50 cells) were imaged and counted after crystal violet staining.
Transwell assay
Transwell chambers (Corning Incorporated, Corning, NY, USA) in a 24-well format were used for transwell assay. For cell migration analysis, transfected HCCLM3 and Huh-7 in serum-free medium cells were seeded in the upper chamber with an 8-μm pore membrane. For cell invasion analysis, the membrane of upper chambers were pre-coated with Matrigel (BD Biosciences). Then, 10% FBS-contained medium (600 μL) was filled into the lower chamber. 24 h later, the number of migrated and invaded cells in five random fields were counted after being fixed and stained.
Tube formation assay
The tumor-conditioned medium (TCM) of transfected HCCLM3 and Huh-7 cells was collected. Each well of a 48-well plate was filled with 75 μL Matrigel (BD Biosciences) and solidified at 37°C for more than 1 h. HUVECs were starved in ECM without serum for 24 h, and then seeded onto the gel with TCM plus 1% FBS for 4–6 h incubation. Tubular structures were observed using a bright-field microscope (Bio-Rad) and tube formation ability was quantified by detecting the number of branches.
Flow cytometry
HCCLM3 and Huh-7 cells, following assigned transfection, were collected, rinsed with cold PBS and fixed in 70% ethanol. After washing, fixed cells were stained with propidium iodide (PI) (Sigma-Aldrich, St. Louis, MO, USA). At least, cell cycle distribution was evaluated using a FACS Calibur flow cytometer (BD Biosciences).
Transfected HCCLM3 and Huh-7 cells were washed with PBS and resuspended in 500 μL Annexin-binding buffer, after which cells were double-stained with 5 μL of Annexin V-fluorescein Isothiocyanate (Abcam, Cambridge, UK) and 5 μL of PI (Sigma-Aldrich) for after 15 min in the darkness. Then, cell apoptosis was analyzed by FACSCalibur II sorter (BD Biosciences) within 1 h.
Western blot
Proteins were isolated through RIPA cleavage, then total protein was separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred onto nitrocellulose membranes. The membranes were then incubated overnight at 4°C with the specific primary antibodies after sealing with 5% nonfat dry milk for 2 h, followed by interaction with anti-rabbit or anti-mouse HRP for 1 h at 37°C with a 1:5000 dilution. Then, protein bands were quantified using enhanced chemiluminescence kits (Beyotime, Shanghai, China). The primary antibodies included: E-cadherin (ab40772, 1:2000, Abcam), vimentin (ab92547, 1:2000, Abcam), N-cadherin (ab18203, 1:2000, Abcam), ACER3 (1:500, Saierbio, Tianjin, China) and GAPDH (ab181602, 1:1000, Abcam).
Dual-luciferase reporter assay
The pMIR-REPORT luciferase reporter plasmid was inserted with the fragments of ACER3 3’UTR or circ_0001955 encompassing the complementary sequence and the mutated sequences in the complementary region in GenePharma. Then, 200 ng of reporter plasmids and 50 ng of pRL-TK Renilla Luciferase vector were transfected into HCCLM3 and Huh-7 cells with miR-655-3p mimic or mimic control at 50 nM. Lastly, luciferase activity was evaluated after 48 h of infection.
RNA immunoprecipitation (RIP) assay
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit together with anti-Argonaute-2 (AGO2) and control IgG antibodies was employed to conduct RIP assay as described previously [Citation12]. Finally, the coprecipitated RNAs were evaluated by qRT-PCR.
Tumor xenograft assay
Six-weeks-old BALB/c nude mice (N = 12, 20–25 g, Charles River Labs, Beijing, China) were injected with Huh-7 cells (5 × 106 cells) stably expressing lentivirus-mediated short hairpin RNA (shRNA) targeting circ_0001955 or nontarget shRNA (GenePharma). The volume of subcutaneous tumor was measured every weeks based on the equation: volume = (length × width2)/2. At day 28, xenograft tumors were isolated, weighed and collected for molecular analysis or for immunohistochemistry (IHC) staining with Ki67 antibody (1:1000, ab15580, Abcam) as described previously [Citation15]. The animal experiment was ratified by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
Statistical analysis
Data from three repeated experiments were presented as mean ± standard deviation. Student’s t test (two-sided) or analysis of variance followed by Tukey’ s post hoc analysis was used for statistical comparisons. P < 0.05 indicated significantly difference.
Results
We hypothesizes that circ_0001955 accelerate the tumorihenesis of HCC through miR-655-3p/ACER3 axis. Through the in vitro and in vivo assays, we confirmed that circ_0001955 silencing suppressed HCC cell growth, metastasis and angiogenesis in vitro, as well as hindered HCC growth in nude mice. Additionally, with dual-luciferase reporter assay and RIP assay, we clarified the network of circ_0001955/miR-655-3p/ACER3 in HCC cells, and circ_0001955 exerted its oncogenic role via miR-655-3p/ACER3 axis.
Circ_0001955 expression is up-regulated in HCC
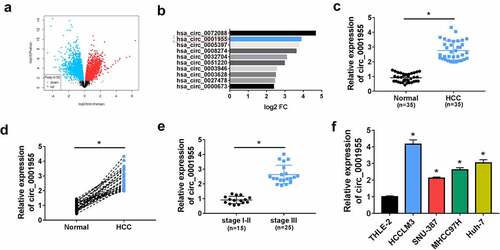
Firstly, public database (www.ncbi.nlm.nih.gov/geo) (GSE97322) was used to analyze the differentially expressed circRNA genes in HCC. According to volcano plot, numerous circRNAs were discovered to be highly expressed in HCC tissues (log2|fold change| ≥ 2 and P < 0.05) including circ_0001955 (). Then the expression profile of circ_0001955 in HCC clinical tissues was investigated. Compared with the adjacent non-cancer tissues, circ_0001955 was highly expressed in HCC tissues (). Furthermore, circ_0001955 expression was dramatically higher in HCC tissues of stage III than those of stage I–II (). Besides that, the correlation between circ_0001955 expression and clinical characteristics in patients with HCC was showed in . The expression of circ_0001955 was significantly correlated with TNM stage (P = 0.0030) and tumor size (cm) (P = 0.0037). Thereafter, an increased circ_0001955 expression was also found in HCC cells relative to the normal THLE-2 cells (). All these data suggested that circ_0001955 might be involved in HCC progression.
Table 2. Correlation between the clinicopathologic variables and hsa_circ_0001955 expression in HCC patients (n = 35)
Figure 1. Circ_0001955 expression is up-regulated in HCC. (a) Volcano plot of the differential circRNA expression analysis. (b) CircRNA expression profiles. (c, d) qRT-PCR analysis of circ_0001955 expression level in HCC tissues and normal tissues (n = 5). (e) qRT-PCR analysis of circ_0001955 expression level in HCC tissues from stage I–II (n = 12) and stage III (n =23). (f) qRT-PCR analysis of circ_0001955 expression level in HCC cells and normal THLE-2 cells. *P < 0.05.
Characterization of molecular structure of circ_0001955
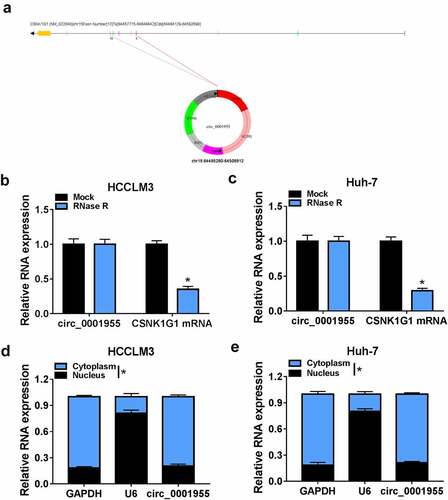
Circ_0001955 is derived from exons 5 to 10 of the CSNK1G1 gene and locates at chr15:64,495,280–64,508,912 (). Results of RNase R digestion suggested that circ_0001955 could resistant to RNase R digestion in HCCLM3 and Huh-7 cells owing to the lack of a free 3’terminus compared to the linear CSNK1G1 mRNA (). Additionally, circ_0001955 was discovered to be predominately distributed in the cytoplasm of HCCLM3 and Huh-7 cells ().
Figure 2. Characterization of molecular structure of circ_0001955. (a) The exonic information of circ_0001955 was illustrated as indicated. (b, c) qRT-PCR analysis of circ_0001955 and linear CSNK1G1 mRNA expression in HCCLM3 and Huh-7 cells treated with RNase R or mock. (d, e) Analysis of the cellular distribution of circ_0001955 by cellular RNA fractionation assay. *P < 0.05.
Knockdown of circ_0001955 restrains HCC tumorigenesis in vitro
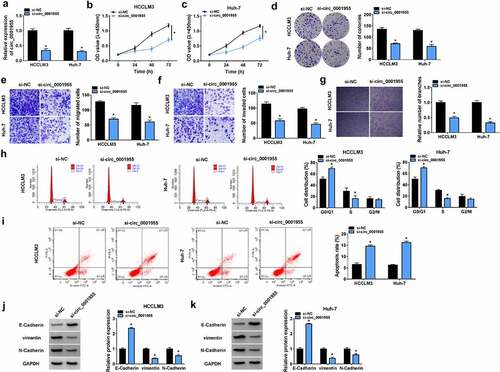
To evaluate the role of circ_0001955 in HCC development, loss-of-function assay was conducted. As shown in , transfection of si-circ_0001955 sufficiently down-regulated circ_0001955 level in HCCLM3 and Huh-7 cells. Functionally, CCK-8 assay showed that circ_0001955 silencing decreased the proliferation of HCCLM3 and Huh-7 cells (). Similarly, results from colony formation assay suggested that silencing of circ_0001955 impaired the colony-forming abilities of HCCLM3 and Huh-7 cells (). Transwell assay indicated circ_0001955 silencing inhibited cell migration and invasion in HCCLM3 and Huh-7 cells (). Moreover, circ_0001955 knockdown led to a decrease of the number of tube branches in HUVECs (). After that, flow cytometry revealed that circ_0001955 knockdown arrested cell cycle in the G1 phase in HCCLM3 and Huh-7 cells (). Moreover, it also showed that apoptosis of circ_0001955-decreased HCCLM3 and Huh-7 cells was significantly increased compared to the control group (). Western blot analysis indicated that circ_0001955 silencing increased the expression of E-cadherin (epithelial markers) and decreased the expression of vimentin and N-cadherin (mesenchymal markers) in HCCLM3 and Huh-7 cells (). Taken together, knockdown of circ_0001955 restrained HCC tumorigenesis in vitro by suppressing cell growth, metastasis and angiogenesis.
Figure 3. Knockdown of circ_0001955 restrains HCC tumorigenesis in vitro. (a-k) HCCLM3 and Huh-7 cells were transfected with si-circ_0001955 or si-NC. (a) qRT-PCR analysis of circ_0001955 expression in cells. (b-d) Cell proliferation analysis using CCK-8 and colony formation assays. (e, f) Transwell assay for cell migration and invasion. (g) Tube formation assay for cell angiogenesis analysis. (h-j) Flow cytometry for cell cycle distribution and cell apoptosis rate. (j, k) Western blot analysis of the protein levels of E-cadherin, vimentin and N-cadherin. *P < 0.05.
Circ_0001955 directly targeted miR-655-3p in HCC cells
Given that circ_0001955 was distributed predominantly in cell cytoplasm, we hypothesized that circ_0001955 might act as a sponge of miRNA to exert its functions. Through the starbase database (http://starbase.sysu.edu.cn/starbase2), we identified that miR-655-3p had the complementary sequences on circ_0001955 (). To confirm the binding potential, the transfection efficiency of miR-655-3p mimic was firstly validated using qRT-PCR, as expected, miR-655-3p mimic significantly increased the expression of miR-655-3p in HCCLM3 and Huh-7 cells (). Next, a dual-luciferase reporter assay was performed. The results showed that miR-655-3p mimic markedly reduced the luciferase activity of the wild-type circ_0001955 reporter vector but not the mutated one (). Further RIP assay suggested that the AGO2 antibody was able to pull down both endogenous circ_0001955 and miR-655-3p (). All these results validated the binding between circ_0001955 and miR-655-3p. Moreover, we found miR-655-3p expression was decreased in HCC tissues and cells (). After confirming the transfection efficiency of miR-655-3p inhibitor using qRT-PCR (), si-circ_0001955 together with miR-655-3p inhibitor were co-transfected into HCCLM3 and Huh-7 cells, then it was proved that circ_0001955 knockdown led to an increase of miR-655-3p expression in HCCLM3 and Huh-7 cells, which was reserved by miR-655-3p inhibition (). Therefore, we confirmed that circ_0001955 targeted miR-655-3p and inversely regulated its expression.
Figure 4. Circ_0001955 directly targeted miR-655-3p in HCC cells. (a) The binding site of miR-655-3p on circ_0001955 was predicted by software. (b) Detection of the transfection efficiency of miR-655-3p mimic or mimic negative control using qRT-PCR. (c-f) The interaction between miR-655-3p and circ_0001955 was analyzed by dual-luciferase reporter assay and RIP assay in HCCLM3 and Huh-7 cells. (g, h) qRT-PCR analysis of miR-655-3p expression level in HCC tissues and normal tissues (n =35), as well as in HCC cells and normal THLE-2 cells. (i) Detection of the transfection efficiency of miR-655-3p inhibitor or inhibitor negative control using qRT-PCR. (j, k) Measurement of miR-655-3p expression in HCCLM3 and Huh-7 cells transfected with si-NC, si-circ_0001955, si-circ_0001955 + anti-miR-NC, or si-circ_0001955 + anti-miR-655-3p using qRT-PCR. *P < 0.05.
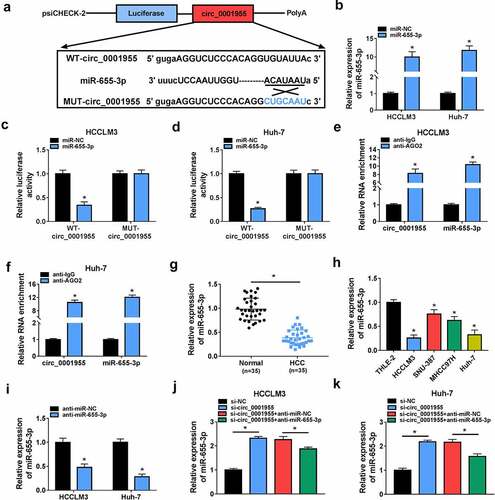
Knockdown of circ_0001955 suppresses HCC tumorigenesis via targeting miR-655-3p
Subsequently, we investigated the functional relationship between circ_0001955 and miR-655-3p using a series of rescue experiments. The results exhibited that co-transfection of circ_0001955 siRNA and miR-655-3p inhibitor led to the increases of HCCLM3 and Huh-7 cell proliferation (), migration and invasion abilities (), enhancement of tube formation ability of HUVECs (), progression of cell cycle in HCCLM3 and Huh-7 (), and decrease of HCCLM3 and Huh-7 cell apoptotic rate (). Besides that, miR-655-3p inhibitor abolished circ_0001955 siRNA-mediated elevation of E-cadherin as well as decrease of N-cadherin and vimentin HCCLM3 and Huh-7 cells (). Taken together, circ_0001955 knockdown attenuated HCC tumorigenesis via targeting miR-655-3p.
Figure 5. Knockdown of circ_0001955 suppresses HCC tumorigenesis via targeting miR-655-3p. (a-k) HCCLM3 and Huh-7 cells were transfected with si-NC, si-circ_0001955, si-circ_0001955 + anti-miR-NC, or si-circ_0001955 + anti-miR-655-3p. (a-c) Cell proliferation analysis using CCK-8 and colony formation assays. (d, e) Transwell assay for cell migration and invasion. (f) Tube formation assay for cell angiogenesis analysis. (g-i) Flow cytometry assay for cell cycle distribution and cell apoptosis rate. (j, k) Western blot analysis of the protein levels of E-cadherin, vimentin and N-cadherin. *P < 0.05.
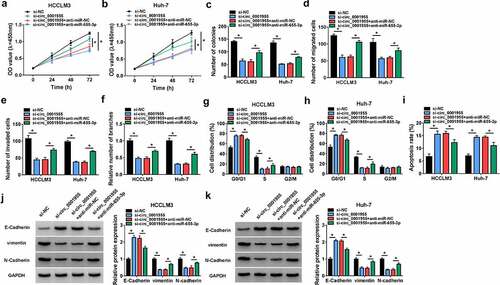
ACER3 is a target of miR-655-3p in HCC cells
Using the TargetScan database (http://www.targetscan.org/vert_72), we found that ACER3 had the complementary sequences on miR-655-3p (). Then, results from dual-luciferase reporter assay indicated that the luciferase intensity markedly decreased in HCCLM3 and Huh-7 cells co-transfected with wild-type ACER3 reporter vector and miR-655-3p mimic (). Furthermore, RIP assay suggested that ACER3 and miR-655-3p were enriched in AGO2-containing microribonucleoproteins relative to control IgG in HCCLM3 and Huh-7cells (). Thus, we validated that miR-655-3p targeted ACER3. Additionally, the expression profile of ACER3 was explored. We discovered that ACER3 was highly expressed in HCC tissues and cells (). The expression of ACER3 was significantly up-regulated by the transfection of ACER3 vector (). Then, it was observed that miR-655-3p overexpression down-regulated ACER3 level in HCCLM3 and Huh-7cells, which was rescued by ACER3 vector transfection (). Thus, we verified that miR-655-3p targetedly and negatively regulated ACER3 expression in HCC cells.
Figure 6. ACER3 is a target of miR-655-3p in HCC cells. (a) The binding site of miR-655-3p on ACER3 was predicted by software. (b-e) The interaction between miR-655-3p and ACER3 was analyzed by dual-luciferase reporter assay and RIP assay in HCCLM3 and Huh-7 cells. (f-h) qRT-PCR and Western blot analysis of ACER3 expression level in HCC tissues and normal tissues (n =35), as well as in HCC cells and normal THLE-2 cells. (i) Detection of the transfection efficiency of ACER3 or vector using Western blot. (j, k) Western blot analysis of ACER3 expression in HCCLM3 and Huh-7 cells transfected with miR-NC, miR-655-3p, miR-655-3p + vector, or miR-655-3p + ACER3. *P < 0.05.
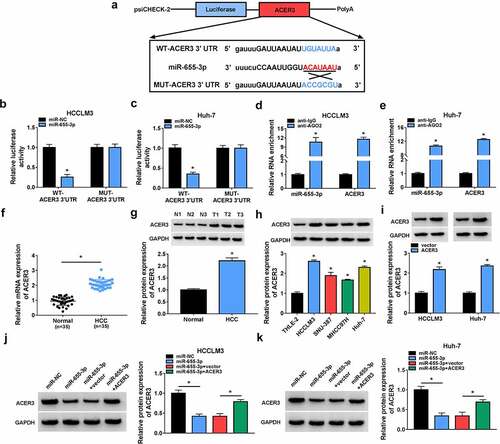
MiR-655-3p impedes HCC tumorigenesis via targeting ACER3
We then elucidated whether miR-655-3p/ACER3 axis was responsible for the tumorigenesis of HCC. HCCLM3 and Huh-7 cells were co-transfected with miR-655-3p mimic and ACER3 vector to conduct rescue assay. Overexpression of miR-655-3p suppressed the proliferation (), migration () and invasion () capacities of HCCLM3 and Huh-7 cells, which were partly counteracted by ACER3 up-regulation. Besides, miR-655-3p overexpression led to a decreased tube formation ability of HUVECs, while co-transfection with ACER3 showed opposite effect (). Flow cytometry analysis suggested that miR-655-3p up-regulation arrested cell cycle in the G1 phase in HCCLM3 and Huh-7 cells () and increased the apoptosis rate of HCCLM3 and Huh-7 cells (), while this condition was reversed by ACER3 up-regulation (). Moreover, overexpression of ACER3 could partly abolished the corresponding increase in E-cadherin and decreases in vimentin as well as N-cadherin induced by miR-655-3p in HCCLM3 and Huh-7 cells (). Collectively, miR-655-3p served as a tumor suppressor to impede HCC tumorigenesis via targeting ACER3.
Figure 7. MiR-655-3p impedes HCC tumorigenesis via targeting ACER3. (a-k) HCCLM3 and Huh-7 cells were transfected with miR-NC, miR-655-3p, miR-655-3p + vector, or miR-655-3p + ACER3. (a-c) Cell proliferation analysis using CCK-8 and colony formation assays. (d, e) Transwell assay for cell migration and invasion. (f) Tube formation assay for cell angiogenesis analysis. (g-i) Flow cytometry assay for cell cycle distribution and cell apoptosis rate. (j, k) Western blot analysis of the protein levels of E-cadherin, vimentin and N-cadherin. *P < 0.05.
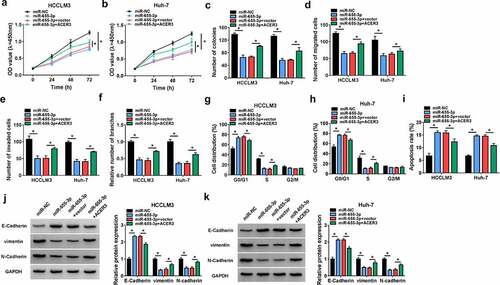
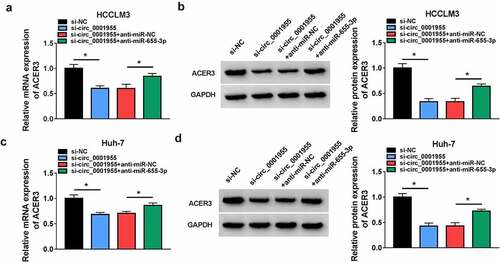
Circ_0001955 up-regulates ACER3 expression via sequestering miR-655-3p
Whether circ_0001955 could regulate ACER3 via miR-655-3p was then explored. As shown in , circ_0001955 knockdown led to a decrease of ACER3 expression in HCCLM3 and Huh-7 cells, which was reverted by miR-655-3p inhibition, suggesting that circ_0001955 could indirectly regulate ACER3 expression via sponging miR-655-3p.
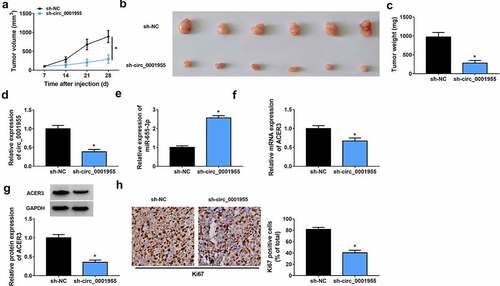
Silencing of circ_0001955 hinders HCC growth in vivo
Next, a subcutaneous xenograft model was establish to investigate the biological function of circ_0001955 in vivo. Consistent with the results in vitro, circ_0001955 silencing significantly decreased tumor volume and weight relative to the control group (). Furthermore, molecular analysis showed that the expression levels of circ_0001955 and ACER3 were decreased (), while miR-655-3p expression level () was increased in tumors of sh-circ_0001955 group than those in tumors of sh-NC groups. In addition, IHC assay demonstrated that circ_0001955 knockdown caused decreased Ki67 expression (), revealing decreased cell proliferation. Altogether, circ_0001955 knockdown could suppress HCC growth in vivo.
Figure 9. Silencing of circ_0001955 hinders HCC growth in vivo. (a) Tumor volume was measured every week. (b) Representative images of xenografts isolated from each group. (c) Tumor weight was measured at day 28. (d-f) qRT-PCR of circ_0001955, miR-655-3p and ACER3 expression in tumors of each group. (g) Western blot analysis of ACER3 protein expression in tumors of each group. (h) Representative IHC staining of Ki67 in the indicated xenografts and the corresponding statistical plots were presented in the right panel. *P < 0.05.
Discussion
HCC is a prevalent malignancy through the world, despite great improvements have been made in the diagnosis and therapeutic options of HCC, HCC remains a major threaten for human health with a high morbidity and mortality [Citation16,Citation17]. The main problems for the limitation of the five-year survival rate of HCC are metastasis and recurrence, thus, further investigations on the mechanism underlying HCC cell growth and metastasis are of great significant.
CircRNAs are one type of RNAs with high cellular stability, they have been revealed to play essential roles in biological processes involved in HCC progression. For instance, Yu et al. showed that circRNA cSMARCA5 acted as a tumor suppressor to restrain HCC growth and migration through positively regulating tissue inhibitor of metalloproteinase 3 via binding to miR-181b-5p and miR-17-3p [Citation18]. CircSETD3 was demonstrated to suppress HCC growth in vitro and in vivo by miR-421/mitogen-activated protein kinase axis [Citation19]. In addition, the transfer of circRNA-100338 via exosomes expedited HCC metastasis through enhancing angiogenesis and invasiveness [Citation20]. In the present study, we found a highly expressed circ_0001955 in HCC, circ_0001955 deletion could affect HCC cell growth and metastasis by reducing cell proliferation, invasion, migration, angiogenesis, EMT, and enhanced cell apoptosis in HCC in vitro. Importantly, xenografts assay also suggested that circ_0001955 silencing impaired tumor growth and proliferation in vivo. Taken together, circ_0001955 acted as an onco-circRNA to promote HCC growth and metastasis.
According to the competitive endogenous RNA (ceRNA) mechanism, circRNAs can function as miRNA sponges in cells to eliminate the suppressive impact of the miRNA on their target gene, thereby elevating the expression level of targeted genes [Citation21,Citation22]. Previous studies have suggested that a specific circRNA-miRNA-mRNA network is related to the carcinogenesis of HCC, which may benefit the development of molecular biomarkers and therapeutic targets for HCC [Citation23,Citation24]. Therefore, the potential miRNAs interacted by circ_0001955 were investigated. The present study verified that circ_0001955 directly targeted miR-655-3p. miR-655-3p was identified as tumor suppressor in numerous cancers, such as ovarian cancer [Citation25], non-small lung cell cancer [Citation26], and glioma [Citation27], by involving in the processes of cell survival and metastasis. In HCC, previous findings also suggested that low miR-655-3p expression was related to the malignancy and worse clinical outcome in cancer patients, miR-655-3p could be of prognostic value in HCC [Citation28]. Besides that, miR-655-3p suppressed HCC cell proliferation and metastasis by targeting A Disintegrin and metalloproteinase domain-containing protein 10 [Citation28]. In this work, we demonstrated that miR-655-3p increase restrained HCC cell growth and metastasis. Importantly, miR-655-3p inhibition attenuated the action of circ_0001955 knockdown on HCC cells. Therefore, a circ_0001955/miR-655-3p network was identified in HCC.
This study also verified that miR-655-3p targeted ACER3 in HCC cells. Furthermore, circ_0001955 could positively regulated ACER3 expression via targeting miR-655-3p. ACER3 is the first alkaline ceramidase to be cloned in mammals [Citation29]. It was revealed to facilitate HCC cell survival via S1P/S1PR2/PI3K/AKT pathway [Citation30]. Additionally, Yang et al. suggested that lncRNA KCNQ1OT1 up-regulated ACER3 via miR-146a-5p to enhance radioresistance and tumorigenesis in HCC [Citation31]. All these results demonstrated that ACER3 exerted oncogenic role in HCC progression. In the current review, we for the first time identified the circ_0001955/miR-655-3p/ACER3 network in HCC cells, moreover, it was proved that ACER3 overexpression reversed the inhibitory effects of miR-655-3p on HCC tumorigenesis. Thus, circ_0001955/miR-655-3p/ACER3 axis was engaged in HCC progression.
Limitation and future direction
Although some interesting results have been found in this work, the effects of circ_0001955/miR-655-3p/ACER3 axis on normal liver cells should be performed in further researches in vitro and in vivo, thus revealing the clinical relevance of circ_0001955 in HCC treatment.
Conclusion
The results of this work suggested that circ_0001955 served as a sponge for miR-655-3p to up-regulate ACER3, thus promoting HCC growth and metastasis, which opened a new window to the development of molecule-based therapy in HCC.
Authors’ contribution
Kai Bai designed research, performed experiments, analyzed data, and wrote the manuscript. Yubo Ma collected and analyzed data. Jian Li edited the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Thanks for all participants involved in this study.
Disclosure statement
The authors declare that they have no financial conflicts of interest.
Additional information
Funding
References
- Thompson AI, Conroy KP, Henderson NC. Hepatic stellate cells: central modulators of hepatic carcinogenesis. BMC Gastroenterol. 2015;15(1):63.
- Perz JF, Armstrong GL, Farrington LA, et al. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538.
- Cao MQ, You AB, Zhu XD, et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol. 2018;11(1):12.
- Qiu L, Xu H, Ji M, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660.
- Wu J, Liu S, Xiang Y, et al. Bioinformatic analysis of circular RNA-associated ceRNA network associated with hepatocellular carcinoma. Biomed Res Int. 2019;2019:8308694.
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90.
- Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129.
- Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):29.
- Liu Y, Yang Y, Wang Z, et al. Insights into the regulatory role of circRNA in angiogenesis and clinical implications. Atherosclerosis. 2020;298:14–26.
- Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11:8182–8203.
- Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;133(1):1487–1503.
- Gao C, Wen Y, Jiang F, et al. Circular RNA circ_0008274 upregulates granulin to promote the progression of hepatocellular carcinoma via sponging microRNA −140-3p. Bioengineered. 2021;12(1):1890–1901.
- Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10:945.
- Ding B, Fan W, Lou W. hsa_circ_0001955 Enhances In vitro proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS axis. Mol Ther Nucleic Acids. 2020;22:445–455.
- Liang Y, Song X, Li Y, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer. 2020;19(1):85.
- Rich NE, Yopp AC, Singal AG. Medical management of hepatocellular carcinoma. J Oncol Pract. 2017;13(6):356–364.
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227.
- Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98.
- Huang XY, Huang ZL, Huang J, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):20.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283.
- Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220.
- Lin X, Chen Y. Identification of potentially functional circRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. 2018;24:70–78.
- Zha JF, Chen DX. MiR-655-3p inhibited proliferation and migration of ovarian cancer cells by targeting RAB1A. Eur Rev Med Pharmacol Sci. 2019;23(9):3627–3634.
- Wang W, Cao R, Su W, et al. miR-655-3p inhibits cell migration and invasion by targeting pituitary tumor-transforming 1 in non-small cell lung cancer. Biosci Biotechnol Biochem. 2019;83(9):1703–1708.
- Xin J, Zhao YH, Zhang XY, et al. LncRNA NFIA-AS2 promotes glioma progression through modulating the miR-655-3p/ZFX axis. Hum Cell. 2020;33(4):1273–1280.
- Zhao XQ, Liang B, Jiang K, et al. Down-regulation of miR-655-3p predicts worse clinical outcome in patients suffering from hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(4):748–752.
- Mao C, Xu R, Szulc ZM, et al. Cloning and characterization of a novel human alkaline ceramidase. A mammalian enzyme that hydrolyzes phytoceramide. J Biol Chem. 2001;276(28):26577–26588.
- Yin Y, Xu M, Gao J, et al. Alkaline ceramidase 3 promotes growth of hepatocellular carcinoma cells via regulating S1P/S1PR2/PI3K/AKT signaling. Pathol Res Pract. 2018;214(9):1381–1387.
- Yang G, Zhou L, Xu Q, et al. LncRNA KCNQ1OT1 inhibits the radiosensitivity and promotes the tumorigenesis of hepatocellular carcinoma via the miR-146a-5p/ACER3 axis. Cell Cycle. 2020;19(19):2519–2529.