ABSTRACT
This study investigated the function of long non-coding RNA (lncRNA) cytoskeleton regulator RNA (CYTOR) in hepatocellular carcinoma (HCC). In HCC, the expression of CYTOR and microRNA (miR)-125a-5p were measured by quantitative real-time PCR (qRT-PCR). The expression of actin skeletal protein 1 (LASP1) was evaluated by Western blot analysis. Flow cytometry assays, transwell assays, colony formation assay, and cell counting kit-8 (CCK-8) assay were used to evaluate the roles of miR-125a-5p and CYTOR in HCC cells. The target genes of CYTOR and miR-125a-5p were identified by bioinformatics analysis and Luciferase assay. CYTOR was upregulated in HCC cell lines, and knockdown of CYTOR inhibited HCC cell growth. MiR-125a-5p was downregulated in HCC cells and a target of CYTOR in regulating HCC progression. Furthermore, LASP1 was a downstream target of miR-125a-5p. Finally, CYTOR was found to be involved in HCC progression in vivo. CYTOR promotes HCC development by regulating the miR-125a-5p/LASP1 axis.
Graphical abstract
Introduction
As the fifth most common malignancy in the world, hepatocellular carcinoma (HCC) is a serious threat to human survival [Citation1,Citation2]. Although the treatment methods for HCC have made great progress, such as radiofrequency and ablation surgical resection, the long-term therapeutic effects of HCC are still unsatisfactory [Citation3,Citation4]. Even in small hepatocellular carcinoma, the recurrence rate after radical resection is still high after 5 years [Citation5]. Most recurrences happen within 2 years after surgery [Citation6]. Therefore, exploring the characteristics of hepatocellular carcinoma stem cells and the mechanisms of recurrence are important to find new HCC treatment methods.
Long non-coding RNAs (lncRNAs) are known to be involved in early embryo development, cell differentiation, and proliferation through modification and recruitment of cytokines [Citation7,Citation8]. LncRNAs can interact with other nucleotide molecules through complementary base pairing, and can also directly bind to proteins [Citation9]. Various lncRNAs play regulatory roles in tumor formation and affect the prognosis, occurrence, and metastasis of HCC [Citation10,Citation11]. For example, lncRNA SNHG22 facilitates HCC tumorigenesis and angiogenesis via DNA methylation of microRNA (miR)-16-5p [Citation12]. LncRNA DARS-AS1 aggravates the growth and metastasis of HCC via regulating the miR-3200-5p-cytoskeleton associated protein 2 (CKAP2) axis [Citation13]. LncRNA DHRS4-AS1 ameliorates HCC by suppressing proliferation and promoting apoptosis via the miR-522-3p/SOCS5 axis [Citation14]. Cytoskeleton regulator RNA (CYTOR) is a newly identified lncRNA and abnormally expressed in malignancies [Citation15]. CYTOR was reported to be significantly upregulated and resistant to oxaliplatin induced apoptosis in colon cancer [Citation15]. However, the mechanism of CYTOR in HCC is elusive.
Recently, the interactions between miRNAs and lncRNAs have attracted extensive attentions and became a research hotspot [Citation16,Citation17]. LncRNAs can function as competing endogenous RNAs (ceRNAs) to regulate the degradation of target miRNAs [Citation18]. MiRNAs are regulated by promoter methylation and deletion of miRNA target sites [Citation19]. In cancer, miRNAs can function as tumor suppressors to inhibit the expression of target genes and downstream pathways to mediate tumorigenesis [Citation20,Citation21]. Studies have shown that miRNAs are involved in HCC progression [Citation22]. In various tumor tissues, miR-125a-5p is abnormally expressed, suggesting its involvement in tumorigenesis, but its role in HCC is still unknown [Citation23].
It was found that miR-125a-5p suppressed tumorigenesis of breast cancer by targeting HDAC4 [Citation24]. Hsa-miR-125a-5p induced cell apoptosis of lung cancer by activating p53 [Citation25]. Moreover, Actin skeletal protein 1 (LASP1) acts as an effector of miR-125a-5p. LASP1 is a LIM protein subfamily. The main domain consists of the amino-terminal LIM and the carboxy-terminal 1 scr homology domain (SH3) [Citation26]. It mainly encodes cGMP and camp-dependent signaling proteins, which bind to cell membrane actin cytoskeleton and participate in the regulation of cytoskeleton recombination [Citation27]. Studies have shown that LASP1 is upregulated in hepatocarcinoma [Citation28]. Therefore, we speculated that CYTOR activated LASP1 by mutagenizing miR-125a-5p to regulate HCC progression.
In this study, we aimed to investigate the role of CYTOR in HCC progression and its potential as a HCC therapeutic target.
Methods
HCC datasets in TCGA
The HCC gene expression datasets and the RPM of miRNAs were searched in TCGA database using the R package TCGAbiolinks. Then, we screened tissues from patients who had cancer and normal tissues. There were 50 patients screened for gene expression and 48 patients screened for miRNAs. In normal and cancer tissues, the expression levels of CYTOR, LASP1 and miR-125a-5p were calculated using R and t-test.
Tissue samples
HCC patients (32) admitted at the First Affiliated Hospital of Dalian Medical University were enrolled in this study. All patients signed the written informed consent. The cancer and adjacent non-cancerous tissues were collected from each patient. This study was approved by the Ethics Review Committee of the aforementioned hospital and was carried out in accordance with the World Medical Association Declaration of Helsinki. During the 2-year follow-up, 25 patients had isolated tumors without metastasis or recurrence. Patients’ clinical data were shown in the Supplementary Table.
Cell culture
HCCLM3, MHCC97H, LO2, HUH7, SK-Hep1 and HepG2 cells were purchased from the American Type Culture Collection (Manassas, VA). DMEM supplemented with 10% FBS (Invitrogen) was used to culture HepG2 cells, and RPMI 1640 medium supplemented with 10% FBS was used to culture the other cell lines. In a humidified incubator, cells were incubated with 5% CO2 at 37°C.
Plasmid construction and transfection
Plasmids were synthesized at GenePharma (Shanghai, China). For plasmid preparation, the shRNAs targeting CYTOR-1 (5ʹ-CAGUCUCUAUGUGUCUUAATT-3ʹ) or CYTOR-2 (5ʹ-CACACUUGAUCGAAUAUGATT-3ʹ) was inserted into pcDNA3.1. Negative control (shNC) (5ʹ-ACAAAGUUCUGUGAUGCACUGA-3ʹ) was used. CYTOR cDNA fragments containing either the wild type (wt) or mutant (mut) miRNA binding site were cloned into pmirGLO. MiR-125a-5p mimic, inhibitor, and their controls were obtained from GenePharma (Shanghai, China). Transfection was performed using Lipofectamine 2000 following the manufacturer’s instructions. In addition, Lentiviral carrying sh-CYTOR or pc-CYTOR was used to select stable cell lines and construct nude mouse tumor xenograft model.
CCK-8 assay
Lentivirus (sh)-CYTOR or negative control (shNC) was transfected into cells in a 96-well plate overnight. Cell counting Kit-8 (CCK-8) reagents (Dojindo Molecular Technologies, Tokyo, Japan) (100 μl) were added at 12, 24, 48 and 72 h. After incubation for 4 h, the absorbance at 450 nm was measured using a microplate reader (Bio-Tek, Winooski, VT).
Transwell assay
HCC cells were infected with shCYTOR, pcCYTOR, miR-125a-5p inhibitor and mimic or their control for 48 h. Using a 24-well Transwell system (Biofavor, Shanghai, China), migration assay was performed. In upper chamber, cell suspension was maintained and tested.
Colony formation assay
Cells were cultured at 500 cells/well for 10 days to form cell colonies. PBS was used to wash cells, which were then fixed by 4% paraformaldehyde for 15 min. Crystal violet was used to stain cell colonies for 10 min. Finally, under microscope, cells were photographed and counted.
Nude mouse tumor xenograft model
Athymic BALB/c nude mice (male) were purchased from Sun Yat-sen University (Guangzhou, China). To evaluate the role of CYTOR in HCC cell growth and migration in vivo, Luciferase labeled HCC cells were transfected the CYTOR lentivirus vector. As previously described [Citation29], tumor cells were inoculated into mice, which were anesthetized intraperitoneally with tribromoethanol. For tumor metastasis analysis, the nude mice accepted the HCC cells injection via the tail vein. Then, the IVIS@ Lumina II system was used to trace the bioluminescence every week. For the evaluation of tumor growth, the right rear flank of mice accepted 1 × 107 HCC cells subcutaneously. Caliper was used to measure the tumor volume. After 4 weeks, tumor weight was measured. Laboratory Animal Care and Use Committee of the First Affiliated Hospital of Dalian Medical University approved the animal experiments. This study was carried out in compliance with the ARRIVE guidelines.
Apoptosis assay
Cells (5 x 105 cells/well) were cultured for apoptosis analysis. When grown to logarithmic growth phase, cells were counted. After centrifugation, 195 μL of Annexin V-FITC binding solution was added in cell resuspension for 10–20 min incubation. Finally, cells were stored in ice bath.
RNA pull-down assay
The EZMagna RIP kit (Millipore) was used to conduct RNA pull-down assay. Cells were lyzed by RIP lysis buffer. To conduct cell extract, magnetic beads conjugated to control anti-IgG or anti-AGO2 antibodies were used. RT-qPCR analysis was conducted after purifying the RNA samples.
RNA-binding protein immunoprecipitation (RIP) assay
RIP assay was performed using the Magna RIP Kit (EMD Millipore). Cells were lyzed using RIP lysis buffer, and the cell lysate was treated with magnetic beads conjugated to human anti‐Ago2 antibody (Millipore) or an isotype matched control antibody (normal mouse immunoglobulin G [IgG]; Millipore). qRT‐PCR was used to detect the expression of CYTRO and miR-125a-5p.
qRT-PCR
Total RNAs were extracted using the RNAiso Plus kit (TaKaRa Biotechnology, Dalian, China). Prime ScriptTM RT Master Mix was used to conduct reverse transcription. SYBR Premix Ex Taq II (TaKaRa Biotechnology) was used to prepare qRT-PCR reactions. Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) was used to conduct qRT-PCR. The relative gene expression levels were calculated using the 2−ΔΔCt method. Primer sequences were: 5′-AGAATGAAGGCTGAGGTGTG-3′ (forward) and 5′-CAGCGACCATCCAGTCATTTA-3′ (reverse) for CYTOR; 5′-TGGATTTGGACGCATTGGTC-3′ (forward) and 5′-TTTGCACTGGTACGTGTTGAT-3′ (reverse) for GAPDH; 5ʹ-ACACTCCAGCTGGGTCCCTGAGACCCTTTAA-3ʹ (forward) and 5ʹ-CTCAACTGGTGTCGTGGAGT-3ʹ (reverse) for miR-125a-5p; 5ʹ-CTCGC TTCGGCAGCACA-3ʹ (forward) and 5ʹ-ACCGCTTCACGAATTTGGGT-3ʹ (reverse) for U6.
Luciferase activity assay
pGL3 Basic vector was used to carry the mutant (MUT) or wild type (WT) CYTOR binding to miR-125a-5p. pLUC-MUT-CYTOR or pLUC-WT-CYTOR and MiR-125a-5p (RiboBio, Guangzhou, China) was co-transfected into HepG2 cells. MiR-125a-5p was co-transfected with 10 μg of pLUC-MUT-LASP1 or pLUC-WT-LASP1. At 48 h post-transfection, luciferase activity was tested using a dual luciferase assay system (Promega Corporation, Fitchburg, WI, USA).
Immunohistochemical (IHC) staining
The tissue sections were treated by deparaffinizing, followed by rehydration and heating. Next, 10% normal goat serum was used to wash and block the sections, which were then incubated by primary antibodies to Ki-67 (Abcam). The Envision™ABC kit (Gene-Tech Co., Ltd., Shanghai, China) was used. Finally, the sections were photographed by Leica DM4000B/M microscope.
Western blot analysis
Western blot was conducted as previously described [Citation30]. Briefly, total proteins were isolated using RIPA lysis buffer (Yaji, Shanghai, China). BCA Protein Assay Kit was used to measure the protein concentrations. Equal amount of protein samples were transferred to PVDF membrane, which was then incubated with anti-GAPDH antibodies (1:1,000, Abcam, UK) and anti-LASP1 (1:100) at 4°C overnight. Then anti-rabbit secondary antibody (1:1,000, Aibixin, Shanghai, China) was used to incubate the membrane for 1 h.
Statistical Methods
Data were presented as the mean ± standard deviation (SD). Data were analyzed using the SPSS19.0 software. All experiments were repeated as least three times. Student’s t-test was used for comparison of two groups. One-way ANOVA and LSD test were used for single-factor comparison of multiple groups, and two-way ANOVA was used for double factor comparison. Spearman correlation analysis was used to evaluate the correlation between groups. P < 0.05 was considered statistically significantly.
Results
The expression of CYTOR and hsa-miR-125a-5p in HCC
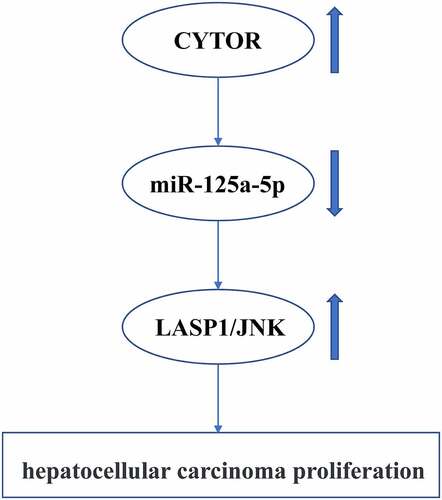
TCGA database was used to investigate the expression of CYTOR in HCC, and it was found that the expression of CYTOR was upregulated in HCC tumors compared with that in normal tissues. Our qRT-PCR results showed that the expression levels of CYTOR were significantly increased in HCC tissues (n = 100) (, p < 0.05), while the expression levels of miR-125a-5p were significantly reduced in HCC tissues (n = 96) compared with that in normal tissues (, p < 0.05). In addition, the expression of CYTOR and miR-125a-5p was negative correlated in HCC tissues (). In addition, the expression levels of CYTOR in HUH7, HCCLM3, MHCC97H, SK-hep1 and HepG2 cells were increased compared with that in LO2 cells (, p < 0.05), while the expression levels of hsa-miR-125a-5p were decreased in HCC cells (). The expression level of CYTOR was the highest and the expression level of miR125 expression was the lowest in HepG2 cells. Therefore, HepG2 cells were selected for further experiments. These results indicated that CYTOR and has-miR-125a-5p might be involved in HCC.
Figure 1. CYTOR and hsa-miR-125a-5p expression in HCC cells. (a) In HCC tissues, CYTOR expression was detected (n = 32). (b) In HCC tissues, hsa-miR-125a-5p expression was detected (n = 32). (c) Negative correlation between CYTOR and miR-125a-5p. (d). CYTOR expression in HCC cells. (e) miR-125a-5p expression in HCC cells. *p < 0.05, ** p < 0.01, n = 3.
CYTOR regulated HCC progression in vitro
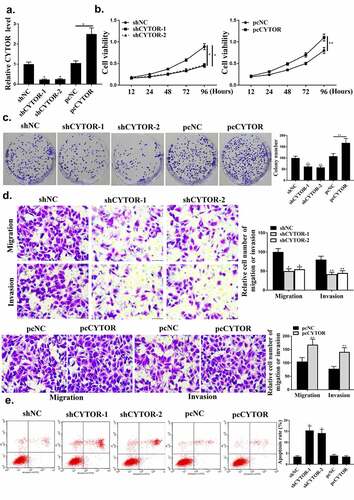
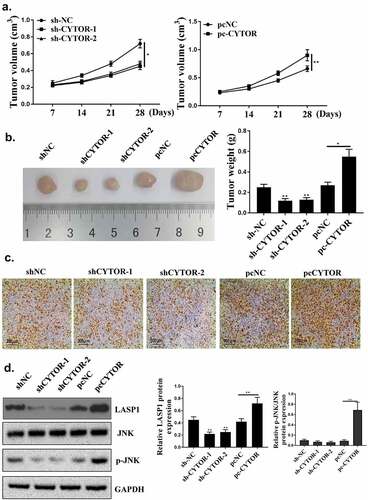
As shown in , CYTOR was regulated by sh-CYTOR or pcCYTOR in HCC cells (P < 0.05). Colony formation and cell viability were significantly reduced in the shCYTOR group compared with that in the sh-NC and pcCYTOR groups (, p < 0.05). And shCYTOR reduced cell invasion and migration (, p < 0.05). And shCYTOR induced apoptosis (, p < 0.05). Taken together, these results indicated that CYTOR was involved in the HCC progression.
MiR-125a-5p regulated HCC progression in vitro
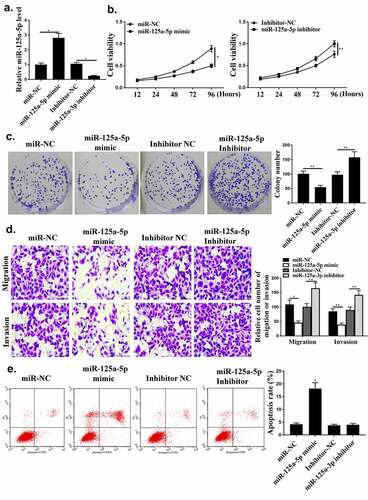
Next, whether miR-125a-5p could affect HCC cell growth was investigated. As shown in , miR-125a-5p was regulated by its mimic or inhibitor in HCC cells (P < 0.05). Compared with miR-NC group, miR-125a-5p mimic reduced the cell viability and colony formation (, p < 0.05). Cell apoptosis, invasion and migration were reduced by miR-125a-5p mimic ( and , p < 0.05).
Hsa-miR-125a-5p was a direct target of CYTOR
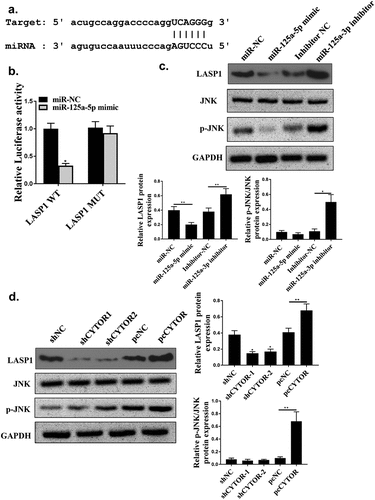
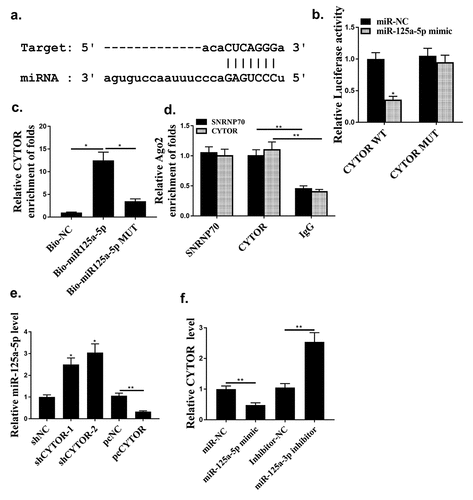
Starbase 2.0 predicted that hsa-miR-125a-5p was a potential target of CYTOR (). MiR-125a-5p mimic reduced luciferase activity of pGL3-REPOR-CYTOR-WT (P < 0.05), but not that of pGL3-REPOR-CYTOR-Mut (, p < 0.05). In addition, the expression levels of CYTOR using the miR-125a-5p-bio probe were significantly raised compared with that of miR-125a-5p-bio mut or NC-bio probe (, p < 0.05). Moreover, RIP assay was performed and it was observed that CYTOR and miR-125a-5p were more abundant in the Ago2 pellet than that in the IgG pellet (, p < 0.01). MiR-125a-5p was up-regulated in shCYTOR-1 and shCYTOR-2 groups compared with that in the control group (, p < 0.05), and reduced in pcCYTOR group (, p < 0.05). In addition, overexpression of miR-125a-5p decreased the expression levels of CYTOR, while knockdown of miR-125a-5p increased the expression levels of CYTOR (P < 0.01, ). These results indicated that miR-125a-5p might be a target of CYTOR in HCC.
Figure 4. Hsa-miR-125a-5p was a direct target for CYTOR. (a) The binding site between miR-125a-5p and CYTOR predicted by Starbase. (b) Relative luciferase activity. (c and d) The effect between has-mir-125a-5p and CYTOR indicated by RNA pull-down analysis and RIP assay. (e) In HepG2 cells, miR-125a-5p expression was detected. (f) In HepG2 cells, CYTOR expression was detected. * p < 0.05, ** p < 0.01, n = 3.
MiR-125a-5p mediated the effect of CYTOR on HCC proliferation
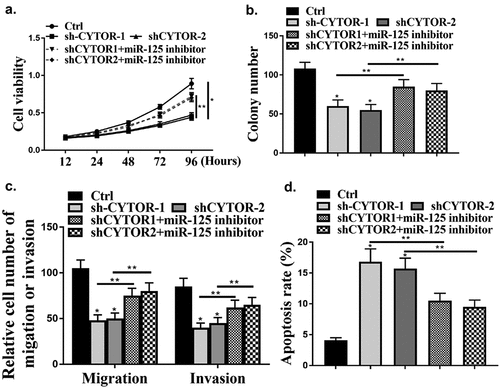
Next, whether CYTOR could affect HCC cells via miR-125a-5p was evaluated. it was found that knockdown of CYTOR significantly reduced cell viability (, p < 0.05), while knockdown of CYTOR and inhibition of miR-125a-5p reversed the effect of CYTOR on cell viability (P < 0.05). In addition, cell invasion and migration were reduced by knockdown of CYTOR (P < 0.05), while the effect was reversed by co-transfection of miR-125a-5p inhibitor with shCYTOR (, p < 0.05). In addition, cell apoptosis was also reduced by knockdown of CYTOR (, p < 0.05), and the effect was reversed by co-transfection of miR-125a-5p inhibitor (, p < 0.05).
LASP1 was a direct target of hsa-miR-125a-5p
Starbase 2.0 also predicted that LASP1 was a potential target of hsa-miR-125a-5p (). Luciferase activity of pGL3-REPOR-LSP1-WT (P < 0.05), but not pGL3-REPOR-LSP1-Mut were reduced by hsa-miR-125a-5p mimic (). In addition, LASP1 was upregulated in HCC tissues (n = 100) (s). MiR-125a-5p mimic significantly decreased expression levels of LASP1 and JNK phosphorylation level (P < 0.05), and its inhibitor upregulated LASP1 and increased JNK phosphorylation level (, p < 0.05). In addition, shCYTOR-1 and shCYTOR-2 significantly decreased the expression levels of LASP1 and JNK phosphorylation level (, p < 0.05), while pcCYTOR significantly increased the expression levels LASP1 and JNK phosphorylation level (, p < 0.05).
Down-regulation of CYTOR restrained HCC progression in vivo
Finally, to explore whether knockdown of CYTOR could inhibit the development of HCC, in vivo experiments were conducted. As shown in , compared with the sh-NC group, sh-CYTOR-1 and sh-CYTOR-2 could inhibit tumor growth, while pc-CYTOR accelerated tumor growth. The number of positive cells in the sh-CYTOR-1 and sh-CYTOR-2 groups was decreased, while pc-CYTOR could increase the number of positive cells (). In addition, the expression levels of LASP1 were reduced in the sh-CYTOR-1 and sh-CYTOR-2 groups (P < 0.05), and the expression levels of LASP1 and JNK phosphorylation level were increased in the pc-CYTOR group (, p < 0.05).
Discussion
The most common histological type in liver tumors is HCC, which accounts for the second highest incidence of malignant tumors in men, and is also the sixth place in malignant tumors in women [Citation31,Citation32]. The risk factors of HCC include age, gender, and cirrhosis, but the main cause could also be excessive drinking [Citation33]. These pathogenic factors often lead to the accumulation of fluid, which also leads to early HCC that lack symptoms [Citation34]. Most patients are diagnosed at late stages and unable to operate for liver transplantation. Molecular targeted therapy has attracted extensive attention and shows a broad application prospect [Citation35,Citation36].
LncRNAs have attracted attention due to the unique structure and potential functions in cancer treatment [Citation37]. LncRNAs have high specificity in tumor tissues [Citation38]. Studies have shown that lncRNAs affect HCC progression and prognosis through various pathyways [Citation39,Citation40]. For example, highly expressed lncRNA ANRIL promotes HCC proliferation by regulating miR-122-5p [Citation41]. CYTOR is abnormally expressed in various cancers. For example, the expression of CYTOR is significantly elevated in colon cancer [Citation15]. In our study, CYTOR was significantly elevated in HCC cells and tissues. Sh-CYTOR could induce cell apoptosis and inhibit cell proliferation, and effectively inhibited tumor in mice, and the results of pc-CYTOR were reversed in in vivo experiments. Therefore, CYTOR might function as an oncogene to control HCC development.
LncRNAs regulate cellular activities and protein translation by regulating miRNAs [Citation42] and involve in many biological functions [Citation43], such as tumor differentiation, migration, and apoptosis. Studies on the relationship between miRNAs and HCC have gained more attention [Citation44]. MiRNAs might be utilized as potential targets for HCC treatment [Citation45]. For example, miR-339 inhibited invasion and prognosis of HCC by regulating ZNF689 [Citation46]. Another study found that miR-429 regulates HCC cell metastasis by targeting RAB23. It has been reported that miR-125a-5p-abundant exosomes derived from mesenchymal stem cells suppress chondrocyte degeneration via targeting E2F2 in traumatic osteoarthritis [Citation47]. MiR-125a-5p inhibits non-small cell lung cancer migration [Citation48]. Overexpression of miR-125a-5p inhibited HCC proliferation and induced HCC apoptosis in vitro and in vivo, and knockdown of miR-125a-5p had the opposite effects. In addition, MAP3K11 and PTPN1 were identified as targets of miR-125a-5p. Knockdown of MAP3K11 and PTPN1 induce HCC cell apoptosis and suppress HCC cell proliferation through the JNK/MAPK signaling pathway [Citation49]. MiR-125a-5p here was downregulated in HCC and negatively correlated with CYTOR. Co-transfection of CYTOR with miR-125a-5p inhibitor reversed the effect of sh-miR-125a-5p on apoptosis, proliferation, tumor volume and weight, suggesting that CYTOR promotes HCC growth by modulating miR-125a-5p.
During tumor formation, miRNAs involve in tumor progression and formation [Citation50]. LASP1 is closely related to tumor proliferation [Citation51]. LASP1 is highly upregulated in breast cancer cells than that of normal breast cells [Citation52]. LASP1 was shown to be up-regulated in nasopharyngeal carcinoma [Citation53]. In this study, miR-125a-5p targets LASP1, based on the observation that its mimic could reduce the expression levels of LASP1, and its inhibitor increased the expression levels of LASP1, while sh-CYTOR could downregulate LASP1, and pc-CYTOR could upregulate LASP1. Also, sh-CYTOR-1 and sh-CYTOR-2 could inhibit tumor growth. Moreover, knockdown of CYTOR reduced the expression levels of LASP1, and overexpression of CYTOR increased the expression levels of LASP1 in vivo.
This study indicated that CYTOR promoted HCC development via the miR-125a-5p/LASP1 axis (graphical abstract). Furthermore, we revealed a functional CYTOR/miR-125a-5p/LASP1 axis that suppresses progression of HCC. There are also a few limitations in this study. There are more other targets that mediate the roles of CYTOR in HCC. Additional experiments are needed to explore more underlying mechanisms of CYTOR in HCC for identification of effective therapeutic targets.
In conclusion, lncRNA CYTOR was found to be upregulated in HCC and able to promote HCC progression via the miR-125a-5p/LASP1 axis. This suggests that the CYTOR/miR-125a-5p/LASP1 axis may act as a new ceRNA regulatory network, participating in the HCC progression, thus accelerating the malignant processes of HCC. These findings suggest that CYTOR may serve as a potential therapeutic target and a novel biomarker for the precise treatment of HCC.
List of abbreviations:
LncRNA CYTOR (CYTOR); Actin Skeletal Protein 1 (LASP1); hepatocellular carcinoma (HCC); Long-chain non-coding RNA (LncRNA); carboxy-terminal 1 scr homology domain (SH3); Wild type (WT); mutant (MUT); cell-counting kit-8 (CCK-8).
Highlights:
1, CYTOR was upregulated in HCC tumors and cell lines.
2, CYTOR promoted HCC progression.
3,MiR-125a-5p/LASP1 axis mediated the function of CYTOR in HCC progression.
Declarations
Ethics approval and consent to participate
This study was approved by the First Affiliated Hospital of Dalian Medical University. Written consent for participation was obtained from each patient.
Author contribution
Guarantor of integrity of the entire study:XG. Study concepts:XG and YL. Study design:XG. Definition of intellectual content: YL. All authors help prepare this manuscript, and the submitted version was approved by all authors.
Data availability
The datasets used analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental Material
Download Zip (5 KB)Acknowledgments
We thank the financial support from Doctoral Scientific Research Foundation of Liaoning Province, 2020-BS-280
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ringelhan M, Pfister D, O’Connor T, et al. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19(Suppl):222–232.
- Wang Z, Wang MD, Zhen-Li LI, et al. Immunotherapy for hepatocellular carcinoma:research advances and clinical prospects. Acad J Second Military Med Univ. 2017;38(8):953–960.
- Chan A, Chan S, Chok K, et al. Treatment strategy for recurrent hepatocellular carcinoma: salvage transplantation, repeated resection or radiofrequency ablation? Liver Transpl. 2013;19(4):411–419.
- Sadiq TS, Shrestha R, Weeks S, et al. Laparoscopic radiofrequency ablation provides local control of hepatocellular carcinoma in patients awaiting liver transplant. J Am Coll Surg. 2004;199(3–supp–S):16.
- Gardini AC, Foschi FG, Conti F, et al. Immune inflammation indicators and ALBI score to predict occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. Digest Liver Dis. 2018;50(1):50–51.
- Young IC, Chuang ST, Gefen A, et al. A novel compressive stress-based osteoarthritis-like chondrocyte system. Exp Biol Med. 2017;242(10):1062–1071.
- Chen W, Jia Y, Qi Z, et al. Downregulation of lncRNA OGFRP1 inhibits hepatocellular carcinoma progression by AKT/mTOR and Wnt/β-catenin signaling pathways. Cancer Manage Res. 2018;10:1817–1826.
- Qi P, Wan-run L, Meng Z, et al. E2F1 induces LSINCT5 transcriptional activity and promotes gastric cancer progression by affecting the epithelial-mesenchymal transition. Cancer Manage Res. 2018;10:2563–2571.
- Qian L, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):9.
- Huang X, Gao Y, Qin J, et al. LncRNA-MIAT promoted proliferation and invasion of HCC cells via sponging miR-214. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2017;314(5):G559.
- Sui J, Miao Y, Han J, et al. Systematic analyses of anovel lncRNA-associated signature as the prognostic biomarker for Hepatocellular Carcinoma . Cancer Medicine. 2018;7:3240–3256.
- Zhang Y, Lu C, Cui H. Long non-coding RNA SNHG22 facilitates hepatocellular carcinoma tumorigenesis and angiogenesis via DNA methylation of microRNA miR-16-5p. Bioengineered. 2021;12(1):7446–7458.
- Feng Y, Wei G, Zhang L, et al. LncRNA DARS-AS1 aggravates the growth and metastasis of hepatocellular carcinoma via regulating the miR-3200-5p-Cytoskeleton associated protein 2 (CKAP2) axis. Bioengineered. 2021;12(1):8217–8232.
- Zhou Y, Li K, Zou X, et al. LncRNA DHRS4-AS1 ameliorates hepatocellular carcinoma by suppressing proliferation and promoting apoptosis via miR-522-3p/SOCS5 axis. Bioengineered. 2021;12(2):10862–10877.
- Yue B, Liu C, Sun H, et al. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol Ther. 2018;26(5):1287–1298.
- Wang P, Jing L, Wei Z, et al. A Novel LncRNA-miRNA-mRNA Triple Network Identifies LncRNA RP11-363E7.4 as An Important Regulator of miRNA and Gene Expression in Gastric Cancer. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2018;47:1025–1041.
- Alenka MI, Mojca T, Bti E, et al. Identifying Novel Glioma-Associated Noncoding RNAs by Their Expression Profiles. Int J Genomics. 2017;2017 :1–18.
- Sun CC, Zhang L, Li G, et al. The lncRNA PDIA3P Interacts with miR-185-5p to Modulate Oral Squamous Cell Carcinoma Progression by Targeting Cyclin D2. Mol Ther Nucleic Acids. 2017;9:100–110.
- Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: an Overview. Methods in Molecular Biology (Clifton, N.J.). 2017;1509:1.
- Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29.
- Lai L, Chen J, Wang N, et al. MiRNA-30e mediated cardioprotection of ACE2 in rats with Doxorubicin-induced heart failure through inhibiting cardiomyocytes autophagy. Life Sciences. 2017;169:69.
- Morishita A, Masaki T. mi RNA in hepatocellular carcinoma. Hepatol Res Off J Jap Soc Hepatol. 2015;45(2):128–141.
- Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–1067.
- Hsieh TH, Hsu CY, Tsai CF, et al. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget. 2014;6(1):494.
- Jiang L, Huang Q, Chang J, et al. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp Lung Res. 2011;37(7):387–398.
- Chiyomaru T, Enokida H, Kawakami K, et al. Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol Sem Orig Invest. 2012;30(4):434–443.
- Traenka C, Remke M, Korshunov A, et al. Role of LIM and SH3 Protein 1 (LASP1) in the Metastatic Dissemination of Medulloblastoma. Cancer Res. 2010;70(20):8003–8014.
- Cao Y, Wang P, Ning S, et al. Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget. 2016;7(27):41737–41747.
- Avram S, Coricovac DE, Pavel IZ, et al. Standardization of A375 human melanoma models on chicken embryo chorioallantoic membrane and Balb/c nude mice. Oncol Rep. 2017;38(1):89–99.
- Toyokuni S, Kawaguchi W, Akatsuka S, et al. Intermittent microwave irradiation facilitates antigen-antibody reaction in Western blot analysis. Pathol Int. 2003;53(4):259–261.
- El-Serag HB. Advances in the management of hepatocellular carcinoma. Clinical Advances in Hematology & Oncology: H&O. 2017;15 Suppl 9(8):2–6.
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227.
- Jiang T, Guan LY, Ye YS, et al. MiR-874 inhibits metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting SOX12. American Journal of Cancer Research. 2017;7(6):1310.
- Zhou W, Wang Q, Xu Y, et al. RMP promotes epithelial-mesenchymal transition through NF-κB/CSN2/Snail pathway in hepatocellular carcinoma. Oncotarget. 2017;8(25):40373–40388.
- Lu M, Kong X, Wang H, et al. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(5):87955–87970.
- Liu L, C X, L E, et al. MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget. 2017;8(40):68263.
- Geng C, Wang Z, Wang D, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. NAR. 2012;41:D983–D986.
- Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329.
- Ye S, Yang L, Zhao X, et al. Bioinformatics method to predict two regulation mechanism: TF–miRNA–mRNA and lncRNA–miRNA–mRNA in pancreatic cancer. Cell Biochem Biophys. 2014;70(3):1849–1858.
- Shi F, Xiao F, Ding P, et al. Long Noncoding RNA Highly Up-regulated in Liver Cancer Predicts Unfavorable Outcome and Regulates Metastasis by MMPs in Triple-negative Breast Cancer. Arch Med Res. 2016;47(6):446–453.
- Ishii N, Ozaki K, Sato H, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–1099.
- Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281(16):3766–3775.
- Taylor MA, Sossey-Alaoui K, Thompson CL, et al. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Investig. 2012;123(1):150–163.
- Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29(11):2126–2131.
- Li C, Pan S, Song Y, et al. Silence of lncRNA MIAT protects ATDC5 cells against lipopolysaccharides challenge via up-regulating miR-132. Artif Cells Nanomed Biotechnol. 2019;47(1):2521–2527.
- Wang YL, Chen C, Wang XM. Effects of miR-339-5p on invasion and prognosis of hepatocellular carcinoma. Clinics and Research in Hepatology and Gastroenterology. 2016;40(1):51–56.
- Xia Q, Wang Q, Lin F, et al. miR-125a-5p-abundant exosomes derived from mesenchymal stem cells suppress chondrocyte degeneration via targeting E2F2 in traumatic osteoarthritis. Bioengineered. 2021;12(2):11225–11238.
- Wang RJ, Zheng YH, Wang P, et al. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(1):765–771.
- Xu X, Tao Y, Niu Y, et al. miR-125a-5p inhibits tumorigenesis in hepatocellular carcinoma. Aging (Albany NY). 2019;11(18):7639–7662.
- Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(39):15472–15477.
- Nishikawa R, Goto Y, Sakamoto S, et al. Tumor‐suppressive micro RNA‐218 inhibits cancer cell migration and invasion via targeting of LASP 1 in prostate cance. Cancer Sci. 2014;105(7):802–811.
- Zhang Y, Wang YY, Wei LI. Expression of Lasp1 protein in invasive breast cancer tissue and its significance in clinical prognosis. Mod Med J. 2016;44:519–522.
- Gao Q, Tang L, Wu L, et al. LASP1 promotes nasopharyngeal carcinoma progression through negatively regulation of the tumor suppressor PTEN. Cell Death Dis. 2018;9(3):393.