ABSTRACT
Acute myocardial infarction (AMI) is characterized by high morbidity and mortality rates. Circular RNAs collectively participate in the initiation and development of AMI. The purpose of this study was to investigate the role of circRbms1 in AMI. Ischemia-reperfusion (I/R) was performed to establish an AMI model. RT-qPCR and Western blotting were performed to detect mRNA and analyze protein expression, respectively. The interaction between miR-92a and circRbms1/BCL2L11 was confirmed by luciferase and RNA pull-down assays. circRbms1 is overexpressed in AMI. However, circRbms1 knockdown alleviated H9c2 cell apoptosis and reduced the release of reactive oxygen species. circRbms1 targeted miR-92a, the downregulation of which alleviated the effects of circRbms1 knockdown and increased oxidative stress and H9c2 cell apoptosis. Moreover, circRbms1 sponged miR-92a to upregulate BCL2L11, which modulated the expression of apoptosis-related genes. circRbms1 participated in myocardial I/R injury by regulating the miR-92a/BCL2L11 signaling pathway, which may provide a new strategy for the treatment of AMI.
Introduction
Acute myocardial infarction (AMI) is a prevalent heart disease characterized by myocardial necrosis caused by persistent and severe myocardial ischemia. Myocardial ischemia disrupts cellular function, morphology, and metabolism. Patients with AMI are conducive to disability worldwide [Citation1]. Restoring tissue blood perfusion is an effective strategy for AMI treatment [Citation2]. However, emerging myocardial injury, myocardial energy metabolism disorders, and dysfunction-inducing ischemia-reperfusion (I/R) injury offset the clinical results [Citation3,Citation4]. Excessive production of oxygen free radicals (such as nitric oxide), lipid peroxidation, and reactive oxygen species (ROS), is one of the leading causes of I/R injury. Apoptosis plays a vital role in the structural and functional damage of myocardial cells [Citation5]. Unveiling the mechanism of cardiomyocyte death is urgently required.
Circular RNAs (circRNAs) are endogenous RNAs that are reverse-spliced by precursor RNA [Citation6]. Recently, circRNAs have attracted considerable attention due to their crucial roles in cellular functions, such as apoptosis, inflammatory response, and oxidative stress [Citation7]. CircRNAs are also involved in cardiovascular diseases [Citation8]. For instance, circRNA-Las1L inhibits the proliferation and migration of cardiac fibroblasts and promotes cell apoptosis [Citation9]. circRNA-0010729 suppresses injuries to human cardiomyocytes [Citation10]. circRNA ACR has a pro-autophagic role to attenuate myocardial I/R injury in mice by Pink1/FAM65B pathway [Citation11]. However, the role of circRbms1 in AMI remains unclear.
CircRNA contains microRNAs (miRNA) response element, which can bind miRNA as competitive endogenous RNA (ceRNA), inhibit the binding of miRNA to mRNA in cytoplasm and regulate gene expression. This function is called ‘miRNA sponge’. Many reports demonstrated circRNAs modulated the AS progression through functioning as ceRNA. MicroRNAs are small RNAs and many microRNAs were reported to participate in the occurrence and progression of myocardial I/R injury [Citation12]. For instance, miR-206 promotes myocardial hypertrophy and cardiomyocyte survival by mediating YAP signaling [Citation13]. Overexpression of miR-21 in the early AMI stage reduces cell apoptosis and infarct size in the infarct margin area [Citation14]. Additionally, overexpression of miRNA-22 inhibits cardiomyocyte apoptosis, reduces myocardial remodeling, and promotes cardiac function recovery by suppressing microcystin-3 [Citation15]. However, the mechanism of miR-92a in AMI remains unclear.
In this study, we investigated the role of circRbms1 in AMI and its underlying mechanisms. We hypothesized that circRbms1 knockdown alleviated I/R injury via the miR-92a/BCL2L11 axis. These findings provide important insights into the pathology of ischemic heart injury.
Material and Methods
Animals
AMI mice model was established according to previous study [Citation16]. Eighteen 8-week-old male C57BL/6 mice were provided by Nanjing Medical University (Nanjing, China). The mice were housed in an SPF environment (room temperature 23°C; humidity 65%; 12 h light/dark cycle), with ad libitum water and food. The hearts of the mice in the model group were exposed by a left thoracotomy. Ligation of the left anterior descending coronary artery (LAD) was then performed. The I/R procedure included ischemia for 1 h and reperfusion for 1, 2, 4, and 8 h. The mice in the sham group underwent a procedure without LAD. Subsequently, the mice were sacrificed and the hearts were collected for the TTC, HE, IHC and TUNEL staining.
TTC staining
According to a previous study [Citation17]. 2% Evans blue was injected into the femoral vein after LAD ligation. Following injection, the heart tissue was collected immediately and rinsed with frozen normal saline. The heart was frozen at 20°C for 30 min and cut into thin slices with 2 mm thi ck. The sections were incubated with trinitrotoluene at 37°C for 15 min. After being fixed with formaldehyde for 24 h. The sections were imaged. White area indicates the infarcted area, and the red zone indicats the dangerous area (AAR). The infarct size and AAR was calculated by ImageJ (version 1.42) software. Firstly, the infarct size and AAR of each slice was calculated, and then the sum of the infarct size and AAR of each slice multiplied by the thickness (2 mm) was the total size and AAR.
HE staining
Morphological observation of mice myocardial tissue was conducted HE staining according to a previous study [Citation18]. The left ventricle of the mice was collected and put in 10% formaldehyde. Next, the samples were dehydrated, embedded and cut down into 4 μm slices. After deparaffinage, the sections were stained with hematoxylin and eosin and observed under a light microscope.
Immunohistochemical (IHC) staining
According to a previous study [Citation19]. The heart slices obtained in HE staining were treated with 3% hydrogen peroxide for 20 min. After that, the slices were incubated with 1% bovine serum albumin and anti-BCL2L11 for 2 h. Then the slices were incubated with horseradish peroxidase (HRP)-conjugated anti-goat IgG for 60 min. Diaminobenzidine (DAB) was selected as chromogen.
TUNEL assay
TUNEL Apoptosis Assay Kit (Beyotimr, Jiangsu, China) was purchased to measure the apoptosis according to a previous study [Citation20]. The heart tissue was washed and fixed with 4% paraformaldehyde (Beyotime). Then, the heart tissues were washed with phosphate buffer solution (PBS) for twice. Next, PBS containing 0.3% Triton X-100 was added to the heart tissues. Finally, 50 µl TUNEL solution were used for staining, and nuclear cells that labeled positively were considered as apoptotic cells. The positive cells in randomly five views were observed by a fluorescence microscope. The apoptosis rates were presented as positive cells/total cells.
Cell culture
H9c2 cells were provided by the American Type Culture Collection (VA, USA) and incubated with DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
The H9c2 cells were exposed to H2O2 to establish an oxidative stress injury model (200 μM) [Citation21].
Construction and transfection of plasmid
The oligonucleotide sequence of the expression vector was designed and synthesized by Hanbio (Shanghai, China) according to a previous study [Citation22]. The sequence was as follows: circRbms1 short interference (si) RNA, 5’-TCCAGGCTCAGATGGCAAA-3’; negative control (NC) siRNA, 5’-AGATGAAATTGTGGCTAAA-3.’ The sequence of miR-92a was as follows: 5’-ACAGGCCGGGACAAGTGCAATA-3’. For circRbms1 knockout structures, siRNA oligonucleotides were inserted into the pLVTHM carrier (System Biosciences, CA, USA) located between the MluI and ClaI limiting sites. The BCL2L11 overexpression vector and NC were provided by Shanghai GenePharma (Shanghai, China). The H9c2 cells were seeded in 12-well plates for 24 h. Transfection was performed using the Lipofectin2000 regent (QIAGEN, Hilden, Germany). Chemically modified oligonucleotides (antagomiR) were used to knock down the miRNAs. The antagomir-92a was purchased from RiboBio (Guangzhou, China). We injected antagomiR-92a into the tail vein of rats three consecutive times to silence miR-92a expression in vivo.
Determination of ROS
Intracellular ROS were detected by DCFH-DA staining (Sigma-Aldrich, MO, USA), as previously described [Citation23].
Flow cytometry
According to a previous study [Citation24], cells were plated in 24-well plates. The H9c2 cells were collected, digested, and centrifuged, and then double-stained with Annexin V-FITC/propidium iodide. Apoptosis was detected by flow cytometry using a BD FACSCalibur (Becton Dickinson, NJ, USA). The gating strategy in flow cytometry was as follows. Firstly, we selected our target group of cells according to FSC and SSC parameters to exclude cell fragments and dead cells. After adjusting fluorescence compensation, we set the gate according to the signal of unstained control cells and single positive stained cells.
RT-qPCR
RNA was isolated from the cells. Reverse transcription was then performed. RT-qPCR was conducted using SYBR Green mixture (Yisheng, Shanghai). The miRNAs and mRNA were normalized to U6 and GAPDH, respectively. The results were measured using the 2−ΔΔCt method [Citation25].
RNA pull-down
Biotin-labeled miR-92a probe and NC were provided by Shanghai Sanggong Biotechnology Co., Ltd (China) according to a previous study [Citation26]. The cells were harvested and dissolved in a lysis buffer. The binding reaction was performed using a microliter of magnetic beads. After washing with lysis buffer, the results were analyzed using RT-qPCR.
Luciferase activity
According to a previous study [Citation27], the binding sites between miR-92a and circRbms1/BCL2L11 were predicted using StarBase 3.0, miRDB, and TargetScan7.2. The wild-type (WT) and mutant type (MUT) of circRbms1 and BCL2L11 were inserted into the psiCHECK vector (Promega, WI, USA). Cells were co-transfected with miR-92a and the 3ʹUTR of WT or MUT of circRbms1/BCL2L11 for 48 h. Luciferase activity was detected by a double luciferase assay (Promega).
Western blot analysis
Total protein was collected from the H9c2 cells. The protein concentration was calculated using a BCA Kit. Electrophoresis was performed to isolate the proteins, which were subsequently transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% skimmed milk and incubated with the primary antibodies (anti-BCL2L11, anti-bid, anti-cyto-c, anti-bcl-2, anti-caspase-3, and anti-β-actin; Boster Biological Technology, Wuhan, China) overnight at 4°C, and then with secondary antibodies. The bands were visualized using a microscope (Bio-Rad, CA, USA).
Statistical analysis
Data were analyzed using SPSS17.0 (SPSS, IL, USA) and represented as the mean ± SD. One-way ANOVA was applied for analyzing the difference among multiple groups, and Student’s t-test for the difference between two groups. Statistical significance was set at P < 0.05.
Result
This research demonstrated that circRbms1 and BCL2L11 was over expressed, and miR-92a was decreased in AMI. CircRbms1 knockdown inhibited cardiomyocyte apoptosis and ROS release via regulation of miR-92a/BCL2L11. The circRbms1/miR-92a/BCL2L11 axis may be a potential target for the treatment of I/R injury.
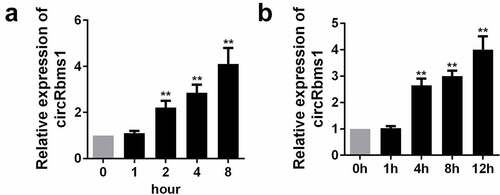
circRbms1 is overexpressed in myocardial I/R
To investigate the role of circRbms1 in AMI in vivo and in vitro model, we determined its expression levels I/R mice and H2O2 treated H9c2 cells. The expression of circRbms1 in the I/R mice was increased (). This was consistent with the results of the in vitro assays. circRbms1 expression was also upregulated in the H9c2 cells after exposure to H2O2 ().
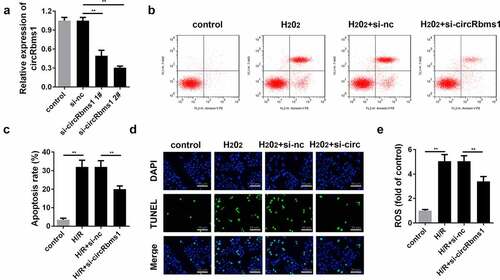
Silencing circRbms1 inhibits H2O2-induced cardiomyocyte apoptosis and the release of ROS
As shown in , the expression of circRbms1 is significantly decreased in cells transfected with circRbms1 knockdown plasmids, which is more potent in the si-circRbms1 2# group. Therefore, si-circRbms1 2# was used in the subsequent experiments. si-circRbms1 knockdown significantly alleviated the increase in apoptosis rates (). Moreover, the increase in ROS induced by H2O2 treatment was significantly attenuated by circRbms1 knockdown ().
Figure 2. Silencing circRbms1 can attenuate apoptosis and oxidative stress injury of H9c2 cells induced by H2O2. (a) circRbms1 expression was determined by RT-PCR. (b, c) Apoptosis of H9c2 cells was detected using flow cytometry. (d) The apoptosis was detected by TUNEL assays. (e) Reactive oxygen species was assessed. Each experiment was repeated three times. **P < 0.01.
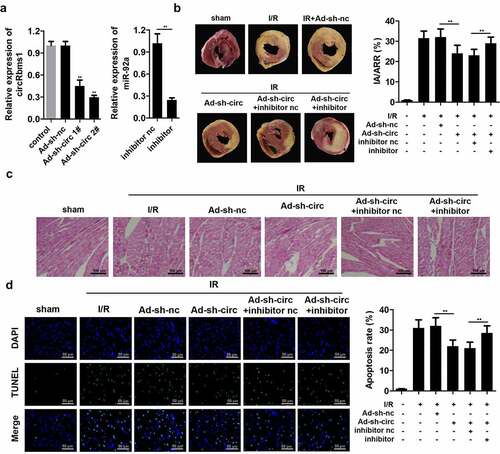
circRbms1 knockdown reduces myocardial I/R injury in vivo via targrting miR-92a
We confirmed the role of circRbms1 in I/R injury using an animal model. circRbms1 expression was remarkably downregulated in mice injected with ad-sh-circRbms1, which was more pronounced in the ad-sh-circRbms1 2# group. Hence, ad-sh-circRbms1 2# was used in the subsequent experiments. Besides, miR-92a inhibitor significantly decreased the miR-92a expression (). Additionally, there was no infarct area in sham group, while the infarct size in the I/R group were significantly increased. CircRbms1 knockdown significantly reduced the infarct size (). Furthermore, HE staining showed that compared to the sham group, clear damage to myocardium could be observed in I/R group. Knockdown of circRbms1 relieved the effects of I/R on myocardial tissue (). Additionally, circRbms1 knockdown also markedly suppressed cardiomyocyte apoptosis (). However, miR-92a knockdown neutralized the sh-circRbms1 effects in the myocardial I/R injury of the mice.
Figure 3. Silencing circRbms1 inhibits I/R injury in mice. (a) circRbms1 and miR-92 expression was determined by RT-PCR. (b) The AMI was analyzed using TTC staining. (c) The histopathology examination of myocardial tissues was performed with HE staining. (d) Primary cardiomyocyte apoptosis was evaluated by TUNEL assay. Each experiment was repeated three times. **P < 0.01.
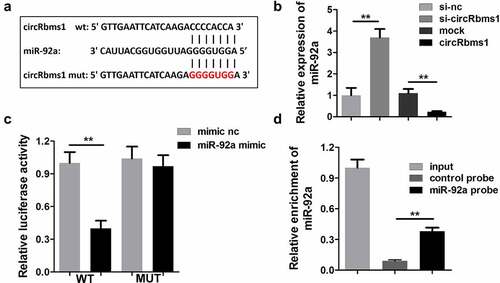
circRbms1 targets miR-92a
The possible target miRNAs of circRbms1 were predicted using miRDB and StarBase (). Moreover, miR-92a expression was remarkably downregulated by circRbms1, while, it was upregulated by circRbms1 knockdown (). The binding sites were further confirmed using luciferase activity (), and RNA pull-down assays ().
miR-92a reverses circRbms1-induced cardiomyocyte apoptosis
circRbms1 acts as a sponge for miR-92a in cardiomyocytes. We performed rescue experiments to study the role of miR-92a in I/R injury. Cardiomyocytes were transfected with si-circRbms1 and an miR-92a-inhibitor. circRbms1 knockdown promoted miR-92a expression and the miR-92a inhibitor notably inhibited this effect (). Rescue experiments indicated that miR-92a downregulation abrogated the effects of circRbms1 knockdown on cardiomyocyte apoptosis (). Meanwhile, the miR-92a inhibitor reversed the decrease in ROS that was induced by circRbms1 silencing ().
Figure 5. MiR-92a inhibition reverses the effect of circRbms1 knockdown. (a) miR92a expression was determined by RT-PCR. (b-d) Apoptosis rates of H9c2 cells were evaluated by flow cytometry and TUNEL staining. (e) The reactive oxygen species in H9c2 cells was detected. Each experiment was repeated three times. **P < 0.01.
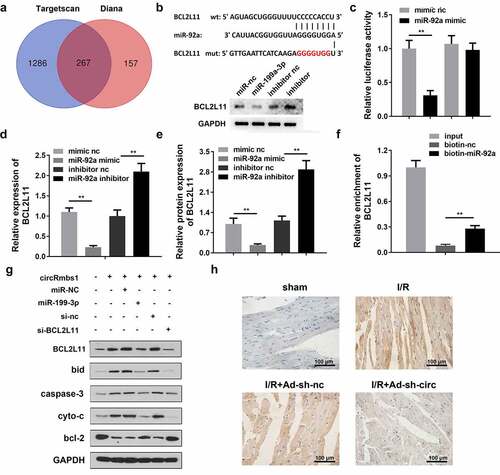
miR-92a directly targets BCL2L11 in cardiomyocytes
TargetScan and Diana databases predicted BCL2L11 as a target of miR-92a (). The target binding regions between miR-92a and BCL2L11 are shown in . miR-92a knockdown improved the protein level of BCL2L11. Moreover, luciferase activity was significantly decreased by co-transfection with the miR-92a mimic and pGL3-3ʹUTR BCL2L11 WT (). RT-qPCR analysis showed that miR-92a markedly negatively regulated BCL2L11 expression (). Additionally, miR-92a overexpression decreased BCL2L11 protein levels, whereas it was promoted by miR-92a knockdown (). An RNA pull-down assay confirmed the interaction between miR-92a and BCL2L11 (). As shown in , BCL2L11 knockdown alleviates the effects of miR-92a on the expressions of BCL2L11, caspase-3, bcl-2, bid, and cyto-c. Additionally, the results of IHC staining showed that the BCL2L11 expressions were significantly decreased in heart tissues of the I/R mice after circRbms1 knockdown ()
Figure 6. MiR-92a inhibits cardiomyocyte apoptosis by regulating BCL2L11. (a) The TargetScan database and Diana database predicted target genes of miR-92a. (b) The binding sites between miR-92a and BCL2L11. (c) The binding sites were confirmed by luciferase report analysis. (d) BCL2L11 expression was determined by RT-PCR. (e) BCL2L11 protein expression was evaluated by Western blot. (f) The binding sites were confirmed by RNA pull-down assay. (g) Protein expression was detected using Western blot. (h) BCL2L11 protein expression was evaluated by IHC staining. Each experiment was repeated three times. **p < 0.01.
Discussion
Myocardial ischemia increases the risk of coronary heart disease and myocardial infarction [Citation28]. Modulation of oxygen consumption, angiogenesis, or cardiomyocyte apoptosis are effective means of treating myocardial ischemia [Citation29]. Therefore, elucidating the underlying molecular mechanisms may provide new strategies for the treatment of AMI. In this study, circRbms1 expression was upregulated in AMI in vivo and in vitro. Silencing circRbms1 displayed a cardioprotective effect by inhibiting cardiomyocyte apoptosis and the release of ROS. Interestingly, circRbms1 modulated the progression of AMI by regulating the miR-92a/BCL2L11 axis. Thus, circRbms1 may be a potential biomarker for AMI.
circRNAs are key regulators in the development of I/R. Yang et al. revealed that circ-008018 protects against I/R injury by targeting miR-99a [Citation30]. circ-100,338 acts as the cavernous body of miR-200a-3p to promote blood tube formation following myocardial I/R injury [Citation31]. circ-0068566 inhibits the occurrence of myocardial I/R injury by regulating the miR-6322/PARP2 signaling pathway [Citation32]. circRbms1 expression is upregulated during heart failure and is associated with cardiomyocyte apoptosis [Citation33]. In this study, circRbms1 expression was increased in AMI models both in vivo and in vitro. However, circRbms1 knockdown inhibited cardiomyocyte apoptosis and the release of ROS, which are the key players in the initiation and progression of AMI. Therefore, circRbms1 knockdown may provide protection against AMI by inhibiting cardiomyocyte apoptosis and ROS accumulation. However, the underlying molecular mechanisms remain unclear.
Numerous reports have confirmed that circRNAs may act as molecular sponges to regulate the expression and biological functions of miRNA [Citation34–36]. circRNA-100269 inhibits the development of gastric cancer by targeting miR-630 [Citation37]. Dysregulated circRNA-100290 promotes the aggressiveness of oral cancer through the sponge miR-29 family [Citation38]. In this study, circRbms1 negatively regulated miR-92a expression, which alleviated ischemic injury. Moreover, miR-92a is a critical protective miRNA during cardiac ischemic injury that inhibits inflammatory response and apoptosis [Citation39]. These results suggested that miR-92a may function as a cardioprotective agent, which is consistent with the findings of Song et al. [Citation40]. However, miR-92a downregulation reversed the effects of circRbms1 knockdown and promoted cardiomyocyte apoptosis and ROS release, suggesting that miR-92a may play a beneficial role in AMI and that circRbms1 may participate in the progression of AMI by suppressing miR-92a expression.
Accumulating evidence has revealed that miRNAs interact with their targets to participate in the development of cardiovascular diseases, including AMI [Citation41–43]. miR-130 can target PPAR-γ to aggravate AMI-induced myocardial injury [Citation44]. In this study, BCL2L11 was proved to be a target of miR-92a. BCL2L11 is a key regulator of protein synthesis and cell growth. Activated BCL2L11 promotes myocardial ischemic injury, and its phosphorylation is a crucial step in the protection of cardiomyocytes [Citation45–47]. In this study, BCL2L11 knockdown downregulated the expression of the apoptosis-related genes, caspase-3, bids, and cyto-c, and upregulated the expression of bcl-2. Therefore, circRbms1 modulated BCL2L11 expression to promote the progression of AMI by sponging miR-92a. The circRbms1/miR-92a/BCL2L11 axis may be a potential biomarker for AMI treatment.
Conclusion
In the present study, circRbms1 was overexpressed in AMI. circRbms1 knockdown inhibited cardiomyocyte apoptosis and ROS release via regulation of miR-92a/BCL2L11. The circRbms1/miR-92a/BCL2L11 axis may be a potential target for the treatment of I/R injury.
Author contributions
Ling Jin grafted the work; worked with Yuan Zhang to collecte and analyze the data. Yun Jiang interpreted the data. Mingjuan Tan and Caidong Liu provided the concept and idea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Li Y, Ren S, Xia J, et al. EIF4A3-induced circ-BNIP3 aggravated hypoxia-induced injury of H9c2 cells by targeting miR-27a-3p/BNIP3. Mol Ther Nucleic Acids. 2020;19:533–545.
- Binder A, Ali A, Chawla R, et al. Myocardial protection from ischemia-reperfusion injury post coronary revascularization. Expert Rev Cardiovasc Ther. 2015;13(9):1045–1057.
- Chen L, Zhang D, Yu L, et al. Targeting MIAT reduces apoptosis of cardiomyocytes after ischemia/reperfusion injury. Bioengineered. 2019;10(1):121–132.
- Shen Y, Liu X, Shi J, et al. Involvement of Nrf2 in myocardial ischemia and reperfusion injury. Int J Biol Macromol. 2019;125:496–502.
- Cadenas S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic Biol Med. 2018;117:76–89.
- Fan X, Weng X, Zhao Y, et al. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int. 2017;2017:5135781.
- Jiao J, Zhang T, Jiao X, et al. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J Cell Physiol. 2020;235(2):1287–1295.
- Jiang J, Yang Y, Jiang R. Regulating mechanisms of circRNA and their relationship with cardiovascular diseases]. Zhonghua Xin Xue Guan Bing Za Zhi. 2016;44(4):364–366.
- Sun LY, Zhao JC, Ge XM, et al. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem Funct. 2020;38(4):443–450.
- Jin Q, Chen Y. Silencing circular RNA circ_0010729 protects human cardiomyocytes from oxygen-glucose deprivation-induced injury by up-regulating microRNA-145-5p. Mol Cell Biochem. 2019;462(1–2):185–194.
- Zhou LY, Zhai M, Huang Y, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26(7):1299–1315.
- Kukreja RC, Yin C, Salloum FN. MicroRNAs: new players in cardiac injury and protection. Mol Pharmacol. 2011;80(4):558–564.
- Yang Y, Del Re DP, Nakano N, et al. miR-206 mediates YAP-induced cardiac hypertrophy and survival. Circ Res. 2015;117(10):891–904.
- Dong S, Cheng Y, Yang J, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284(43):29514–29525.
- Chen Z, Qi Y, Gao C. Cardiac myocyte-protective effect of microRNA-22 during ischemia and reperfusion through disrupting the caveolin-3/eNOS signaling. Int J Clin Exp Pathol. 2015;8(5):1299–1315.
- Yang J, Huang X, Hu F, et al. LncRNA ANRIL knockdown relieves myocardial cell apoptosis in acute myocardial infarction by regulating IL-33/ST2. Cell Cycle. 2019;18(23):3393–3403.
- Tanaka J, Kiyoshi K, Kadokura T, et al. Elucidation of the enzyme involved in 2,3,5-triphenyl tetrazolium chloride (TTC) staining activity and the relationship between TTC staining activity and fermentation profiles in Saccharomyces cerevisiae. J Biosci Bioeng. 2021;131(4):396–404.
- Feldman AT, Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol. 2014;1180:31–43.
- Akturk G, Sweeney R, Remark R, et al. Multiplexed Immunohistochemical Consecutive Staining on Single Slide (MICSSS): multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol Biol. 2020;2055:497–519.
- Kyrylkova K, Kyryachenko S, Leid M, et al. Detection of apoptosis by TUNEL assay. Methods Mol Biol. 2012;887:41–47.
- Cheng Y, Liu X, Zhang S, et al. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47(1):5–14.
- Yu T, Zhao C, Hou S, et al. Exosomes secreted from miRNA-29b-modified mesenchymal stem cells repaired spinal cord injury in rats. Braz J Med Biol Res. 2019;52(12):e8735.
- Weintraub WS, Hartigan PM, Mancini GBJ, et al. Effect of coronary anatomy and myocardial ischemia on long-term survival in patients with stable ischemic heart disease. Circ Cardiovasc Qual Outcomes. 2019;12(2):e005079.
- Spurgeon BEJ, Naseem KM. Platelet flow cytometry: instrument setup, controls, and panel performance. Cytometry B Clin Cytom. 2020;98(1):19–27.
- Nagura-Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58(9):e01438–20.
- Bierhoff H. Analysis of lncRNA-Protein Interactions by RNA-protein pull-down assays and RNA Immunoprecipitation (RIP). Methods Mol Biol. 2018;1686:241–250.
- Xu YZ, Kanagaratham C, Jancik S, et al. Promoter deletion analysis using a dual-luciferase reporter system. Methods Mol Biol. 2013;977:79–93.
- Shao Y, Zhong P, Sheng L, et al. Circular RNA circDENND2A protects H9c2 cells from oxygen glucose deprivation-induced apoptosis through sponging microRNA-34a. Cell Cycle. 2020;19(2):246–255.
- Yuan Y, Zheng Z. Geniposide protects PC-12 cells against oxygen and glucose deprivation-induced injury by up-regulation of long-noncoding RNA H19. Life Sci. 2019;216:176–182.
- Yang X, Ji H, Yao Y, et al. Downregulation of circ_008018 protects against cerebral ischemia-reperfusion injury by targeting miR-99a. Biochem Biophys Res Commun. 2018;499(4):758–764.
- Chang H, Li ZB, Wu JY, et al. Circ-100338 induces angiogenesis after myocardial ischemia-reperfusion injury by sponging miR-200a-3p. Eur Rev Med Pharmacol Sci. 2020;24(11):6323–6332.
- Zhou HF, Xu LL, Xie B, et al. Hsa-circ-0068566 inhibited the development of myocardial ischemia reperfusion injury by regulating hsa-miR-6322/PARP2 signal pathway. Eur Rev Med Pharmacol Sci. 2020;24(12):6980–6993.
- Wu HJ, Zhang CY, Zhang S, et al. Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell Physiol Biochem. 2016;39(1):205–216.
- Zhao Y, Alexandrov PN, Jaber V, et al. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s Disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel). 2016;7(12):116.
- Dori M, Bicciato S. Integration of bioinformatic predictions and experimental data to identify circRNA-miRNA associations. Genes (Basel). 2019;10(9):642.
- Liu K, Guo Y, Zheng K, et al. Identification of the circRNA-miRNA-mRNA regulatory network of Hsp90 inhibitor-induced cell death in colorectal cancer by integrated analysis. Gene. 2020;727:144232.
- Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9(6):1585–1594.
- Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561.
- Yin F, Zhou H, Fang Y, et al. Astragaloside IV alleviates ischemia reperfusion-induced apoptosis by inhibiting the activation of key factors in death receptor pathway and mitochondrial pathway. J Ethnopharmacol. 2020;248:112319.
- Song YS, Joo HW, Park IH, et al. Bone marrow mesenchymal stem cell-derived vascular endothelial growth factor attenuates cardiac apoptosis via regulation of cardiac miRNA-23a and miRNA-92a in a rat model of myocardial infarction. PLoS One. 2017;12(6):e0179972.
- Yin Q, Wang P, Wu X. MicroRNA −148 alleviates cardiac dysfunction, immune disorders and myocardial apoptosis in myocardial ischemia-reperfusion (MI/R) injury by targeting pyruvate dehydrogenase kinase (PDK4). Bioengineered. 2021;12(1):5552–5565.
- Qi H, Zhang J, Shang Y, et al. Argon inhibits reactive oxygen species oxidative stress via the miR-21-mediated PDCD4/PTEN pathway to prevent myocardial ischemia/reperfusion injury. Bioengineered. 2021;12(1):5529–5539.
- Zheng HF, Sun J, Zou ZY, et al. MiRNA-488-3p suppresses acute myocardial infarction-induced cardiomyocyte apoptosis via targeting ZNF791. Eur Rev Med Pharmacol Sci. 2019;23(11):4932–4939.
- Chu X, Wang Y, Pang L, et al. miR-130 aggravates acute myocardial infarction-induced myocardial injury by targeting PPAR-gamma. J Cell Biochem. 2018;119(9):7235–7244.
- Huang J, Huang Y, Feng Z, et al. MiR-1247-3p protects rat cardiomyocytes against hypoxia/reoxygenation-induced injury via targeting BCL2L11 and caspase-2. J Recept Signal Transduct Res. 2020;1–9. DOI:10.1080/10799893.2020.1859533
- Zhai C, Hu H, Tang G, et al. MicroRNA-101a protects against the H2O2-induced injury on cardiomyocytes via targeting BCL2L11. Am J Transl Res. 2020;12(6):2760–2768.
- Yang W, Han Y, Yang C, et al. MicroRNA-19b-1 reverses ischaemia-induced heart failure by inhibiting cardiomyocyte apoptosis and targeti-ng Bcl2 l11/BIM. Heart Vessels. 2019;34(7):1221–1229.