ABSTRACT
Previous studies indicated that long noncoding RNA (lncRNA) serine peptidase inhibitor, Kunitz type 1 antisense RNA1 (SPINT1-AS1) could function as an oncogenic gene in various human cancers. However, the regulatory mechanisms of SPINT1-AS1 in the tumorigenesis of colorectal cancer (CRC) remain unclear. It was found that SPINT1-AS1 was upregulated in CRC and contributed to the poor prognosis of CRC patients. Silencing of SPINT1-AS1 inhibited proliferation and metastasis but increased apoptosis of CRC cells. Furthermore, we found that SP1 could activate SPINT1-AS1 by acting as a transcription factor. Meanwhile, we identified miR-214 was negatively regulated by SPINT1-AS1. Furthermore, miR-214 repression restored the suppressive effects on malignant biological behaviors of CRC caused by SPINT1-AS1 silencing. In addition, SPINT1-AS1 mediated HDGF expression through targeting miR-214. Finally, overexpressed heparin-binding growth factor (HDGF) overturned the effects on viability, metastasis, and apoptosis of CRC cells induced by SPINT1-AS1 depletion or miR-214 upregulation. In conclusion, our results demonstrated that SP1-induced SPINT1-AS1 could facilitate CRC progression by inhibiting miR-214 and increasing HDGF expression. These findings might provide a new approach for CRC treatment.
Introduction
Colorectal cancer (CRC) is the third most common malignancy and the fourth leading cause of cancer-related mortality in the world [Citation1,Citation2]. Since no obvious symptoms during the early stage, about 70% of patients with CRC are diagnosed at an advanced stage [Citation3]. Despite great progress in CRC treatment strategies, the overall survival rate of CRC patients remains low due to the high frequency of metastasis and recurrence [Citation4]. Hence, the exploration of new possible biomarkers for diagnosis and prognosis prediction and identification of novel molecular mechanisms governing CRC malignancy may offer benefits for CRC patients [Citation5].
Long noncoding RNAs (LncRNAs) are a group of RNAs transcripts that are greater than 200 nt and lack protein-coding ability [Citation6]. Accumulating studies have reported that lncRNAs play vital roles in regulating cellular biological processes, such as cell growth, apoptosis, and differentiation, especially in cancer cells. For instance, LINC00116 promoted cervical cancer tumorigenesis via sponging miR-106a and increasing c-Jun level [Citation7]. NR2F1 antisense RNA 1 (NR2F1-AS1) contributed to the angiogenesis of breast cancer via regulating insulin-like growth factor 1 (IGF-1)/ insulin-like growth factor 1 receptor (IGF-1 R) signaling [Citation8]. MCM3AP antisense RNA 1 (MCM3AP-AS1) exhibited a carcinogenic effect on hepatocellular carcinoma by decreasing miR-194-5p and increasing forkhead box A1 (FOXA1) level [Citation9]. Previous studies indicated that high expression of Serine peptidase inhibitor, Kunitz type 1 antisense RNA1 (SPINT1-AS1) was associated with regional lymph node metastasis, distant metastasis, and shorter relapse-free survival time in CRC [Citation10]. Nevertheless, the role and molecular mechanisms of SPINT1-AS1 in CRC need to be further elucidated.
Amounting studies have indicated that microRNAs (miRNAs) participate in the tumorigenesis of human malignancies [Citation11]. For instance, miR-328 inhibited the carcinogenesis of bladder cancer by regulating PI3K/AKT signaling [Citation12]. Overexpression of miR-381 acted as a suppressor against endometrial carcinoma by targeting IGF-1 R [Citation13]. Depletion of miR-4443 and miR-5139-3p facilitated the malignant biological behaviors of ovarian cancer [Citation14]. Notably, miR-214 was demonstrated to act as a tumor suppressor in various cancers, including CRC [Citation15]. Previous studies have identified that lncRNAs can mediate a ‘sponge’ regulatory network by sequestering miR-214. For example, HOXA11-AS accelerated the tumorigenesis of liver cancer through decreasing miR-214 expression [Citation16]. Hepatocellular carcinoma up-regulated EZH2-associated long non-coding RNA (HEIH) served as a miR-214 sponge to accelerate gastric cancer progression [Citation17]. Deletion of HCP5 inhibited the proliferative and metastatic abilities of renal cell carcinoma cells via binding miR-214 [Citation18]. However, the regulatory axis of SPINT1-AS1 and miR-214 in CRC is unclear.
This study aimed to investigate the biological role of SPINT1-AS1 in CRC, and we hypothesized that SPINT1-AS1 exerted an oncogenic effect on CRC. Our findings might provide a promising therapeutic strategy for CRC.
Materials and methods
Clinical sample
Sixty paired tissues and adjacent normal tissues were collected from CRC patients at the Affiliated Yangming Hospital of Ningbo University and the clinical characteristics of CRC patients are presented in . All samples were instantly transferred to liquid nitrogen for frozen storage at −80°C. All patients signed the written informed consent and this research was approved by the Ethics Committee of the Affiliated Yangming Hospital of Ningbo University.
Table 1. Clinicopathological features of CRC patients
Cell culture and transfection
CRC cells (HCT116, LoVo, SW480, and Caco-2) and normal colonic epithelial cell line (NCM460), bought from ATCC (USA), were cultured in DMEM (Corning Life Sciences) with 10% FBS (Thermo Fisher Scientific) at 37°C with 5% CO2.
The short hairpin RNA (shRNAs) against SPINT1-AS1 (shSPINT1-AS1#1, 5ʹ-GCGAAUGAGCUGCACUGACUA-3ʹ; shSPINT1-AS1#2, 5ʹ-GCUCAGACAUCGAUGCUGACU-3ʹ; shSPINT1-AS1#3, 5ʹ-GCUCAUGCGCACUAUGCUGUA-3ʹ) and SP1 (shSP1; 5ʹ-GCAUAUUUGCCACAUCCAAGG-3ʹ) with negative control (shNC; 5ʹ-UUCUCCGAACGUGUCACGU-3ʹ), miR-214 mimics and miR-214 inhibitor with their corresponding controls (NC mimics and NC inhibitor) were acquired from GenePharma (Shanghai). To overexpress SPINT1-AS1, HDGF, and SP1, the full-length of SPINT1-AS1, HDGF, and SP1 were cloned into the pcDNA3.1 (GenePharma). The transfection was performed using Lipofectamine 2000 (Invitrogen).
RT-qPCR
The RNA extraction was performed with Trizol reagent (Invitrogen). Total RNAs were reversed into complementary DNA (cDNA) using PrimeScriptTM RT reagent kit. The SYBR® Premix Ex TaqTM II reagent kit (TaKaRa) was used for RT-qPCR on a 7500 Real-Time PCR System. Relative expression was calculated using the 2−ΔΔCq method with GAPDH or U6 as the control genes.
CCK-8 assay
HCT116 and SW480 cells were seeded into 96-well plates for 0, 24, 48, 72 h. Then, 10 μl CCK-8 reagent (Dojindo) was added to each well and incubated for 4 h at room temperature. The absorbance at 450 nm was measured using a microplate reader (Bio-Rad) [Citation19].
Transwell
Transwell chambers pre-coated with Matrigel (BD Biosciences) were used to evaluate cell invasive ability [Citation20]. CRC cells (5 × 104 cells/well) were seeded into the upper chambers after transfection and suspended in 200 μL serum-free DMEM (Corning Life Sciences). Then, 600 μL DMEM with 10% FBS was added into the lower chambers. After 48 h, the invaded cells in the lower chamber were fixed and stained. The number of invaded cells was measured with a microscope. Cell migration was performed using the protocol described above, except that the transwell chambers were not coated with Matrigel.
Luciferase reporter assay
Wild-type and mutant sequences of SPINT1-AS1 or HDGF were cloned into pmirGLO reporter vectors to establish SPINT1-AS1-WT/Mut or HDGF-WT/Mut reporters. Then, miR-214 mimics or NC mimics were co-transfected with the above reporters into cells. The luciferase activity was evaluated at 48 h after co-transfection by the Dual-Luciferase Reporter Assay System (Promega) [Citation21].
Western blotting
Total protein was extracted using radioimmunoprecipitation lysis buffer (Beyotime), followed by separation of 10% SDS-PAGE and transfer to the PVDF membrane. The membranes were incubated with primary antibodies against HDGF and GAPDH. Then, the membranes were incubated with secondary antibodies. The protein bands were quantified using an ECL reagent [Citation22].
TUNEL assay
The apoptosis of CRC cells was evaluated via InSitu Cell Death Detection kit (Roche, Germany) [Citation23]. After rising twice with PBS, cells were fixed with 4% paraformaldehyde and permeabilized in 0.25% Triton-X 100. Subsequently, the cells were incubated with the TUNEL enzyme, and then counterstained with DAPI for 10 min. Finally, TUNEL-positive cells were counted using a cell counter (BD Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS 19.0 (IBM). The differences between groups were analyzed by Student's t-test or one-way ANOVA. The correlation between gene expression was determined by Pearson’s correlation analysis. p < 0.05 was considered statistically significant.
Results
This study explored the role and mechanism of SPINT1-AS1 in CRC and our data indicated that SP1-induced SPINT1-AS1 contributed to CRC progression by regulating miR-214 and HDGF expression. Our findings uncovered the oncogenic role of SPINT1-AS1 in CRC and might provide a promising therapeutic strategy for CRC treatment.
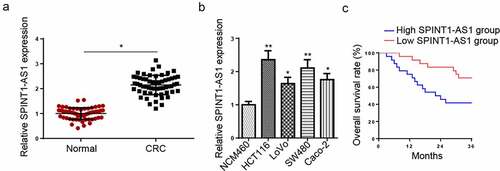
SPINT1-AS1 was upregulated in CRC
Firstly, RT-qPCR indicated that SPINT1-AS1 was upregulated in CRC tissues and cells ( and b). In addition, CRC patients with high SPINT1-AS1 levels exhibited a shorter survival time than those with low SPINT1-AS1 levels (). These data suggested that SPINT1-AS1 might be associated with the carcinogenesis of CRC.
Figure 1. SPINT1-AS1 was upregulated in CRC. (a) RT-qPCR showed the relative expression of SPINT1-AS1 in CRC tissues (n = 60) and adjacent healthy tissues (n = 60). (b) RT-qPCR detected the expression of SPINT1-AS1 in CRC cell lines (HCT116, LoVo, SW480, and Caco-2) and normal colon mucosal epithelial cell line (NCM460). (c) Kaplan–Meier analysis of the association between SPINT1-AS1expression and the overall survival of patients with CRC. *p < 0.05, **p < 0.01.
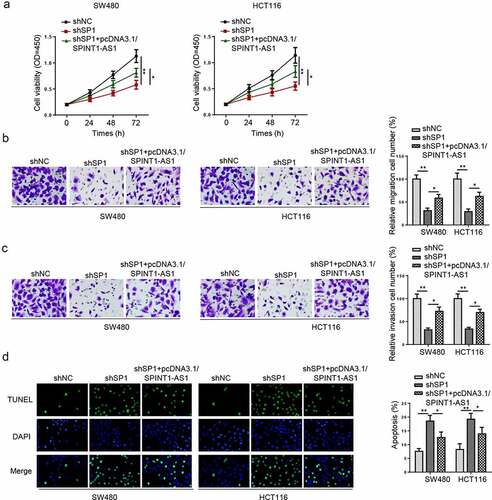
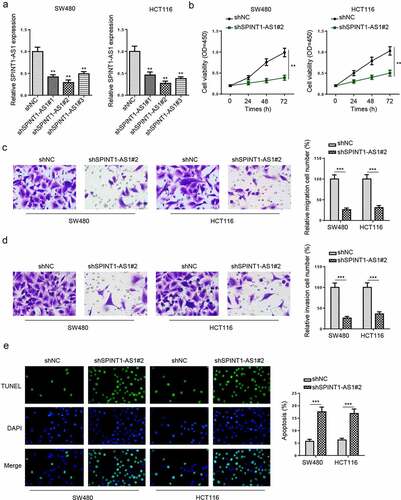
Knockdown of SPINT1-AS1 inhibited the malignant behaviors of CRC cells
To elaborate the effect of SPINT1-AS1 on CRC, shSPINT1-AS1#1, shSPINT1-AS1#2, or shSPINT1-AS1#3 were transfected into SW480 and HCT116 cells, and the knockdown efficiencies were confirmed by RT-qPCR (). shSPINT1-AS1#2 was used for the following experiments due to the highest transfection efficiency. CCK-8 and transwell assays uncovered that SPINT1-AS1 silencing suppressed the viability, migration, and invasion (), while TUNEL assay revealed the silencing of SPINT1-AS1 increased cell apoptosis of SW480 and HCT116 cells (). The above results uncovered that knockdown of SPINT1-AS1 repressed the CRC progression.
Figure 2. Knockdown of SPINT1-AS1 inhibited the malignant behaviors of CRC cells. (a) RT-qPCR indicated the SPINT1-AS1 expression in SW480 and HCT116 cells transfected with shSPINT1-AS1#1, shSPINT1-AS1#2, or shSPINT1-AS1#3. (b) CCK-8 assay evaluated the cell proliferation of SW480 and HCT116 cells transfected with shSPINT1-AS1#2. (c and d) The migration and invasion abilities of shNC or shSPINT1-AS1#2 transfected SW480 and HCT116 cells were assessed via transwell assays. (e) TUNEL assay showed the apoptosis rate of SW480 and HCT116 cells transfected with shSPINT1-AS1#2 or shNC. ** p < 0.01, ***p < 0.001.
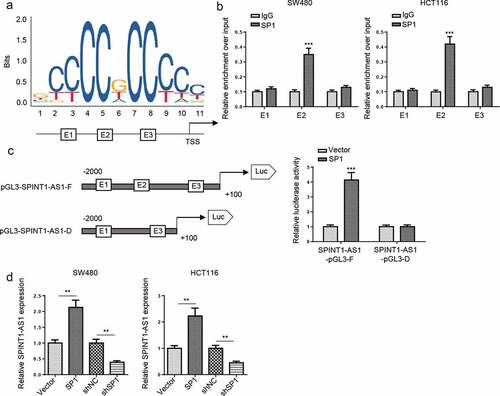
SP1 activated SPINT1-AS1 expression by functioning as a transcription factor
Since several transcription factors (TFs), such as YY1 [Citation24], EZH2 [Citation25], STAT3 [Citation26], were found to participate in the regulation of lncRNAs expression. We then explore the upstream regulation of SPINT1-AS1. The promoter region of SPINT1-AS1 was identified by JASPAR database, and found that three SP1 binding sites with scores >9 at the regions E1 (−1693bp to −1703bp, CCCCCACCCCA), E2 (−73 to −64 bp, GTGGCGGGGA), and E3 (−19 to −9 bp, GGCCCTCCCAC) in the SPINT1-AS1 promoter (). ChIP assay indicated that SP1 could bind to E2 site (). Furthermore, full promoter region of SPINT1-AS1 and E2 deleted promoter region were cloned into pGL3-basic reporter, and the data indicated that the deletion of E2 sites substantially impaired the effect of SP1 on SPINT1-AS1 transcription activation (). Moreover, overexpression of SP1 upregulated SPINT1-AS1 expression, while knockdown of SP1 downregulated SPINT1-AS1 expression in CRC cells (). In addition, SP1 knockdown inhibited cell proliferation, migration, and invasion, but promoted the cell apoptosis in CRC, while SPINT1-AS1 overexpression partially restored these effects (), suggesting that SP1 upregulated SPINT1-AS1 expression promoted the tumorigenesis of CRC.
Figure 3. SP1 activated SPINT1-AS1 expression by functioning as a transcription factor. (a) The predicted positions of putative SP1 binding motif in SPINT1-AS1 promoter. (b) ChIP assays were performed to show the direct binding of SP1 to SPINT1-AS1 promoter regions. (c) A luciferase reporter assay was used by cotransfecting the full SPINT1-AS1 promoter (SPINT1-AS1-pGL3-F) or deleted SPINT1-AS1 promoter fragment E2 (SPINT1-AS1-pGL3-D) with SP1 expression plasmid or blank vector in 293 T cells. (d) RT-qPCR analysis of SPINT1-AS1 expression levels following the treatment of shSP1 in CRC cells. ** p < 0.01, ***p < 0.001.
SPINT1-AS1 directly targets miR-214
Through using Starbase, the potential interacting sequences between miR-214 and SPINT1-AS1 are presented in . The luciferase activities of SPINT1-AS1-WT were remarkably decreased by miR-214 upregulation and increased by miR-214 downregulation in CRC cells, while the luciferase activities of SPINT1-AS1-Mut showed no significant difference (). Moreover, miR-214 expression was increased by SPINT1-AS1 knockdown but decreased by SPINT1-AS1 overexpression (). In addition, SPINT1-AS1 and miR-214 expression were negatively correlated in CRC tissues (). These findings revealed that miR-214 was negatively regulated by SPINT1-AS1 in CRC.
Figure 5. SPINT1-AS1 directly targets miR-214. (a) StarBase presented the binding site between SPINT1-AS1 and miR-214. (b) Luciferase activity of SPINT1-AS1-WT and SPINT1-AS1-Mut reporters in CRC cells was determined in the NC mimics or miR-214 mimics groups using a dual-luciferase reporter assay. (c) RT-qPCR showed the relative miR-214 expression in SW480 and HCR116 cells transfected with shNC, shSPINT1-AS1, pcDNA3.1, or pcDNA3.1/SPINT1-AS1. (d) Pearson’s correlation analysis showed the correlation between SPINT1-AS1 expression and miR-214 in CRC tissues. ** p < 0.01.
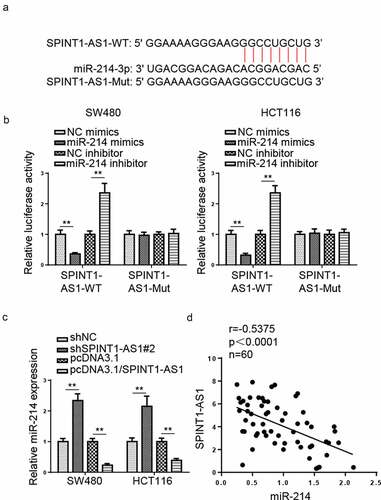
SPINT1-AS1 accelerated CRC progression by targeting miR-214
To figure out whether SPINT1-AS1 could regulate CRC progression via miR-214, shNC, shSPINT1-AS1#2, and shSPINT1-AS1#2+ miR-214 inhibitor were transfected into CRC cells. RT-qPCR showed that miR-214 inhibitor partially neutralized the effect caused by SPINT1-AS1 knockdown on the miR-214 expression (). Functional assays indicated that SPINT1-AS1 inhibition repressed the viability, migration, and invasion of CRC cells, which was counteracted by inhibition of miR-214 (). Furthermore, silencing of SPINT1-AS1 increased cell apoptosis, while miR-214 inhibition reversed this effect (). In sum, the above data demonstrated that SPINT1-AS1 facilitated CRC progression via miR-214.
Figure 6. SPINT1-AS1 accelerated CRC progression by targeting miR-214. (a) RT-qPCR showed miR-214 expression in SW480 and HCT116 cells transfected with shNC, shSPINT1-AS1#2, and shSPINT1-AS1#2 + miR-214 inhibitor. (b-e) CCK-8, transwell, and TUNEL assays showed the proliferation, migration, invasion and apoptosis of CRC cells transfected with shNC, shSPINT1-AS1#2, and shSPINT1-AS1#2 + miR-214 inhibitor. *p < 0.05, ** p < 0.01, ***p < 0.001.
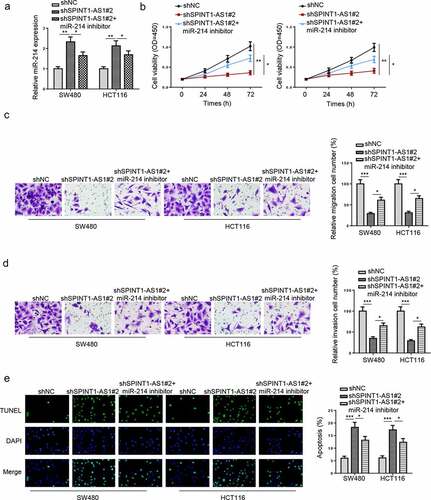
SPINT1-AS1 regulated HDGF expression through targeting miR-214
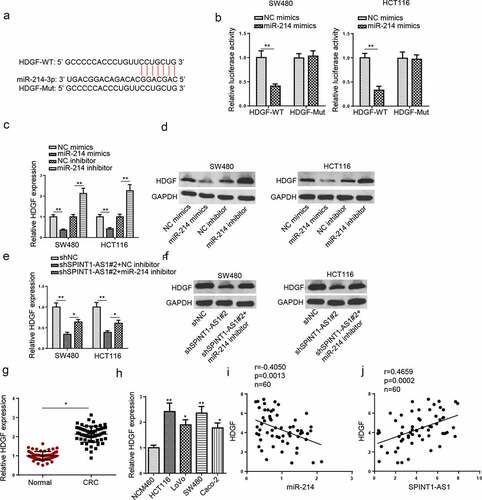
To further investigate the downstream target of miR-214, Starbase website was used to screen the downstream genes and the potential binding site between miR-214 and HDGF is presented in . Luciferase activities of HDGF-WT were dramatically reduced by transfecting with miR-214 mimics in CRC cells, while no difference was found in HDGF-Mut (). In addition, HDGF expression was remarkably downregulated by miR-214 upregulation, whereas inhibition of miR-214 increased HDGF level in CRC cell lines ( and d). Besides, to investigate whether SPINT1-AS1 regulated HDGF expression via binding miR-214, we transfected shNC, shSPINT1-AS1#2, or shSPINT1-AS1#2+ miR-214 inhibitor into the CRC cells. Silencing of SPINT1-AS1 reduced HDGF expression, while this effect was restored by the inhibition of miR-214 ( and f). In addition, HDGF was upregulated in CRC tissues and cells ( and h). Moreover, a negative correlation between HDGF and miR-214 level and a positive correlation between HDGF and SPINT1-AS1 levels were found in CRC tissues ( and j). These findings demonstrated that SPINT1-AS1 could positively regulate HDGF by absorbing miR-214.
Figure 7. SPINT1-AS1 regulated HDGF expression through targeting miR-214. (a) StarBase website predicted the binding site between HDGF and miR-214. (b) Dual-luciferase reporter assay was adopted to analyze the luciferase activity of HDGF-WT or HDGF-Mut reporter in CRC cells transfected with NC mimics or miR-214 mimics. (c and d) RT-qPCR and Western blot showed the mRNA and protein level of HDGF in SW480 and HCT116 cells transfected with NC mimics, NC inhibitor, miR-214 mimics and miR-214 inhibitor. (e and f) RT-qPCR and Western blot assays showed HDGF expression in CRC cells transfected with shNC, shSPINT1-AS1#2, and shSPINT1-AS1#2+ miR-214 inhibitor. (g and h) RT-qPCR showed the expression of HDGF in CRC tissues and cells. (i and j) Pearson’s correlation analysis was used to analyze the correlation between HDGF and miR-214 or SPINT1-AS1 in CRC. *p < 0.05, ** p < 0.01.
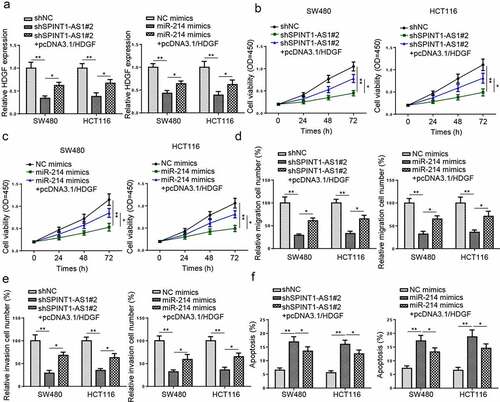
SPINT1-AS1 contributed to the tumorigenesis of CRC via sponging miR-214 and upregulating HDGF expression
To investigate whether SPINT1-AS1 or miR-214 regulated CRC progression via HDGF, rescue experiments were performed. RT-qPCR revealed that HDGF overexpression restored the inhibitory effect of shSPINT1-AS1#2 or miR-214 upregulation on HDGF expression (). CCK-8 and transwell assays uncovered that SPINT1-AS1 downregulation and miR-214 upregulation suppressed the cell proliferation, migration, and invasion, which could be counteracted by HDGF mimics (). Besides, TUNEL assay showed elevated cell apoptosis caused by SPINT1-AS1 knockdown or miR-214 overexpression was then partially neutralized by HDGF overexpression (). All these data indicated that SPINT1-AS1 promoted viability and metastasis of CRC cells and decreased cell apoptosis through upregulating HDGF expression and sponging miR-214.
Figure 8. SPINT1-AS1 contributed to the tumorigenesis of CRC via sponging miR-214 and upregulating HDGF expression. (a-f) CCK-8, transwell, and TUNEL assays showed the viability, migration, invasion, and apoptosis of CRC cells transfected with shNC, shSPINT1-AS1#2, and shSPINT1-AS1#2+ pcDNA3.1/HDGF, or transfected with NC mimics, miR-214 mimics, miR-214 mimics + pcDNA3.1/HDGF. *p < 0.05, **p < 0.01.
Discussion
CRC is the most common malignancy of the digestive system, and approximately 20% of CRC patients are diagnosed in the metastatic stage [Citation27]. Previous studies have identified that lncRNAs might act as diagnostic and prognostic biomarkers for CRC. For instance, SNHG11 might be a new therapeutic target for CRC treatment and a potential biomarker for the early diagnosis of CRC [Citation28]. High expression of MALAT1 could act as a negative prognostic biomarker for CRC patients at stage II/III [Citation29]. SPINT1-AS1 has been reported to act as an oncogene in various human cancers, such as breast cancer [Citation30], esophageal squamous cell carcinoma [Citation31], and cervical cancer [Citation32]. In this research, we demonstrated that SPINT1-AS1 was upregulated in CRC, and silencing of SPINT1-AS1 suppressed the viability, migration, and invasion, and promoted the apoptosis of CRC cells.
Increasing evidence has indicated that TFs are the vital regulator to altering the expression of lncRNAs. For example, YY1-induced activation of double homeobox A pseudogene 8 (DUXAP8) promoted triple-negative breast cancer progression by regulating suppressor APC domain containing 2 (SAPCD2) [Citation33]. ELK1-mediated upregulation of LBX2-AS1 contributed to the tumorigenesis of CRC via the miR-491-5p/S100 calcium-binding protein A11 (S100A11) axis [Citation34]. SP1, as a TF, has been reported to participate in the dysregulation of lncRNAs, such as LINC00514 [Citation35], CTBP1 divergent transcript (CTBP1-AS2) [Citation36], and cancer susceptibility 11 (CASC11) [Citation37], which further modulate the progression of various cancers. Herein, we identified that SP1 could activate SPINT1-AS1 expression by acting as a TF in CRC cells.
LncRNA may act as competitive endogenous RNA (ceRNA) by competitively binding with miRNAs to modulate the expression of target genes [Citation38,Citation39]. Previous studies suggested that the lncRNA-related ceRNA network was associated with the malignant behaviors of CRC. For example, LINC00239 functioned as a ceRNA in regulating Kruppel like factor 12 (KLF12) by competitively binding to miR-484 in CRC [Citation40]. Prostate androgen-regulated transcript 1 (PART-1) functioned as a ceRNA by sponging miR-143 and promoted tumor progression in CRC [Citation41]. Therefore, we hypothesized that SPINT1-AS1 might act as a ceRNA to modulate its downstream target gene in CRC. Herein, miR-214 was verified as a target of SPINT1-AS1. Previous studies identified miR-214 functioned as a tumor-suppressive gene in CRC. For example, the inhibition of miR-214 abolished the inhibitory effects of LINC00324 knockdown on cell proliferation and metastasis in CRC [Citation42]. Silencing of PVT1 hindered the viability and invasion of CRC cells via upregulating miR-214. In our study, we demonstrated that the attenuated cell proliferation and metastasis and increased cell apoptosis induced by SPINT1-AS1 silencing were restored by miR-214 inhibitor, suggesting that SPINT1-AS1 facilitated the tumorigenesis of CRC via miR-214.
The hepatoma-derived growth factor (HDGF), a heparin-binding protein, was reported to be overexpressed in bladder cancer [Citation43], liver cancer [Citation44], glioma [Citation45], and osteosarcoma [Citation46], which further promoted malignant tumor progression. Similarly, we demonstrated that HDGF was upregulated in CRC. Several miRNAs, such as miR-4323, miR-95-3p, miR-610, and miR-511, participated in CRC progression by targeting HDGF [Citation47–50]. In this study, we identified that miR-214 negatively regulated HDGF expression by direct interaction. Rescue experiments demonstrated that the suppression of HOXA11-AS knockdown and miR-214 mimics on cell viability, migration, invasion were abolished by HDGF overexpression.
Conclusion
Our study demonstrated that SPINT1-AS1 activated by SP1 promoted the progression of CRC via the miR-214/HDGF axis. These results suggested that SPINT1-AS1 might be a novel therapeutic target for CRC treatment. There were several limitations in this study. For example, the present study lacked in vivo experiments, which should be performed in future studies to further validate these findings.
Authors’ contributions
Ying Fang and Qianqian Yang designed the study, analyzed the data, prepared the figures, and wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Lemoine L, Sugarbaker P, Van der Speeten K. Pathophysiology of colorectal peritoneal carcinomatosis: role of the peritoneum, World. J Gastroenterol. 2016;22:7692–7707.
- Moyano A, Serrano-Pertierra E, Duque JM, et al. Magnetic lateral flow immunoassay for small extracellular vesicles quantification: application to colorectal cancer biomarker detection. Sensors (Basel). 2021;21(11):3756.
- Ye S-P, Zhu W-Q, Liu D-N, et al. Robotic-vs laparoscopic-assisted proctectomy for locally advanced rectal cancer based on propensity score matching: short-term outcomes at a colorectal center in China. J Gastrointest Oncol. 2020;12(4):424–434.
- Wang L, Jiang X, Zhang X, et al. Prognostic implications of an autophagy-based signature in colorectal cancer. Medicine (Baltimore). 2021;100(13):e25148.
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
- Huang L, Wang Y, Chen J, et al. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10(7):513.
- Lai Y, Zhou B, Tan Q, et al. LINC00116 enhances cervical cancer tumorigenesis through miR-106a/c-Jun pathway. J Cell Biochem. 2020;121(3):2247–2257.
- Zhang Q, Li T, Wang Z, et al. Lin, lncRNA NR2F1-AS1 promotes breast cancer angiogenesis through activating IGF-1/IGF-1R/ERK pathway. J Cell Mol Med. 2020;24(14):8236–8247.
- Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18(1):28.
- Li C, Li W, Zhang Y, et al. Increased expression of antisense lncRNA predicts a poor prognosis in colorectal cancer and is negatively correlated with its sense transcript. Onco Targets Ther. 2018;11:3969–3978.
- Zhang N, Hu X, Du Y, et al. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed Pharmacother. 2021;134:111099.
- Ge X, Li W, Huang S, et al. Increased miR-21-3p in injured brain microvascular endothelial cells after traumatic brain injury aggravates blood-brain barrier damage by promoting cellular apoptosis and inflammation through targeting MAT2B. J Neurotrauma. 2019;36(8):1291–1305.
- Tu C, Wang F, Wan J. MicroRNA-381 inhibits cell proliferation and invasion in endometrial carcinoma by targeting the IGF-1R. Mol Med Rep. 2018;17(3):4090–4098.
- Ebrahimi SO, Reiisi S. Downregulation of miR-4443 and miR-5195-3p in ovarian cancer tissue contributes to metastasis and tumorigenesis. Arch Gynecol Obstet. 2019;299(5):1453–1458.
- Zhou Z, Wu L, Liu Z, et al. MicroRNA-214-3p targets the PLAGL2-MYH9 axis to suppress tumor proliferation and metastasis in human colorectal cancer. Aging (Albany NY). 2020;12(10):9633–9657.
- Zhan M, He K, Xiao J, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018;22(8):3758–3767.
- Jiang L, Zhang L, Chen Q, et al. LncRNA HEIH promotes cell proliferation, migration and invasion by suppressing miR-214-3p in gastric carcinoma. J Biochem. 2021;169(5):535–542.
- Hao J-F, Chen P, Li H-Y, et al. Effects of LncRNA HCP5/miR-214-3p/MAPK1 molecular network on renal cell carcinoma cells. Cancer Manag Res. 2020;12:13347–13356.
- Zhou X, Wang Z, Xu B, et al. Long non-coding RNA NORAD protects against cerebral ischemia/reperfusion injury induced brain damage, cell apoptosis, oxidative stress and inflammation by regulating miR-30a-5p/YWHAG. Bioengineered. 2021;12(2):9174–9188.
- Zhang M, Yang L, Hou L, et al. LncRNA SNHG1 promotes tumor progression and cisplatin resistance through epigenetically silencing miR-381 in breast cancer. Bioengineered. 2021;12(2):9239–9250.
- Li H, Xuan J, Zhang W, et al. Long non-coding RNA SNHG5 regulates ulcerative colitis via microRNA-375/Janus kinase-2 axis. Bioengineered. 2021;12:4150–4158.
- Zhang Q, Wang Z, Cheng X, et al. lncRNA DANCR promotes the migration an invasion and of trophoblast cells through microRNA-214-5p in preeclampsia. Bioengineered. 2021;12(2):9424-9434 .
- Sun M, Geng D, Li S, et al. LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol Chem. 2018;399(4):387–395.
- Yang R, Liu Z, Cao H, et al. LINC01089, suppressed by YY1, inhibits lung cancer progression by targeting miR-301b-3p/HPDG axis. Cell Biol Toxicol. 2021. doi:10.1007/s10565-021-09643-8.
- Gai Q, Guo W, Liu J. EZH2-mediated long-chain non-coding RNA LINC00963 promotes proliferation and invasion of glioma cells through inhibiting p21 expression. J Buon. 2021;26(2):380–387.
- Huo N, Cong R, Sun ZJ. STAT3/LINC00671 axis regulates papillary thyroid tumor growth and metastasis via LDHA-mediated glycolysis. Cell death & disease. 2021;12:799.
- Silva-Fisher JM, Dang HX, White NM, et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun. 2020;11(1):2156.
- Xu W, Zhou G, Wang H, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2020;146(10):2901–2912.
- Zheng H-T, Shi D-B, Wang Y-W, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181.
- Zhou T, Lin K, Nie J, et al. LncRNA SPINT1-AS1 promotes breast cancer proliferation and metastasis by sponging let-7 a/b/i-5p. Pathol Res Pract. 2021;217:153268.
- Qi-Dong X, Yang X, Lu J-L, et al. Development and validation of a nine-redox-related long noncoding RNA signature in renal clear cell carcinoma. Oxid Med Cell Longev. 2020;2020:6634247.
- Song H, Liu Y, Liang H, et al. SPINT1-AS1 drives cervical cancer progression via repressing miR-214 biogenesis. Front Cell Dev Biol. 2021;9:691140.
- Yang Z, Ding H, Pan Z, et al. YY1-induced activation of lncRNA DUXAP8 promotes proliferation and suppresses apoptosis of triple negative breast cancer cells through upregulating SAPCD2. Cancer biology & therapy. 2021;22:216–224.
- Ma G, Dai W, Zhang J, et al. ELK1‑mediated upregulation of lncRNA LBX2‑AS1 facilitates cell proliferation and invasion via regulating miR‑491‑5p/S100A11 axis in colorectal cancer. Int J Mol Med. 2021;48(1). DOI:10.3892/ijmm.2021.4971
- Mi LD, Sun CX, He SW, et al. SP1-Induced upregulation of lncRNA LINC00514 promotes tumor proliferation and metastasis in osteosarcoma by regulating miR-708. Cancer Manag Res. 2020;12:3311–3322.
- Liu LX, Liu B, Yu J, et al. SP1-induced upregulation of lncRNA CTBP1-AS2 accelerates the hepatocellular carcinoma tumorigenesis through targeting CEP55 via sponging miR-195-5p. Biochem Biophys Res Commun. 2020;533(4):779–785.
- Jin J, Zhang S, Hu Y, et al. SP1 induced lncRNA CASC11 accelerates the glioma tumorigenesis through targeting FOXK1 via sponging miR-498. Biomed Pharmacother. 2019;116:108968.
- Zeng ZL, Lu JH, Wang Y, et al. The lncRNA XIST/miR-125b-2-3p axis modulates cell proliferation and chemotherapeutic sensitivity via targeting Wee1 in colorectal cancer. Cancer medicine. 2021;10:2423–2441.
- Chen W, Du M, Hu X, et al. Long noncoding RNA cytoskeleton regulator RNA promotes cell invasion and metastasis by titrating miR-613 to regulate ANXA2 in nasopharyngeal carcinoma. Cancer medicine. 2020;9:1209–1219.
- Luo X, Yue M, Li C, et al. Long Non-Coding RNA LINC00239 functions as a competitive endogenous RNA by sponging microRNA-484 and enhancing KLF12 expression to promote the oncogenicity of colorectal cancer. Onco Targets Ther. 2020;13:12067–12081.
- Hu Y, Ma Z, He Y, et al. PART-1 functions as a competitive endogenous RNA for promoting tumor progression by sponging miR-143 in colorectal cancer. Biochem Biophys Res Commun. 2017;490(2):317–323.
- Ni X, Xie JK, Wang H, et al. Knockdown of long non-coding RNA LINC00324 inhibits proliferation, migration and invasion of colorectal cancer cell via targeting miR-214-3p. Eur Rev Med Pharmacol Sci. 2019;23(24):10740–10750.
- Zhang C, Chang X, Chen D, et al. Downregulation of HDGF inhibits the tumorigenesis of bladder cancer cells by inactivating the PI3K-AKT signaling pathway. Cancer Manag Res. 2019;11:7909–7923.
- Enomoto H, Nakamura H, Nishikawa H, et al. Hepatocellular carcinoma-associated microRNAs induced by hepatoma-derived growth factor stimulation. Vivo. 2020;34(5):2297–2301.
- Xu W, Che DD, Liu Q, et al. The inhibitory effect of miR-345 on glioma progression is closely related to circRNA-hsa_circ_0073237 and HDGF. Cells Tissues Organs. 2021;210(5–6):368–379.
- Liu X, Ma W, Ma J, et al. Upregulation of miR‑95-3p inhibits growth of osteosarcoma by targeting HDGF. Pathol Res Pract. 2019;215(8):152492.
- Xia C, Li Q, Cheng X, et al. miR-4323 targets hepatoma-derived growth factor (HDGF) to suppress colorectal cancer cell proliferation. Pathol Res Pract. 2021;225:153544.
- Hong Y-G, Huang Z-P, Liu Q-Z, et al. MicroRNA-95-3p inhibits cell proliferation and metastasis in colorectal carcinoma by HDGF. Biomed J. 2020;43(2):163–173.
- Sun B, Gu X, Chen Z, et al. MiR-610 inhibits cell proliferation and invasion in colorectal cancer by repressing hepatoma-derived growth factor. Am J Cancer Res. 2015;5(12):3635–3644.
- He S, Wang G, Ni J, et al. MicroRNA-511 inhibits cellular proliferation and invasion in colorectal cancer by directly targeting hepatoma-derived growth factor. Oncol Res. 2018;26(9):1355–1363.