ABSTRACT
The function of long non-coding RNA LHFPL3 antisense RNA 1 (LHFPL3-AS1) in cancer progression has been studied, while its role in nasopharyngeal carcinoma (NPC) remains unclear. This study aims to unravel the effects of LHFPL3-AS1 on NPC progression via microRNA (miR)-143-5p/homeobox A6 (HOXA6) axis. NPC tissues were collected and NPC cells were cultured. NPC cells were subjected to radiation therapy to construct the radiation therapy resistance NPC cell line. The levels of LHFPL3-AS1, miR-143-5p and HOXA6 in NPC cells and tissues were examined. LHFPL3-AS1, miR-143-5p or HOXA6 expression was changed and then transfected into radiation-resistant NPC cells to detect cell proliferation, colony formation, migration, invasion and cell apoptosis in vitro. The tumorigenesis in nude mice in vivo was conducted to detect tumor growth. The targeting relations among LHFPL3-AS1, miR-143-5p and HOXA6 were validated. It was discovered that LHFPL3-AS1 and HOXA6 expression was elevated while the miR-143-5p level was depleted in radiation-resistant NPC cells and NPC tissues. The silenced LHFPL3-AS1 or augmented miR-143-5p repressed the proliferation, colony formation, migration and invasion of radiation-resistant NPC cells, while accelerated cell apoptosis in vitro. Silenced LHFPL3-AS1 hindered tumor growth in vivo. MiR-143-5p deletion reversed the effects of reduced LHFPL3-AS1; while HOXA6 upregulation reversed the effects of enriched miR-143-5p. LHFPL3-AS1 sponged miR-143-5p that targeted HOXA6. It is concluded that the down-regulated LHFPL3-AS1 retards the development of radiation-resistant NPC cells via sponging miR-143-5p to modulate HOXA6. This study reveals novel therapeutic targets for NPC treatment.
Introduction
Nasopharyngeal carcinoma (NPC) is defined as an epithelial carcinoma stemming from the nasopharyngeal mucosal lining. The tumor is often observed at the pharyngeal recess (fossa of Rosenmüller) [Citation1]. Predisposing genetic factors, environmental carcinogens, and Epstein-Barr virus infection lead to the occurrence of genetic and epigenetic alterations that accelerate NPC development [Citation2]. There are substantial advances for NPC treatment recently, for instance, liquid biopsies with plasma Epstein-Barr virus DNA, minimally invasive surgery of nasopharynx, and induction chemotherapy and immunotherapy with a PD-1 inhibitor [Citation3]. Nevertheless, the initial misdiagnosis of NPC is common and its presenting symptoms are variable. More severely, the diagnosis for NPC encounters great obstacles due to its anatomic isolation [Citation4]. Therefore, it is pivotal to gain a better understanding of NPC treatment via exploring novel therapeutic targets.
Long non-coding RNAs (LncRNAs) have been revealed to alter the efficacy of NPC therapy and serve as novel therapeutic targets for NPC treatment [Citation5]. For instance, lncRNA plasmacytoma variant translocation 1 (PVT1) is found to exist high level in NPC cells and is implicated in shorter survival time of NPC patients [Citation6]. LncRNA FAM225A is also implicated in tumorigenesis and metastasis in NPC [Citation7]. LncRNAs play oncogenic or oncogenic roles by regulating the biological behavior of tumor cells, and the dysregulated lncRNAs may affect apoptosis, growth, metabolism or metastasis in cancer progression [Citation8,Citation9]. Among them, the function of LHFPL3-AS1 has been reported in both melanoma and oral squamous cell carcinoma [Citation10,Citation11], but its role was not explored in NPC, which has attracted our attention. In addition, it was predicted that LHFPL3-AS1 had binding sites with microRNA (miR)-143-5p by the RNA22 website (https://cm.jefferson.edu/rna22/Interactive/). The aberrantly expressed miRs exert crucial effects on numerous neoplasms, including NPC. Additionally, miR signatures also function as potential predictors for response and clinical outcomes of NPC [Citation12]. For instance, miR-143 has been validated to retard the biological activity of NPC cells in vitro and repress xenograft tumor growth in vivo [Citation13,Citation14]. Furthermore, the tumor invasion depth and lymph node metastasis in NPC are both hindered by the enforced miR-143 expression [Citation15]. MiR-143-5p has been reported to alleviate malignant phenotypes of lung adenocarcinoma cells [Citation16], while its function in NPC remained obscure. Moreover, the bioinformatics website Targetscan predicted that there existed binding sites between miR-143-5p and homeobox A6 (HOXA6). HOXC6 is verified to display high level in NPC cells, and its robust expression is correlated to accelerated tumor stage, advanced nodal status and worse prognosis [Citation17]. As for HOXA6, it has been revealed that the high-expressed HOXA6 in colorectal cancer cells is accounted for repressed apoptosis of CC cells and accelerated biological activities of colorectal cancer cells [Citation18].
As stated above, LHFPL3-AS1, miR-143-5p and HOXA6 exert pivotal effects on regulating cancer progression, while their functions in NPC progression remained unknown. Given that we hypothesized in the current research that LHFPL3-AS1 might modulate NPC progression via the miR-143-5p/HOXA6 axis. By unraveling the potential mechanism of LHFPL3-AS1/miR-143-5p/HOXA6 axis, the study was committed to reveal novel therapeutic targets for NPC treatment.
Material and methods
Ethics statement
The study was approved by the ethics committee of Hainan Cancer Hospital. All participants signed the informed consent form and enjoyed the right to know. The animal protocol for this study was approved by the Institutional Animal Care and Use Committee of Hainan Cancer Hospital.
Clinical sample collection
One-hundred and thirty NPC tissues were collected from patients in Hainan Cancer Hospital. None of NPC patients had ever received radiotherapy or chemotherapy before biopsy; while after biopsy, all patients were received radiotherapy with complete clinical data. There were 96 males and 34 females at median age of 48. The follow-up duration was 5 years. The patients’ survival time was started from the day of radiotherapy and ended with patients’ death, lost contact or end of follow-up.
Cell culture
Human NPC cell lines C666-1, HNE-3, HK-1 (Ningbo Mingzhou Biotechnology Co. Ltd, Zhejiang, China) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, CA, USA) containing 10% fetal bovine serum (FBS, Gibco, CA, USA) in a humid incubator at 37°C with 5% CO2. Human immortalized nasopharyngeal epithelial cell line NP69 (Ningbo Mingzhou Biotechnology Co. Ltd., Zhejiang, China) was cultured in a serum-free medium (Invitrogen) containing the bovine pituitary extract.
Establishment of radiation-resistant cell lines
The NPC cell lines C666-1 and HNE-3 were irradiated with 2 Gy X-rays from a linear gas pedal (6 MV X-ray, UNIQUE™, Varian, Palo Alto, CA, USA) at a rate of 4 Gy/min to the top of the plates. Then, the cells were placed back into the incubator. When the cells reached 70% confluence, they were irradiated again at a certain dose, with each fraction gradually increasing until 10 Gy was reached. The total dose was 60 Gy. Radiation-resistant NPC cells were obtained at the indicated time points. Sub-lineage cells were named C666-1/R or HNE-3/R [Citation19].
Cell transfection and grouping
Cells at the logarithmic growth phase were divided into the following groups: small interfering RNA (si)-negative control (NC) group (transfected with negative control siRNA), si-LHFPL3-AS1-1 group (transfected with LHFPL3-AS1-1 siRNA-1), si-LHFPL3-AS1-2 group (transfected with LHFPL3-AS1-2 siRNA-2), mimic-NC group (transfected with mimic NC), mimic-miR-143-5p group (transfected with miR-143-5p mimic), si-LHFPL3-AS1 + inhibitor-NC group (transfected with si-LHFPL3-AS1 and inhibitor NC), si-LHFPL3-AS1 + miR-143-5p inhibitor group (transfected with si-LHFPL3-AS1 and miR-143-5p inhibitor), miR-143-5p mimic + overexpression (oe)-NC group (transfected with miR-143-5p mimic and empty expressionvectors), and miR-143-5p mimic + oe-HOXA6 group (transfected with miR-143-5p mimic and high-expressed HOXA6 vectors). The constructs used above were all constructed by GenePharma Co. Ltd (Shanghai, China). Cell transfection was performed under the manufacturer’s instructions of Lipofectamine 3000 reagent (Life Technologies Corporation, Carlsbad, CA, USA).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The Trizol reagent (Thermo Fisher Scientific, Carlsbad, CA, USA) was used for total RNA extraction, and RNA purity was analyzed by the ultraviolet spectrophotometer. An All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, Md, USA) was used to determine the miR-143-5p expression. HOXA6 mRNA and LHFPL3-AS1expression levels were measured using a PrimeScript RT Reagent Kit and SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Japan). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was the standardized control. Gene expression level was calculated by 2−ΔΔCt method and the primer sequences were shown in Supplementary Table 1 [Citation10].
Western blot assay
The r cells were lysed in radio-immunoprecipitation assay (RIPA) cell lysis buffer (Invitrogen) containing 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, CA, USA) and proteinase inhibitor (Sigma-Aldrich). The protein extract was quantified by the bicinchoninic acid protein assay kit (Thermo Fisher Scientific), separated with 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a 0.2 μm polyvinylidene fluoride membrane (Merck Millipore, MA, USA). After sealing with 5% bovine serum albumin (Sigma-Aldrich) in tris-buffered saline with tween 20 for 2 hours, the membrane was incubated with the primary antibodies HOXA6 (1:3,000, BIOSS, MA, USA), GAPDH (1:2500, Abcam, MA, USA) overnight at 4°C, and then followed by a 2-h incubation with the horseradish peroxidase-coupled secondary antibody. The reaction blots were examined by the enhanced chemiluminescence kit (Merck Millipore).
Cell counting kit (CCK)-8 assay
Cell viability was analyzed using a CCK-8 kit (Dojindo MolecularTechnologies, Shanghai, China). Cells were seeded into 96-well plates at a density of 5000 cells per well and treated with different Gy radiation. The CCK-8 solution was added to each well at the indicated time points after radiation (day 1, day 2, day 3, day 4) concerning the manufacturer’s instructions. Absorbance was measured at 490 nm using the microplate reader (ELx800, BioTek, Winooski, VT, USA).
Colony formation assay
The transfected cells were cultured in six-well plates (300 cells/well) for 14 days. The medium was then discarded, and each well was supplemented with 2 mL 4% paraformaldehyde, and fixed for 15 minutes. Then, 4% paraformaldehyde was discarded, and each well was added with 2 mL crystalline violet working solution and stained for 30 minutes. The excess staining solution was washed with running water, and then the sample was dried. The number of colonies with 50 or more cells was calculated.
Flow cytometry
Cells were seeded in 6-well plates at 1 × 105/well. After cell collection and digestion, the cells were then stained with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide using a FITC Annexin V cell apoptosis assay kit (BD Biosciences, NJ, USA) and then immediately analyzed by flow cytometer (BD Biosciences). The percentage of apoptotic cells was calculated using Cell QuestPro software (BD Biosciences) [Citation11]
Transwell assay
The cells were resuspended in the serum-free RPMI 1640 medium. Then, the cell confluence was adjusted to 5 × 105 cells/mL and 200 μL cells were added into the upper Transwell chamber. The RPMI-1640 containing 10% FBS was added to the bottom chamber and cultured for 24 hours. Thereafter, the cells were treated with formaldehyde fixation and crystal violet staining, and observed with a microscope. As for the detection of cell invasion, 50 μg Matrigel was coated in the bottom of the Transwell chamber, and gelatinized at 37°C for 30 minutes. The other steps were the same as the process above.
Tumorigenesis in nude mice
Male BALB/c nude mice (n = 5 each group; aging 5 weeks; weighing 16–18 g) were purchased from Guangdong Laboratory Animal Monitoring Institute (Guangzhou, China). Mice were fed at constant temperature (20–25°C) and humidity (40–70%) under specific pathogen-free circumstances, and supplied with water and food and a 12-hour light/dark cycle. One week later, mice were anesthetized through isoflurane at 4–5% induction and 1–3% maintenance. The right anterior limb of nude mice was subcutaneously injected with cells that were steadily transfected with si-NC or si-LHFPL3-AS1. Each mouse was injected with 3.0 × 106 cells which were suspended in 200 μL normal saline. Tumors grew from the 5th day, and the tumor weight, length (L) and width (W) were measured every other day. After injection of the cells for 21 days, mice were euthanized by cervical dislocation after anesthesia with 4–5% isoflurane. Tumor tissues were excised, photographed and weighed. Tumor volume = 0.52 × L × W2 [Citation20].
Dual luciferase reporter gene assay
The mutant (MUT) and wild type (WT) sequences (MUT-LHFPL3-AS1 and WT-LHFPL3-AS1, MUT-HOXA6 and WT-HOXA6) were amplified and cloned into the luciferase reporter gene vector pGL3-vectors (Promega, Madison, WI, USA). The MUT or WT luciferase reporter gene vectors were then co-transfected into NPC cells with miR-143-5p mimic or mimic NC . After 48-h transfection, the luciferase activity of firefly and renilla was assessed by the dual luciferase reporter gene assay kit (Promega). The renilla luciferase activity was set as the endogenous reference, and the relative activity was expressed as the ratio of the luciferase activity of firefly to that of renilla.
RNA immunoprecipitation (RIP) assay
The Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, MA, USA) was used to determine the relationship between LHFPL3-AS1 and miR-143-5p. Antibodies used for RIP analysis included anti-AGO2 and control IgG (Millipore, USA). The co-precipitated RNA was used for complementary DNA synthesis and evaluation of RT-qPCR.
Statistical analysis
All statistical analysis was performed using GraphPad Prism 6 (GraphPad, La Jolla, CA). The measurement data were expressed as mean ± standard deviation. The t-test was adopted for comparison between two groups; and analysis of variance (ANOVA) and the Tukey’s test were used for comparison among multiple groups. Survival curves were plotted using the Kaplan–Meier method. The count data were expressed by frequency and percentage and analyzed using Fisher's exact test or chi-square test. P < 0.05 was considered as an indicator of statistical significance.
Results
LHFPL3-AS1 is highly expressed in NPC tissues and cells and is associated with shorter overall survival (OS)
LHFPL3-AS1 is up-regulated in OSCC tissues and cell lines and exerts crucial effects on modulating the development of OSCC [Citation10]. In light of this, we first examined LHFPL3-AS1 expression in human NPC tissues, suggesting that LHFPL3-AS1 expression was elevated in NPC tissues ()). Subsequently, we examined LHFPL3-AS1 level in several NPC cell lines, and the results indicated that LHFPL3-AS1 exhibited a high level in NPC cell lines (C666-1, HNE-3 and HK-1), and its high expression was most significant in HNE-3 and C666-1 cell lines ()), so we screened HNE-3 and C666-1 cell lines for subsequent experiments.
Figure 1. LHFPL3-AS1 is high-expressed in NPC tissues and cells and is associated with shorter OS. (a) LHFPL3-AS1 level in NPC tissues was detected by RT-qPCR; (b) LHFPL3-AS1 levels in NPC cell lines were examined by RT-qPCR; (c) Relation curve between LHFPL3-AS1 expression and OS of NPC patients. The data was expressed as mean ± standard deviation. * P < 0.05 vs. the NP69 cells.
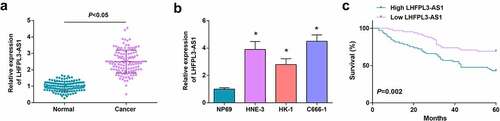
Thereafter, we investigated the clinical association between LHFPL3-AS1 expression and NPC patients’ characteristics. According to the median level of LHFPL3-AS1 in NPC samples, we classified all samples into the high-expressed group (n = 65) and the low-expressed group (n = 65). The outcome of Fisher’s exact test suggested that the high-expressed LHFPL3-AS1 might be associated with TNM and lymph node metastasis (Supplementary Table 2). We then further probed the effect of LHFPL3-AS1 on prognosis in NPC patients. Through Kaplan–Meier analysis, it turned out that the OS was saliently shortened in patients with high LHFPL3-AS1 ()).
Silenced LHFPL3-AS1 inhibits radiation-resistant NPC cell growth in vitro and tumor growth in vivo
To further explore the impacts of LHFPL3-AS1 on radiation-resistant NPC cells, we treated HNE-3 and C666-1 cell lines with X-ray irradiation radiotherapy to construct radiotherapy-resistant strains HNE-3/R and C666-1/R. We continued to verify the expression of LHFPL3-AS1 in C666-1/R and HNE-3/R after X-ray irradiation treatment by RT-qPCR, which suggested that LHFPL3-AS1 expression was significantly elevated in C666-1/R and HNE-3/R cells after X-ray treatment (); Supplementary fig. 1A). The CCK-8 assay was performed to determine the radiotherapy resistance. The CCK-8 results revealed that the viability was significantly reduced of C666-1 and HNE-3 cells exposed to X-rays under the same conditions, while C6661/R and HNE-3/R cells treated with X-ray irradiation displayed no significant change in viability. Such results validated that the constructed radiation-resistant NPC cells C666-1/R and HNE-3/R were both resistant to radiotherapy (), Supplementary fig. 1B).
Figure 2. Silenced LHFPL3-AS1 inhibits radiation-resistant NPC cell growth in vitro and tumor growth in vivo. (a) RT-qPCR was used to detect the expression of LHFPL3-AS1 in both parent and resistant strains; (b). cell proliferation ability of parental and resistant strains was evaluated by CCK-8 assay; (c) LHFPL3-AS1 expression in NPC cells after the down-regulation of LHFPL3-AS1 was detected by RT-qPCR; (d) Proliferation of NPC cells after the down-regulation of LHFPL3-AS1 was examined by CCK-8 assay; (e) Colony formation ability of NPC cells after the down-regulation of LHFPL3-AS1 was assessed by colony formation assay; (f,g) Migration and invasion of NPC cells after the down-regulation of LHFPL3-AS1 were detected by Transwell assay; (h) Apoptosis of NPC cells after the down-regulation of LHFPL3-AS1 was detected by flow cytometry; (i) The weight and volume of the tumor were detected in nude mice. The data iwas expressed as mean ± standard deviation. * P < 0.05 vs. C666-1 cells; # P < 0.05 vs. the si-NC group.
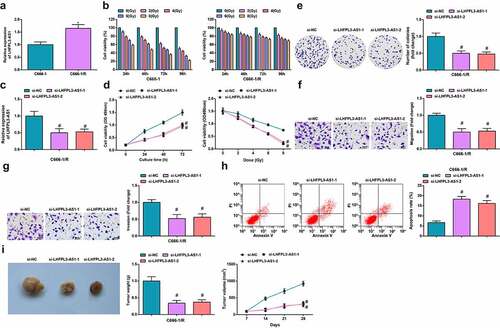
Subsequently, after the construction of interfering sequences that specifically targeted LHFPL3-AS1 and its negative control, we transfected si-NC and si-LHFPL3-AS1 into HNE-3/R and C666-1/R cell lines, respectively. A deletion in LHFPL3-AS1 expression after transfection with si-LHFPL3-AS1 was detected through RT-qPCR, which reflected that the interfering sequences that specifically targeted LHFPL3-AS1 were effective and the transfection was successful (), Supplementary fig. 1C). In response to the silenced LHFPL3-AS1, the proliferation of HNE-3/R and C666-1/R cells was constrained as shown in the CCK-8 assay, and the radiation therapy resistance was also repressed (); Supplementary fig. 1D); the number of colonies was reduced as suggested in colony formation assay (); Supplementary fig. 1E); the migration and invasion capacity were suppressed as reflected in Transwell assay (; Supplementary fig. 1F, G); while the apoptosis rate of HNE-3/R and C666-1/R cells was elevated, thus dampening cell survival as shown in flow cytometry (), Supplementary fig. 1H).
Meanwhile, to unravel the impacts of LHFPL3-AS1 on the biological progress of NPC in vivo, cells with stably expressed si-NC and si-LHFPL3-AS1 were injected into nude mice to establish xenograft models. The results showed that, after the injection of NPC cells that transfected with si-LHFPL3-AS1, the tumor volume and weight of nude mice were drastically decreased ()).
These outcomes above reflected that the silencing of LHFPL3-AS1 could hinder the biological function of radiation-resistant NPC cells in vitro and repressed tumor growth in vivo, thus exerting positive effects on the radiation therapy of NPC.
LHFPL3-AS1 binds to miR-143-5p
To understand the relation between LHFPL3-AS1 and miR-143-5p, we used the RNA22 website, which predicted that there were binding sites between LHFPL3-AS1 and miR-143-5p ()).
Figure 3. LHFPL3-AS1 binds to miR-143-5p. (a) The presence of binding sites between LHFPL3-AS1 and miR-143-5p was predicted by Jefferson website; (b) Binding relation between LHFPL3-AS1 and miR-143-5p was validated by dual luciferase reporter gene assay; (c) enrichment levels of LHFPL3-AS1 and miR-143-5p were detected by RIP assay; (d) miR-143-5p expression in NPC cell lines after the silencing of LHFPL3-AS1 was examined by RT-qPCR. The data was expressed as mean ± standard deviation. & P < 0.05 vs. the si-NC group.
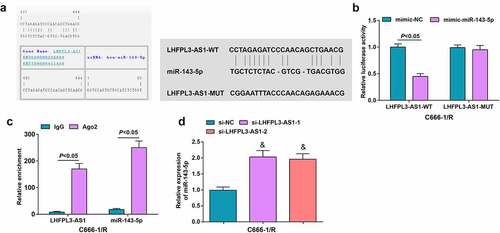
To further explore whether LHFPL3-AS1 directly regulates miR-143-5p, we conducted the dual luciferase reporter gene assay. It came out that the luciferase activity was impaired in cells co-transfected with LHFPL3-AS1-WT and mimic-miR-143-5p ()). Meanwhile, as reflected by the RIP assay, LHFPL3-AS1 and miR-143-5p were significantly enriched after Ago2 treatment ()), suggesting that there was specific targeting relation between LHFPL3-AS1 and miR-143-5p.
To further demonstrate the regulatory relation between LHFPL3-AS1 and miR-143-5p, RT-qPCR was performed to examine miR-143-5p levels in cells transfected with si-LHFPL3-AS1 or si-NC, showing obviously amplified miR-143-5p expression after the decrement of LHFPL3-AS1 ()).
Up-regulated miR-143-5p retards the biological function of radiation-resistant NPC cells
We first observed the changes of miR-143-5p expression in C666-1/R and HNE-3/R cells after X-ray irradiation treatment, and the results disclosed that X-ray irradiation significantly decreased the expression of miR-143-5p (); Supplementary fig. 2A)
Figure 4. Up-regulated miR-143-5p retards the biological function of NPC cells. (a) miR-143-5p expression in NPC cells after X-ray irradiation treatment was detected by RT-qPCR; (b) miR-143-5p expression in NPC cells after the up-regulation of miR-143-5p was detected by RT-qPCR; (c) proliferation of NPC cells after the up-regulation of miR-143-5p was examined by CCK-8 assay; (d) colony formation ability of NPC cells after the up-regulation of miR-143-5p was assessed by colony formation assay; (e,f) Migration and invasion of NPC cells after the up-regulation of miR-143-5p were detected by Transwell assay; (g) Apoptosis of NPC cells after the up-regulation of miR-143-5p was detected by flow cytometry. The data was expressed as mean ± standard deviation.# P < 0.05 vs. the mimic-NC group; * P < 0.05 vs. C666-1 cells.
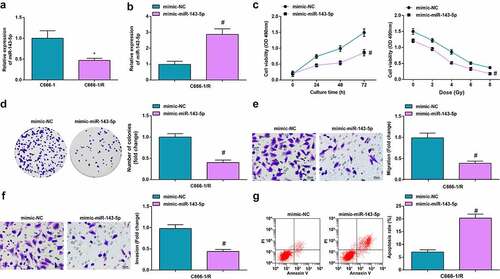
We transfected mimic-miR-143-5p and mimic-NC into C666-1/R and HNE-3/R cell lines, respectively, and the outcome of RT-qPCR unearthed that miR-143-5p was robustly expressed after transfected with mimic-miR-143-5p (); Supplementary fig. 2B).
Subsequently, in response to the up-regulated miR-143-5p, the proliferation of HNE-3/R and C666-1/R cells was constrained as shown in the CCK-8 assay, and the radiation therapy resistance was also repressed (); Supplementary fig. 2C); the number of colonies was reduced as suggested in colony formation assay (); Supplementary fig. 2D); the migration and invasion capacity were suppressed as reflected in Transwell assay (; Supplementary fig. 2E, F); while the apoptosis rate of HNE-3/R and C666-1/R cells was elevated, thus dampening cell survival as shown in flow cytometry (), Supplementary fig. 2G).
Taken together, miR-143-5p amplification could block the biological activities of NPC cells and alleviate the radiation resistance of NPC cells.
MiR-143-5p targets HOXA6
HOXA6 encodes a transcription factor that regulates gene expression and cell differentiation. HOXA6 has been identified to play an important role in cell proliferation, differentiation and invasion, and it has been reported that HOXA6 might be radiation‑sensitive [Citation21,Citation22].
Initially, the targeting relation between miR-143-5p and HOXA6 was predicted through the bioinformatic website TargetScan (http://www.targetscan.org/vert_72/), and it was unveiled that there were binding sites between HOXA6 and miR-143-5p ()).
Figure 5. MiR-143-5p targets HOXA6. (a) Binding sites between miR-143-5p and HOXA6 were predicted by Targetscan; (b) Targeting relation between miR-143-5p and HOXA6 was validated by dual luciferase reporter gene assay; (c) HOXA6 expression was detected byWestern blot analysis. The data was expressed as mean ± standard deviation. % P < 0.05 vs. the mimic-NC group; $ P < 0.05 vs. the si-NC group.
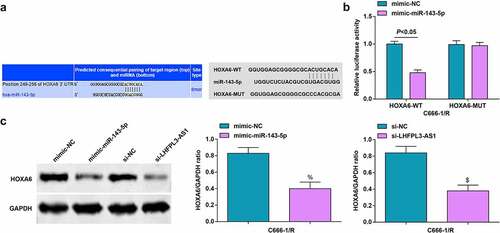
To validate the regulatory relation between HOXA6 and miR-143-5p, we also performed the dual luciferase reporter gene assay. It was uncovered that the luciferase activity was restrained after co-transfection with mimic-miR-143-5p and HOXA6-WT ()).
In radiation-resistant cells transfected with miR-143-5p mimic, HOXA6 level was downregulated, while HOXA6 level was amplified in cells transfected with si-LHFPL3-AS1 ())
These discoveries evidenced that miR-143-5p could target HOXA6 and regulate HOXA6 expression.
Down-regulated LHFPL3-AS1 hinders the development of radiation-resistant NPC cells via miR-143-5p/HOXA6 axis
Finally, to determine whether LHFPL3-AS1 could affect the biological progress of radiation-resistant NPC cells via miR-143-5p/HOXA6 axis, we transfected si-LHFPL3-AS1 + inhibitor NC, si-LHFPL3-AS1 + miR-143-5p inhibitor, miR-143-5p mimic + oe-NC, and miR-143-5p mimic + oe-HOXA6 into radiation-resistant C666-1/R cells, respectively. The cells were then also subjected to CCK-8 assay, colony formation assay, Transwell assay, and flow cytometry (–e)). It turned out miR-143-5p silencing could reverse the inhibitory effect of depleted LHFPL3-AS1 on the development of radiation-resistant NPC cells, while the upregulation of HOXA6 could reverse the impacts of miR-143-5p augmentation on suppressing the progression of C666-1/R cells. The results of HNE-3/R cells were the same as those in C666-1/R cells (Supplementary fig. 3).
Figure 6. Down-regulated LHFPL3-AS1 hinders the development of NPC via miR-143-5p/HOXA6 axis. (a) Proliferation of NPC cells was examined by CCK-8 assay; (b) Colony formation ability of NPC cells was assessed by colony formation assay; (c,d) Migration and invasion of NPC cells were detected by Transwell assay; E, apoptosis of NPC cells was detected by flow cytometry. The data was expressed as mean ± standard deviation.# P < 0.05 vs. the si-LHFPL3-AS1 +inhibitor NC group; * P < 0.05 vs. the miR-143-5p mimic + oe-NC group.
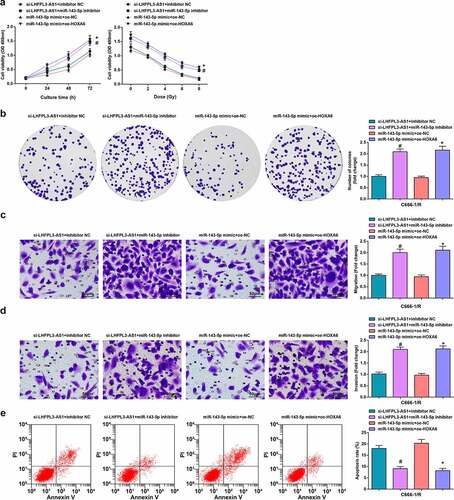
To sum up, decreased LHFPL3-AS1 could retard radiation-resistant NPC cell development through regulating the miR-143-5p/HOXA6 axis.
Discussion
NPC is a specific head and neck squamous cell carcinoma whose etiology is associated with Epstein-Barr virus infection, covert anatomical location, distinct racial and geographical difference, and frequent locoregional recurrence or metastasis [Citation23]. This research focused on the regulatory mechanism of LHFPL3-AS1 on NPC development. Collectively, it was manifested that down-regulated LHFPL3-AS1 could retard radiation-resistant NPC development via sponging miR-143-5p to modulate HOXA6.
Initially, this study uncovered that LHFPL3-AS1 was highly expressed in NPC cells and tissues, and the enforced expression of LHFPL3-AS1 was associated with short OS, TNM and lymph node metastasis of NPC patients, while the silencing of LHFPL3-AS1 could hinder the biological activities of NPC cells in vitro and tumor growth in vivo. The studies for exploring LHFPL3-AS1 function in NPC were insufficient, yet in other cancers, shreds of evidence indicated that LHFPL3-AS1 was involved in tumor pathophysiology and cancer progression. For instance, Peng et al. have elucidated that LHFPL3-AS1 is robustly expressed in melanoma tissues and cells, and is related to unfavorable prognosis to melanoma patients [Citation24]. Furthermore, LHFPL3-AS1 is also associated with accelerated tumorigenesis of melanoma stem cells via elevating B-cell lymphoma 2 protein [Citation11]. More precisely, LHFPL3-AS1 has been revealed to exhibit a high level in OSCC and is implicated with a low survival rate of OSCC patients; LHFPL3-AS1 deficiency can effectively retard the biological behavior of OSCC cells [Citation10].
Thereafter, it was predicted that there existed binding sites between LHFPL3-AS1 and miR-143-5p. Mounting evidences have unveiled the crucial regulatory mechanism of miR-143 in multiple cancers, especially in NPC. As a member of miR-143, miR-143-5p also displayed great research value. For instance, it has been reported that miR-143-5p overexpression contributes to restraining cervical cancer progression [Citation25] and attenuating the malignant phenotypes of esophageal squamous cell carcinoma [Citation26]; on the contrary, miR-143-5p deficiency is associated with larger tumor size and poor survival rate of patients with galbladder cancer [Citation27]. However, the function of miR-143-5p in NPC was rarely explored. Therefore, we chose miR-143-5p to further explore the novel therapeutic targets for NPC treatment. In the current study, it was demonstrated that miR-143-5p was depleted in NPC cells and tissues, and the biological function of NPC cells was impaired after the amplification of miR-143-5p. As similarly reported by Liu et al., miR-143-5p also displays a low level in pancreatic cancer cells, miR-143-5p elevation facilitates cell apoptosis and autophagy, thus restraining pancreatic cancer cell growth [Citation28]. In addition, miR-143-5p is also decreased in gastric cancer cells, the induction of miR-143-5p exerts a tumor-suppressive effect via blocking cell proliferation and accelerating apoptosis of gastric cancer cells [Citation29]. Such finding has also evidenced in lung adenocarcinoma, Wada et al. have unraveled that the robustly expressed miR-143-5p relieves the malignant phenotype of lung adenocarcinoma cells, thus exerting tumor-suppressive effects in lung adenocarcinoma [Citation15]. Furthermore, down-regulated miR-143-5p is also identified in non-small cell lung cancer (NSCLC) cells, and the ablated miR-143-5p expression is associated with carcinogenesis of NSCLC [Citation30]
Afterward, it was also predicted that miR-143-5p had a targeting relation with HOXA6. Multiple homeobox members have been identified as pivotal modulators in cancer progression, such as HOXB13, which was validated as novel marker for laryngeal squamous cell carcinoma clinical screening and treatment [Citation31]. Moreover, cumulative reports have confirmed the regulatory role of HOXA6 in cancers. For instance, Wu et al. have validated that HOXA6 is enriched in colorectal cancer tissues, and depleted HOXA6 hinders the development of colorectal cancer cells via modulating epithelial-mesenchymal transition process [Citation18]. Moreover, it has been reported HOXA6 expression is significantly accounted for inducing unfavorable OS rates and increasing mortality in cervical cancer patients [Citation32]. Additionally, in colorectal cancer, HOXA6 overexpression has been confirmed by Lin et al., who have clarified that HOXA6 ablation abrogates the migration and invasion capacity of colorectal cancer cells [Citation18]. Furthermore, Shao et al have elucidated that HOXA6 can regulate radiation resistance in A549 cells in lung cancer [Citation22]. In light of previous findings, the research further illustrated that HOXA6 was high-expressed in NPC cells, and the deletion of HOXA6 could inhibit the growth of radiation-resistant NPC cells.
Conclusion
In summary, this study manifests that LHFPL3-AS1 and HOXA6 expression is enriched while the miR-143-5p level is decreased in NPC tissues and cells. Silenced LHFPL3-AS1 hinders NPC cell growth in vitro and tumor growth in vivo via sponging miR-143-5p to target HOXA6. The current study defines a mechanism for the silenced LHFPL3-AS1 in NPC progression via regulating miR-143-5p/HOXA6 axis, reflecting LHFPL3-AS1/miR-143-5p/HOXA6 may function as novel therapeutic targets that applied to clinical treatment of NPC. However, the current study has only validated the regulatory mechanism of the LHFPL3-AS1/miR-143-5p/HOXA6 axis in NPC, while the specificity of LHFPL3-AS1/miR-143-5p/HOXA6 signaling axis in different diseases remains further exploration, which may be further investigated in our future works.
Highlights
LHFPL3-AS1 and HOXA6 are enriched while miR-143-5p is depleted in NPC cells and tissues.
Silenced LHFPL3-AS1 inhibits radiotherapy resistance of NPC cells.
High-expressed miR-143-5p suppresses the biological functions of NPC cells.
LHFPL3-AS1 binds to miR-143-5p that targets HOXA6.
The study provides a theoretical basis for LHFPL3-AS1/miR-143-5p/HOXA6 axis on NPC.
Supplemental Material
Download Zip (10.6 MB)Supplemental Material
Download Zip (11.6 MB)Supplemental Material
Download Zip (10.4 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Chen Y-P, Chan ATC, Le Q-T, et al. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80.
- Dai W, Zheng H, Cheung AKL, et al. Genetic and epigenetic landscape of nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):16.
- Lam WKJ, Chan JYK. Recent advances in the management of nasopharyngeal carcinoma. F1000Res. 2018;7:1829.
- Wang KH, Austin, SA, Chen, SH, et al. Nasopharyngeal carcinoma diagnostic challenge in a nonendemic setting: our experience with 101 patients. Perm J. 2017;21:16–180.
- Dong Q, Zhou L, Liu F, et al. Long non-coding RNAs in the development, diagnosis and prognosis of nasopharyngeal carcinoma. Int J Clin Exp Pathol. 2017;10(8):8098–8105.
- Wang Y, Chen W, Lian J, et al. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2020;27(2):695–710.
- van Ahlen H, Porst H, Löcherbach-Zawadzky A, et al. Disorders of the hypothalamo-hypophyseal-gonadal axis and erectile impotence. Dtsch Med Wochenschr. 1988;113(26):1047–1052.
- Dai J, Mu J-W, Mu H, et al. Long non-coding RNA CRNDE regulates cell proliferation, migration, invasion, epithelial-mesenchymal transition and apoptosis in oral squamous cell carcinoma. Oncol Lett. 2019 Mar;17(3):3330–3340.
- Du Y, Yang H, Li Y, et al. Long non-coding RNA LINC01137 contributes to oral squamous cell carcinoma development and is negatively regulated by miR-22-3p. Cell Oncol (Dordr). 2021 Jun;44(3):595–609.
- Li J, Xu, X, Zhang, D, et al. LncRNA LHFPL3-AS1 promotes oral squamous cell carcinoma growth and cisplatin resistance through targeting miR-362-5p/CHSY1 pathway. Onco Targets Ther. 2021 Mar 31;14:2293–2300.
- Zhang S, Wan H, Zhang X, et al. LncRNA LHFPL3-AS1 contributes to tumorigenesis of melanoma stem cells via the miR-181a-5p/BCL2 pathway. Cell Death Dis. 2020 Nov 4;11(11):950.
- Spence T, et al. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):17.
- Cui F, Ji Y, Wang M, et al. miR-143 inhibits proliferation and metastasis of nasopharyngeal carcinoma cells via targeting FMNL1 based on clinical and radiologic findings. J Cell Biochem. 2019;120(10):16427–16434.
- Xu YF, Li YQ, Guo R, et al. Identification of miR-143 as a tumour suppressor in nasopharyngeal carcinoma based on microRNA expression profiling. Int J Biochem Cell Biol. 2015;61:120–128.
- Sanada H, Seki N, Mizuno K, et al. Involvement of dual strands of miR-143 (miR-143-5p and miR-143-3p) and their target oncogenes in the molecular pathogenesis of lung adenocarcinoma. Int J Mol Sci. 2019;20(18):4482.
- Chen JH, Yang R, Zhang W, et al. Functions of microRNA-143 in the apoptosis, invasion and migration of nasopharyngeal carcinoma. Exp Ther Med. 2016;12(6):3749–3755.
- Chang SL, Chan TC, Chen TJ, et al. HOXC6 overexpression is associated with Ki-67 expression and poor survival in NPC patients. J Cancer. 2017;8(9):1647–1654.
- Wu S, Wu F, Jiang Z. Effect of HOXA6 on the proliferation, apoptosis, migration and invasion of colorectal cancer cells. Int J Oncol. 2018;52(6):2093–2100.
- Zhou D, Ye C, Pan Z, et al. SATB1 knockdown inhibits proliferation and invasion and decreases chemoradiation resistance in nasopharyngeal carcinoma cells by reversing EMT and suppressing MMP-9. Int J Med Sci. 2021;18(1):42–52.
- Cheng X, Li F, Tao Z. Tenascin-C promotes epithelial-to-mesenchymal transition and the mTOR signaling pathway in nasopharyngeal carcinoma. Oncol Lett. 2021;22(1):570.
- Wu F, Wu S, Tong H, et al. HOXA6 inhibits cell proliferation and induces apoptosis by suppressing the PI3K/Akt signaling pathway in clear cell renal cell carcinoma. Int J Oncol. 2019;54(6):2095–2105.
- Shao L, Zhang Y, Gong X, et al. Effects of MLL5 and HOXA regulated by NRP1 on radioresistance in A549. Oncol Lett. 2021;21(5):403.
- Xiao Z, Chen Z. Deciphering nasopharyngeal carcinoma pathogenesis via proteomics. Expert Rev Proteomics. 2019;16(6):475–485.
- Peng Q, Liu L, Pei H, et al. A LHFPL3-AS1/miR-580-3p/STAT3 feedback loop promotes the malignancy in melanoma via activation of JAK2/STAT3 signaling. Mol Cancer Res. 2020;18(11):1724–1734.
- Jin X, Chen X, Hu Y, et al. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med. 2017;6(6):1409–1423.
- Wada M, Goto Y, Tanaka T, et al. RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J Hum Genet. 2020;65(11):1019–1034.
- He M, Zhan M, Chen W, et al. MiR-143-5p deficiency triggers EMT and metastasis by targeting HIF-1alpha in gallbladder cancer. Cell Physiol Biochem. 2017;42(5):2078–2092.
- Liu C, Cheng B, Li PY, et al. Long non-coding RNA LINC01207 silencing suppresses AGR2 expression to facilitate autophagy and apoptosis of pancreatic cancer cells by sponging miR-143-5p. Mol Cell Endocrinol. 2019;493:110424.
- Wu XL, Cheng, B, Li, PY, et al. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol. 2013;19(43):7758–7765.
- Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol. 2017;15(1):175.
- Mo B-Y, Li G-S, Huang S-N, et al. The underlying molecular mechanism and identification of transcription factor markers for laryngeal squamous cell carcinoma. Bioengineered. 2021;12(1):208–224.
- Eoh KJ, Kim HJ, Lee J-Y, et al. Upregulation of homeobox gene is correlated with poor survival outcomes in cervical cancer. Oncotarget. 2017;8(48):84396–84402.
