ABSTRACT
Circular RNA (circRNA) hsa_circ_0077837 inhibits colorectal cancer. Our research studied the participation of hsa_circ_0077837 in non-small cell lung cancer (NSCLC). Hsa_circ_0077837 and phosphatase and tensin homolog (PTEN) expression in cancer and paired non-cancer tissues from a total of 64 NSCLC patients were studied with RT-qPCR. To evaluate the prognostic value of hsa_circ_0077837 for NSCLC, these 64 patients were monitored for 5 years. Expression of PTEN in NSCLC cells with hsa_circ_0077837 overexpression was determined by RT-qPCR and Western blot. The methylation of PTEN gene in cells transfected with hsa_circ_0077837 expression vector was analyzed by methylation specific PCR (MSP). The roles of hsa_circ_0077837 and PTEN in NSCLC cell proliferation were evaluated using cell apoptosis assay. Our data showed that hsa_circ_0077837 was upregulated in NSCLC and predicted poor survival. Besides, hsa_circ_0077837 expression level was higher in 36 advanced cases (stage III and IV) than in 28 early-stage cases (stage I and II). Hsa_circ_0077837 was inversely correlated with PTEN across cancer tissues. In NSCLC cells, hsa_circ_0077837 overexpression decreased PTEN expression, increased PTEN gene methylation, and reduced HCC827 cell apoptosis via PTEN. Overall, hsa_circ_0077837 is upregulated in NSCLC and downregulates PTEN by increasing its gene methylation to suppress cell apoptosis.
List of abbreviations:
Non-small cell lung cancer (NSCLC); circRNAs (circular RNAs); methylation-specific PCR (MSP)
Introduction
Lung cancer is the most common cancer in terms of incidence and mortality among males and females in most countries [Citation1–3]. Compared to the 6% 5-year overall survival rate of small cell lung cancer patients (SCLC), more non-small cell lung cancer (NSCLC) patients (about 24%) can survive more than 5 years [Citation4]. However, the overall survival is still poor and needs to be further improved. Although smoking contributes to NSCLC [Citation5], a considerable number of NSCLC is also diagnosed in never-smokers [Citation6], suggesting the complicated pathogenesis of NSCLC. Without timely and effective treatment, NSCLC may transform to SCLC, leading to even worse prognosis [Citation7].
Although efforts have been made to prevent and treat NSCLC, patients’ survival has been significantly improved in recent decades [Citation8]. Therefore, novel therapies are still needed. Studies have identified numerous molecular factors involved in the malignancy of NSCLC [Citation9,Citation10]. Although many of them could be targets for NSCLC treatment [Citation11,Citation12], more targets are needed to improve targeted therapy. Circular RNAs (CircRNAs) have no protein-coding capacity, but they regulate cancers by affecting protein production, suggesting their potentials as targets for targeted cancer therapy [Citation13]. Moreover, circRNAs in lung cancer have been reported to be related to cancer progression, thereby affecting patients’ survival [Citation13]. Some circRNAs with critical functions in NSCLC have been proven to be potential prognostic biomarkers [Citation13]. CircRNA hsa_circ_0077837 inhibits colorectal cancer [Citation14]. Our preliminary microarray data revealed that hsa_circ_0077837 is upregulated in NSCLC and inversely correlated with phosphatase and tensin homolog (PTEN), a critical tumor suppressor [Citation15]. Therefore, it is reasonable to hypothesize that hsa_circ_0077837 might interact with PTEN to participate in NSCLC. Therefore, we investigated the potential interaction between hsa_circ_0077837 and PTEN in NSCLC.
Methods
Patients and tissue collection
From July 2013 to July 2015, 64 NSCLC patients (24 females and 40 males, 61.4 ± 9.8 years) were enrolled in Qilu Hospital of Shandong University after Ethics approval was obtained from the Ethics Committee. NSCLC was diagnosed by computed tomography (CT) scan and confirmed by histopathological biopsy. Patients were included if they were newly diagnosed and had not been treated previously. Patients were excluded if they were recurrent cases and complicated with other diseases. Primary tumors were resected from patients and dissected to isolate paired NSCLC and non-tumor tissues. All participants signed informed consent. The clinical characteristics of NSCLC patients are summarized in .
Table 1. Correlation between hsa_circ_0077837 expression and clinical characteristics in NSCLC patients (n = 64)
Treatment and follow-up
The 64 NSCLC patients were grouped into the stage I or II (n = 28) and stage III or IV (n = 36) according to the 7th edition of the American Joint Committee on Cancer (AJCC) staging systems. All patients did not undergo treatments prior to the surgery. From the day of admission, patients were visited every month for 5 years to monitor their survival. The 64 patients either died of NSCLC during the follow-up or completed the follow-up.
NSCLC cells and cell culture
NSCLC cell line HCC827 was obtained from ATCC and cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS) to about 80% confluence for subsequent assays.
Vectors and transfections
To overexpress hsa_circ_0077837 or PTEN, cells were transfected with hsa_circ_0077837 or PTEN pcDNA3.1 vector using Lipofectamine 2000 (Invitrogen). In each transfection, 1 µg hsa_circ_0077837 or PTEN expression vector was transfected into 108 HCC827 cells.
RNA preparations
Total RNAs were isolated from paired tissue samples and HCC827 cells using EZ RNA Miniprep Kit (EZ BioResearch) and treated with DNase I (Invitrogen) to remove genomic DNAs. RNA purity was checked by measuring OD 260/280 ratios.
RT-qPCR
RNA samples were subject to reverse transcriptions to synthesize cDNA. hsa_circ_0077837 and PTEN mRNA levels were determined using RT-qPCRs with GAPDH as the internal control. The primer sets used for RT-qPCR were 5’-CCTGGAGAAACATGCCAAGGG-3’ and 5’-TCACTTCAGACACAGAGCCTACT-3’ for hsa_circ_0077837, 5’-GTTTACCGGCAGCATCAAA-3’ and 5’-CCCCCACTTTAGTGCACAG-3’ for PTEN, and 5’-AGCCTCCCGCTTCGCTCTC-3’ and 5’-GCGCCCAATACGACCAAATCCG-3’ for GAPDH. PCR data were processed using the 2−ΔΔCT method.
Immunohistochemical assay
Following formalin fixation and dehydration in ethanol, tissues were embedded in paraffin and prepared as 5 µm thick sections. The sections were blocked in 5% normal goat serum and were incubated in turn with rabbit anti-PTEN polyclonal antibody (1:800; ab31392, Abcam, Cambridge, UK) and goat anti-rabbit IgG (Alexa Fluor 594, Invitrogen). The sections were observed under a phase-contrast light microscope (Olympus) and photographed. Integrated optical density (IOD) was calculated using Image-Pro Plus software.
Methylation-specific PCR (MSP)
A total of 5 μg isolated genomic DNAs were used for bisulfite modification using DNA Methylation-GoldTM kit (ZYMO RESEARCH). After that, genomic DNA samples were used as templates to perform both routine PCRs and MSP using 5’-TAGATAGGTGCCCTTTGGGCCCTTG-3’ and 5’-CCCCCAAATCTGTGTCCTCATGGTGT-3’ for routine PCR and 5’-TAGATAGGTGTTTTTTGGGTTTTTG-3’ and 5’-CCCCCAAATCTATATCCTCATAATAT-3’ for MSP.
Western blot analysis
Total proteins were isolated using RIPA solution (Invitrogen) and quantified using BCA assay. After denaturation, proteins were separated by 10% SDS-PAGE gel electrophoresis and transferred onto PVDF membranes. After blocking, the membranes were incubated in turn with GAPDH (ab8245, Abcam) or PTEN (ab31392, Abcam) primary antibodies and HRP IgG secondary antibody (ab6721, Abcam). Signals were then developed using ECL Western blotting Substrate Kit (ab65623, Abcam). Data were normalized using QuantityOne software.
Cell apoptosis assay
HCC827 cells were cultured in a 96-well plate with 3,000 cells per well. Three wells were set for each experiment. After culturing for 48 h, cells were harvested, washed, and stained with Annexin-V FITC and PI. Finally, apoptotic cells were analyzed using FACSCalibur instrument.
Subcellular fractionation analysis
Both nucleus and cytoplasm fractions of HCC827 cells were prepared using a Cell Fractionation Kit from Abcam (ab109719). In brief, cells were washed using ice-cold PBS and counted. A total of 107cells were incubated with cell lysis buffer on ice for at least 20 min. The cell lysates were centrifuged for 10 min at 1200 g. The supernatants, which were the cytoplasm fractions, were collected, transferred to a new tube, and subjected to RNA isolation. Cell pellets, which were the nucleus fractions, were further incubated with nucleus lysis buffer for 10 min on ice and subjected to RNA isolation. Both RNA samples were prepared as cDNA samples and used to determine the expression of hsa_circ_0077837 using PCRs. GAPDH was included in this assay as a cytoplasm marker.
Statistical analyses
Data were expressed as mean ± standard deviation (SD). Differences among multiple groups were analyzed using ANOVA Tukey’s test. The 64 patients were divided into high and low hsa_circ_0077837 level groups (n = 32, cutoff value = median hsa_circ_0077837 expression level in cancer tissue). Survival curves were plotted based on follow-up analysis and compared using log-rank test. The associations between patients’ clinical characteristics and hsa_circ_0077837 expression were analyzed using Chi-squared test. Correlations were analyzed by Pearson’s correlation coefficient. P < 0.05 was statistically significant.
Results
Altered hsa_circ_0077837 and PTEN expression in NSCLC
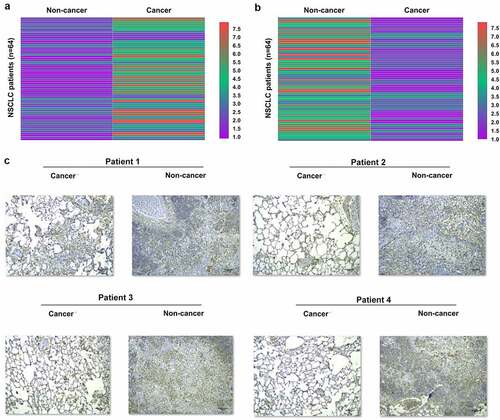
The expression levels of genes may indicate their functions. To this end, Hsa_circ_0077837 and PTEN expression levels in cancer and paired non-cancer tissue samples from 64 NSCLC patients were detected by RT-qPCR. The expression data of hsa_circ_0077837 and PTEN in paired tissues were used to plot heatmaps using Heml 1.0 software. The results showed that hsa_circ_0077837 was upregulated (), and PTEN was downregulated () in cancer tissues. Besides, in these 64 cases of NSCLC, hsa_circ_0077837 expression levels were higher in the 36 advanced cases (stage III and IV) than in the 28 early-stage cases (stage I and II) (data not shown). Immunohistochemical analysis performed on paired tissues from four NSCLC patients showed that PTEN was downregulated in NSCLC tissues because the IOD value of cancer tissues was below 30% of the IOD value of non-cancer tissues in all cases (). Therefore, altered hsa_circ_0077837 and PTEN expression may participate in NSCLC.
Figure 1. Hsa_circ_0077837 and PTEN expression in NSCLC. Hsa_circ_0077837 and PTEN mRNA expression levels in paired cancer and non-cancer tissue samples from 64 NSCLC patients were measured using RT-qPCR and used to plot heatmaps using Heml 1.0 software to reflect differential hsa_circ_0077837 expression in different TNM stages (data not shown). Immunohistochemical analysis was performed on paired tissues from four NSCLC patients to further analyze the differential PTEN expression in NSCLC (c). **, p < 0.01.
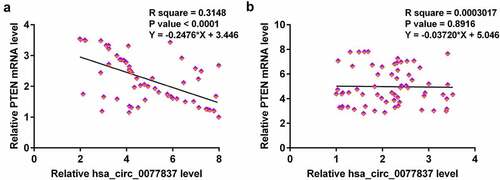
Correlations between hsa_circ_0077837 and PTEN
Correlations suggest interactions. Therefore, correlations between hsa_circ_0077837 and PTEN were explored with Pearson’s correlation coefficient. Hsa_circ_0077837 and PTEN were inversely correlated across cancer tissues () but not across non-cancer tissues (). Therefore, hsa_circ_0077837 may interact with PTEN in NSCLC.
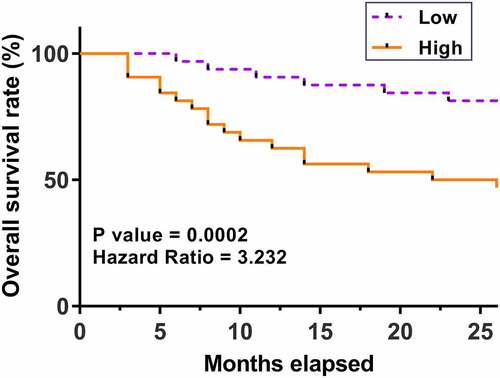
Predictive value of hsa_circ_0077837 for patients’ survival
The prognostic value of hsa_circ_0077837 for NSCLC was explored by plotting survival curves. Compared to patients with low hsa_circ_0077837 levels, patients with high hsa_circ_0077837 levels experienced worse survival (). Therefore, high expression levels of hsa_circ_0077837 in cancer tissues may predict poor survival of NSCLC patients.
Hsa_circ_0077837 overexpression decreased PTEN expression through methylation
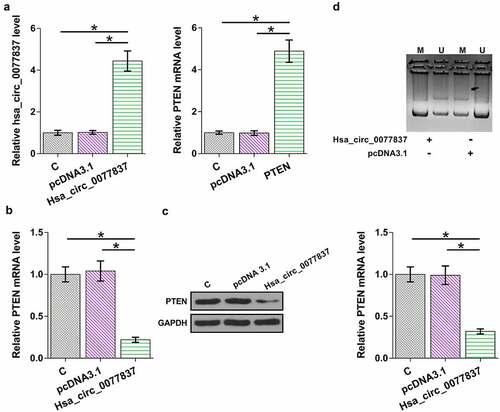
The inverse correlation between hsa_circ_0077837 and PTEN indicated their possible interaction. To further explore their relationship, HCC827 cells were overexpressed with hsa_circ_0077837 or PTEN. hsa_circ_0077837 or PTEN overexpression was confirmed by RT-qPCR (, p < 0.05). Hsa_circ_0077837 overexpression decreased PTEN mRNA () and protein () expression (p < 0.05). The effect of hsa_circ_0077837 overexpression on PTEN gene methylation was evaluated by MSP. Compared to the empty vector group, cells transfected with hsa_circ_0077837 showed increased PTEN gene methylation (). Subcellular fractionation assay was used to determine the subcellular localization of hsa_circ_0077837. Hsa_circ_0077837 can be detected in both nucleus and cytoplasm fractions (Supplemental Fig.1), consistent with its function in regulating DNA methylation. Therefore, hsa_circ_0077837 may downregulate PTEN by increasing its gene methylation.
Figure 4. Hsa_circ_0077837 regulated PTEN methylation. HCC827 cells were overexpressed with hsa_circ_0077837 or PTEN (a). The effects of hsa_circ_0077837 overexpression on PTEN mRNA (b) and protein (c) levels were analyzed. The effect of hsa_circ_0077837 on PTEN gene methylation was analyzed by MSP (d). M, methylated PCR product; U, un-methylated PCR product. *, p < 0.05.
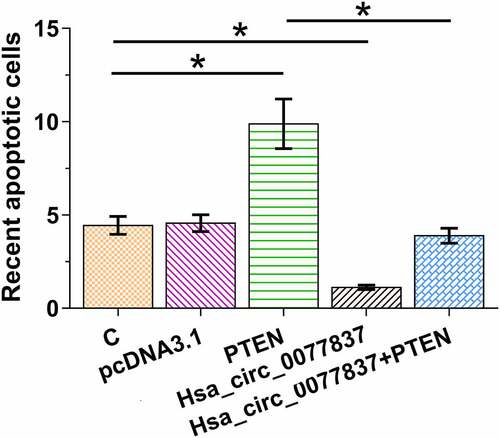
Hsa_circ_0077837 overexpression reduced HCC827 cell apoptosis via PTEN
The role of hsa_circ_0077837 and PTEN in HCC827 cell apoptosis was analyzed. It was observed that hsa_circ_0077837 overexpression decreased HCC827 cell apoptosis, while PTEN overexpression increased HCC827 cell apoptosis. Moreover, PTEN overexpression reduced the inhibitory effect of hsa_circ_0077837 overexpression on cell apoptosis (, p < 0.05). Therefore, hsa_circ_0077837 may suppress NSCLC cell apoptosis via PTEN.
Discussion
This research studied the involvement of hsa_circ_0077837 in NSCLC. We found that hsa_circ_0077837 was upregulated in NSCLC and may downregulate PTEN by increasing PTEN gene methylation to suppress NSCLC cell apoptosis.
Hsa_circ_0077837 was under-expressed in colorectal cancer (CRC), and hsa_circ_0077837 overexpression suppressed CRC cell proliferation, suggesting the role of hsa_circ_0077837 as a tumor suppressor in CRC [Citation14]. Our research revealed that hsa_circ_0077837 could suppress cell apoptosis, indicating it played an oncogenic role in NSCLC. Therefore, hsa_circ_0077837 may play different roles in different cancers, and the functions of hsa_circ_0077837 in other cancers should be investigated.
It has been well established that treatment strategies are closely related to the survival of patients [Citation16]. Hsa_circ_0077837 expression was found to be closely correlated with the poor prognosis of NSCLC patients. Therefore, monitoring changes in hsa_circ_0077837 expression levels in NSCLC patients may improve the design of treatment approaches, thereby improving patients’ survival. However, the prognostic value of hsa_circ_0077837 for NSCLC remains to be further validated by more studies with a larger sample size.
PTEN is a tumor suppressor that participates in cancer biology by inducing cancer cell apoptosis via suppressing the PI3K-Akt pathway, a main cell survival pathway in cancers [Citation15]. Consistently, our results showed downregulation of PTEN in NSCLC and its enhancing effect on NSCLC cell apoptosis. Recent advances in functional characterization of circRNAs have shown that circRNAs interact with methylation-related factors, such as TET1 and DNMT1, to regulate DNA methylation [Citation17]. This study showed that hsa_circ_0077837 could increase PTEN gene methylation to downregulate its expression. Our findings enriched our understanding of the functionality of circRNAs. It is worth noting that hsa_circ_0077837 and PTEN levels were closely correlated across cancer tissues but not non-cancer tissues. Therefore, the interaction between hsa_circ_0077837 and PTEN may be mediated by certain pathological factors. The molecular mechanism that mediates their interaction remains to be further studied.
The roles of circRNAs in NSCLC have been extensively explored in recent years [Citation18–20]. However, the functions of most circRNAs in NSCLC remain unclear and need to be investigated.
Conclusions
Hsa_circ_0077837 is upregulated in NSCLC and predicts poor survival of NSCLC patients. Moreover, hsa_circ_0077837 may downregulate PTEN through methylation to suppress cancer cell apoptosis.
Ethical approval and consent to participate
All patients signed written informed consent. All procedures were approved by the Ethics Committee of Qilu Hospital of Shandong University and operated in keeping with the standards set out in the Announcement of Helsinki and laboratory guidelines of research in China.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author contribution statement
Dezhi Li, Hongying Lv, Huijiang Gao, Lin Li: experimental work, manuscript writing, research design, literature search and data analysis; Zizong Wang, Dongfei Wang, Kaihua Tian: data analysis and statistical analysis. All authors read and approved the final manuscript.
Supplemental Material
Download Zip (949.1 KB)Acknowledgements
We thank the Young Medical Key Talents Project in Jiangsu Province (Grant No. QNRC2016193), the Precision Medicine Project of Wuxi Municipal Commission of Health and Family Planning (Grant No. J201805), and the Youth scientific research project of Wuxi municipal health commission (Grant No. Q201951) for approval.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Team NLSTR. Lung cancer incidence and mortality with extended follow-up in the national lung screening trial. J Thorac Oncol. 2019;14(10):1732–1742.
- Barta JA, Powell CA, and Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(1):8.
- Perloff T, King JC, Rigney M, et al. Survivor guilt: the secret burden of lung cancer survivorship. J Psychosoc Oncol. 2019;37(5):573–585.
- Reddy KP, Kong CY, Hyle EP, et al. Lung cancer mortality associated with smoking and smoking cessation among people living with HIV in the United States. JAMA Intern Med. 2017;177(11):1613–1621.
- Pirie K, Peto R, Green J, et al. Lung cancer in never smokers in the UK million women study. Int J Cancer. 2016;139(2):347–354.
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–72.
- Vijayvergia N, Shah PC, Denlinger CS. Survivorship in non-small cell lung cancer: challenges faced and steps forward. J Natl Compr Canc Netw. 2015;13(9):1151–1161.
- Cheng L, Alexander RE, Maclennan GT, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25(3):347–369.
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454.
- Mayekar MK, Bivona TG. Current landscape of targeted therapy in lung cancer. Clin Pharmacol Ther. 2017;102(5):757–764.
- Noor ZS, Cummings AL, Johnson MM, et al. Targeted therapy for non-small cell lung cancer. Semin Respir Crit Care Med. 2020;41(3):409–434.
- Di X, Jin X, Li R, et al. CircRNAs and lung cancer: biomarkers and master regulators. Life Sci. 2019;220:177–185.
- Sheng WZ, Dong TG, and Zhang B, et al. Novel circular RNA hsa_circ_0077837 suppresses colorectal cancer cell proliferation by regulating the microRNA 21/phosphatase and tensin homolog axis. 2020. doi:10.21203/rs.2.19904/v1.
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4(1):127–150.
- Gainor JF, Tan DS, De Pas T, et al. Progression-free and overall survival in ALK-positive NSCLC patients treated with sequential crizotinib and ceritinib. Clin Cancer Res. 2015;21(12):2745–2752.
- Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218.
- Zhang Y, Zeng S, Wang T. Circular RNA hsa_circ_0002360 promotes non-small cell lung cancer progression through upregulating matrix metalloproteinase 16 and sponging multiple micorRNAs. Bioengineered. 2021. DOI:10.1080/21655979.2021.1999370
- Hu X, Wang P, Qu C, et al. Circular RNA Circ_0000677 promotes cell proliferation by regulating microRNA-106b-5p/CCND1 in non-small cell lung cancer. Bioengineered. 2021;12(1):6229–6239.
- Zhang CC, Li Y, Feng XZ, et al. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered. 2021;12(1):414–425.