ABSTRACT
Circular RNA (circRNA) is considered to be an important regulator that mediates cancer chemoresistance. But whether circ-LPAR3 is involved in ovarian cancer (OC) cisplatin (DDP) resistance is unclear. The circ-LPAR3, miR-634 and pyruvate dehydrogenase kinase 1 (PDK1) expression was measured by quantitative real-time PCR (qRT-PCR). Cell cisplatin resistance and viability were measured by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) assay. In addition, cell colony number, apoptosis, and metastasis were assessed by colony formation assay, flow cytometry and transwell assay. Furthermore, in vivo experiments were performed by constructing mice xenograft models. RNA interaction was confirmed by dual-luciferase reporter assay, and PDK1 protein expression was examined by Western blot analysis. Our results showed that circ-LPAR3 was markedly upregulated in DDP-resistant OC tissues and cells. Silencing of circ-LPAR3 enhanced the DDP sensitivity of OC cells and tumors. MiR-634 could interact with circ-LPAR3, and its inhibitor overturned the regulation of si-circ-LPAR3 on cell DDP resistance. Additionally, PDK1 was targeted by miR-634, and its overexpression inverted the effect of miR-634 on cell DDP resistance. To sum up, circ-LPAR3 might contribute to the DDP resistance of OC via the miR-634/PDK1 axis.
Graphical abstract
Introduction
Ovarian cancer (OC) is a gynecological cancer, and its mortality rate is the highest among the malignant tumors of the female reproductive tract [Citation1,Citation2]. Due to the lack of effective treatment for OC recurrence and chemoresistance, the overall prognosis for OC patients is very poor [Citation3,Citation4]. The occurrence of chemoresistance not only seriously affects the life quality of OC patients, but also makes the chemotherapy window period shorter and shorter [Citation5,Citation6]. Therefore, finding potential biomarkers that affect the chemoresistance of OC is of significance for OC treatment.
As a special kind of non-coding RNA, the unique closed-loop structure of circular RNA (circRNA) makes it extremely stable in organisms [Citation7,Citation8]. In recent years, many studies have confirmed that circRNA is related to cancer malignant progression [Citation9,Citation10]. More importantly, circRNA function in cancer chemoresistance has been confirmed in many research [Citation11,Citation12]. A recent study showed that circ_0074027 served as miR-379-5p sponge to upregulate IGF1, thereby promoting the chemoresistance of non-small cell lung cancer [Citation13]. Besides, circANKS1B might target the miR-515-5p/TGF-β1 axis to promote the cisplatin (DDP) resistance of oral squamous cell carcinoma [Citation14]. CircCELSR1 was reported to be overexpressed in paclitaxel (PTX)-resistant OC cells, and its knockdown significantly promoted the PTX sensitivity of OC cells via regulating miR-1252/FOXR2 network [Citation15]. CircRNA Cdr1as was found to restrain the chemoresistance of OC to DDP by targeting miR-1270 [Citation16].
In the previous study, Xu et al. discovered that circ_0004390 (derived from lysophosphatidic acid receptor 3 (LPAR3) gene, also called circ-LPAR3) was notably upregulated in OC tissues and was proved to be a potential target of OC treatment [Citation17]. However, whether circ-LPAR3 was involved in the regulation of OC chemoresistance has not been studied. In our study, we found that circ-LPAR3 expression was significantly upregulated in DDP-resistant patient’s and cells. Therefore, we speculated that circ-LPAR3 might be involved in the DDP resistance process of OC. The purpose of our study was to reveal the role of circ-LPAR3 in the DDP resistance of OC and its possible molecular mechanism. We hope to provide reliable potential molecular targets for the treatment of OC chemoresistance.
Materials and methods
Samples
In this study, 66 OC patients were recruited from Jinan City People’s Hospital. The OC tissues of patients after hysterectomy were collected and stored with −80°C. After surgery, 66 OC patients were divided into DDP-sensitive patients (n = 33) and DDP-resistant patients (n = 33) according to their sensitivity to DDP. All patients signed written informed consent. The clinicopathologic features of OC patients were shown in . Our study was approved by the Ethics Committee of Jinan City People’s Hospital.
Table 1. The clinicopathologic features of OC patients
Cell culture and transfection
Human OC cell lines (SKOV3 and A2780), DDP-resistant OC cell lines (SKOV3/DDP and A2780/DDP) and ovarian epithelial cells were obtained from Procell (Wuhan, China). OC cells were grown in DMEM medium (Hyclone, Logan, UT, USA) containing 10% FBS (Hyclone) and 1% penicillin/streptomycin (Hyclone). In addition, the medium used for DDP-resistant OC cells was added with 8 µM DDP (Meilunbio, Dalian, China). Ovarian epithelial cells were cultured in the complete medium of human ovarian epithelial cells (Procell).
SKOV3/DDP and A2780/DDP cells were seeded into 6-well plates and transfected with circ-LPAR3 siRNA or shRNA (si-circ-LPAR3 or sh-circ-LPAR3), miR-634 mimic or inhibitor (anti-miR-634), circ-LPAR3 or pyruvate dehydrogenase kinase 1 (PDK1) overexpression vector, and corresponding negative controls using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time PCR (qRT-PCR)
The RNA was extracted by RNAiso Plus reagent (Takara, Dalian, China) and cDNA was obtained by PrimeScript RT Reagent Kit (Takara). SYBR Green (Takara) was mixed with specific primers to perform qRT-PCR. Data were calculated using the 2−ΔΔCT method and normalized by β-actin or U6. All primers were presented in .
Table 2. The primer sequences used for qRT-PCR
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) assay
MTT Kit (Meilunbio) was used to measure cell DDP resistance and viability. For DDP resistance, OC cells were harvested after transfection or treatment with 20 mM 2-DG (Sigma-Aldrich, St. Louis, MO, USA), and then the cells were seeded in 96-well plates. The cells were treated with different concentrations of DDP for 24 h. Afterward, cells were incubated with MTT solution for 4 h. After incubated with formazan dissolved buffer, the absorbance was detected at 570 nm to assess the half-maximal inhibitory concentration (IC50) according to the previous report [Citation13]. For cell viability, cells were cultured for 48 h in 96-well plates. After treated with MTT solution and formazan dissolved buffer, the absorbance was measured to evaluate cell viability.
RNase R assay
In brief, extracted RNA was incubated with RNase R and then the circ-LPAR3 and Linear-LPAR3 expression levels were determined by qRT-PCR according to the previous report [Citation17].
Subcellular fractionation location assay
According to the kit instructions, the nuclear and cytoplasm RNAs of A2780/DDP cells were separated using Nuclear or Cytoplasmic RNA Purification Kit. The circ-LPAR3, U6 and GAPDH expression levels were examined by qRT-PCR, as previously described [Citation17].
Colony formation assay
SKOV3/DDP and A2780/DDP cells were plated into 6-well plates and cultured for 2 weeks. After stained with crystal violet, the colonies were counted by microscope, as previously described [Citation17].
Flow cytometry
OC cells were digested, and the cell suspensions were collected. After stained with Annexin V-FITC and PI (Keygen Biotech, Nanjing, China), the apoptosis rate was evaluated by a flow cytometer according to the previous report [Citation14].
Transwell assay
Transwell chambers (8 μm pore) were bought from Corning Inc. (Corning, New York, USA). As previously described [Citation14], The upper chamber was additionally coated with Matrigel dilution (Corning Inc.) to detect cell invasion. SKOV3/DDP and A2780/DDP cells with serum-free medium were seeded in the upper chamber. 24 h later, the cells on the lower chamber surface were counted by a microscope (100 ×).
Cell glycolysis measurement
Glucose consumption and lactate production in cell supernatants were measured by Glucose Assay Kit and Lactate Assay Kit (Sigma-Aldrich) according to the kit instructions.
Mice xenograft models
Animal experiments were performed according to the previous report [Citation18]. BALB/c nude mice (Vital Rival, Beijing, China) were randomly divided into 4 groups (n = 3). SKOV3/DDP cells (4 × 106) transfected with sh-circ-LPAR3 or sh-NC were subcutaneously injected into mice. After 7 days, tumor length and width were determined to measure tumor volume. Additionally, 5 mg/kg DDP or PBS was intraperitoneally injected into mice every 3 days. After 22 days, the tumors were removed for further assays. Animal assay was approved by the Animal Ethics Committee of Jinan City People’s Hospital.
Dual-luciferase reporter assay
According to the previous report [Citation19], The fragments of circ-LPAR3 or PDK1 3ʹUTR containing the wild-type or mutant-type sequences for miR-634 were cloned into the pmirGLO vector. The constructed vectors were co-transfected with miR-634 mimic/miR-NC into OC cells. Relative luciferase activity was measured using Dual-Luciferase Reporter Kit (Beyotime, Shanghai, China).
Western blot (WB) analysis
As previously described [Citation14], WB analysis was used to detect protein expression. RIPA Lysis Buffer (Beyotime) was used to extract protein. The proteins were resolved by SDS-PAGE gel and transferred to PVDF membrane. The membrane was incubated with anti-PDK1 (1:2,000, Sigma-Aldrich) or anti-β-actin (1:1,000, Sigma-Aldrich), and then incubated with goat anti-mouse IgG (1:20,000, Biovision). Using the ECL luminescent solution (Meilunbio), the protein signals were visualized.
Statistical analysis
Data were displayed as mean ± standard deviation. All statistical analysis was performed using GraphPad Prism 7.0 software. Student’s t-test or analysis of variance was used for evaluating the differences between groups. P < 0.05 was considered statistically significant.
Results
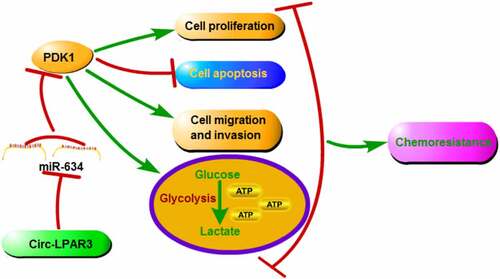
In this study, we investigated the role and potential molecular mechanism of circ-LPAR3 in DDP resistance of OC. Our results suggested that circ-LPAR3 knockdown could enhance DDP sensitivity, which was mainly achieved by regulating the miR-634/PDK1 axis. The proposed circ-LPAR3/miR-634/PDK1 axis revealed a new potential mechanism for regulating chemoresistance in OC, and provided a theoretical basis for improving the chemsensitivity of OC.
Circ-LPAR3 was upregulated in DDP-resistant OC tissues and cells
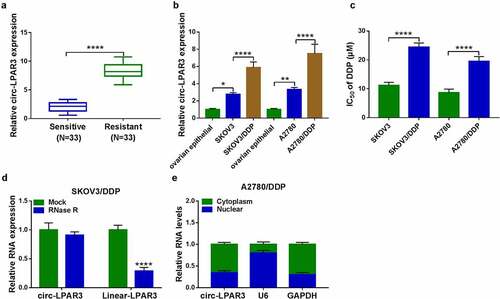
Firstly, we detected circ-LPAR3 expression in OC tissues. As presented in (), we found that circ-LPAR3 was notably overexpressed in DDP-resistant OC tissues compared with that in DDP-sensitive OC tissues. Also, circ-LPAR3 was more highly expressed in OC cells, and was higher in DDP-resistant OC cells than in normal OC cells (). By measuring the IC50 of OC cells, we confirmed the resistance of SKOV3/DDP and A2780/DDP cells to DDP (). In addition, circ-LPAR3 could well resist the digestion of RNase R (), and was mainly distributed in the cytoplasm of OC cells ().
Figure 1. The circ-LPAR3 expression in OC. (a) QRT-PCR was used to measure circ-LPAR3 expression in OC tissues. (b) The circ-LPAR3 expression in cells was detected by qRT-PCR. (c) The IC50 was determined by MTT assay. RNase R assay (d) and subcellular fractionation location assay (e) were used to confirm the circular characteristic of circ-LPAR3. *P < 0.05, **P < 0.01, ****P < 0.0001.
Circ-LPAR3 knockdown enhanced DDP sensitivity of OC cells
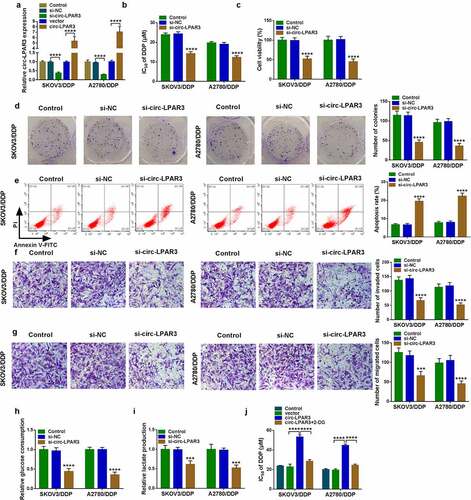
To assess the function of circ-LPAR3 on cell DDP resistance, we performed the functional assays. Firstly, we confirmed that circ-LPAR3 expression could be suppressed by si-circ-LPAR3 and enhanced by circ-LPAR3 overexpression vector (). Using the MTT assay, circ-LPAR3 knockdown reduced the IC50 of OC cells to DDP, and decrease cell viability (). Meanwhile, the number of colonies was suppressed by circ-LPAR3 silencing (). These indicated that downregulated circ-LPAR3 could suppress the proliferation of DDP-resistant OC cells. Furthermore, silenced circ-LPAR3 also enhanced cell apoptosis, while inhibited migration and invasion (). In addition, we measured cell glucose consumption and lactate production to evaluate cell glycolysis ability. As presented in , we discovered that circ-LPAR3 knockdown reduced cell glucose consumption and lactate production. In OC cells overexpressing circ-LPAR3, we uncovered that circ-LPAR3 overexpression markedly enhanced the IC50 of cells to DDP, while this effect was reversed by glycolysis inhibitor 2-DG (), confirming that cell glycolysis process was related to the DDP resistance of OC cells.
Figure 2. Circ-LPAR3 knockdown enhanced cell DDP sensitivity. SKOV3/DDP and A2780/DDP cells were transfected with si-NC, si-circ-LPAR3, vector or circ-LPAR3 as described. (a) QRT-PCR was used to detect circ-LPAR3 expression. (b) The IC50 was measured by MTT assay. MTT assay (c), colony formation assay (d), flow cytometry (e), and transwell assay (f-g) were performed to determine cell viability, colony number, apoptosis and metastasis. (h-i) Glucose consumption and lactate production were tested to assess cell glycolysis. (j) MTT assay was used to assess the IC50. ***P < 0.001, ****P < 0.0001.
Circ-LPAR3 knockdown improved the sensitivity of OC tumors to DDP
To determine the role of circ-LPAR3 in vivo, we constructed a stable SKOV3/DDP cell line that downregulated circ-LPAR3. By measuring circ-LPAR3 expression, we confirmed that circ-LPAR3 expression in the sh-circ-LPAR3 group was remarkably decreased (). Subsequently, we built the mice xenograft model of OC cells. Silencing of circ-LPAR3 could inhibit OC tumor volume and tumor weight (). Under DDP treatment, the tumor volume and tumor weight of the sh-circ-LPAR3 group were reduced, indicating that circ-LPAR3 knockdown might promote the sensitivity of the tumors to DDP (). Furthermore, we confirmed that circ-LPAR3 expression was decreased in the sh-circ-LPAR3 group with or without DDP treatment ().
Figure 3. Effects of circ-LPAR3 knockdown on OC tumor growth and DDP sensitivity. (a) The circ-LPAR3 expression in SKOV3/DDP cells was detected by qRT-PCR. Tumor volume (b) and tumor weight (c) were determined in mice. (d) QRT-PCR was used to measure circ-LPAR3 expression in the tumors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
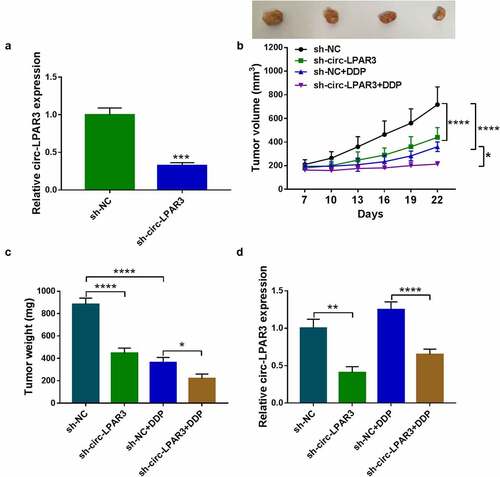
Circ-LPAR3 could negatively regulate miR-634
Many studies confirms that circRNA can be used as the endogenous competitive RNA (ceRNA) of miRNA [Citation20,Citation21]. To determine the molecular mechanism of circ-LPAR3, Circinteractome software was used. The results showed that circ-LPAR3 could be complementary to miR-634 (). MiR-634 mimic specifically inhibited the luciferase activity of WT-circ-LPAR3 vector, confirming the interaction between the two (). Moreover, we discovered that miR-634 was underexpressed in DDP-resistant OC tissues and cells, which was contrary to circ-LPAR3 expression (). Additionally, we found that the expression of circ-LPAR3 and miR-634 showed a significant negative correlation ().
Figure 4. Circ-LPAR3 sponged miR-634. (a) The predicted binding sites and mutant sequence between circ-LPAR3 and miR-634 were shown. (b-c) Dual-luciferase reporter assay was performed to verify the interaction between circ-LPAR3 and miR-634. (d) The miR-634 expression in OC tissues was tested by qRT-PCR. (e) The miR-634 expression in cells was determined by qRT-PCR. (f) The correlation between circ-LPAR3 and miR-634 in DDP-resistant OC tissues was analyzed by Pearson correlation analysis. *P < 0.05, ****P < 0.0001.
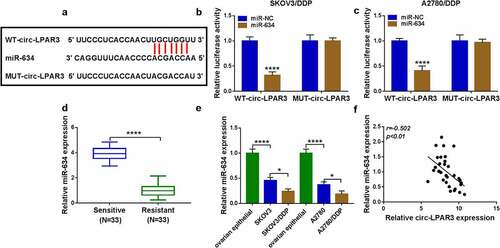
Circ-LPAR3 regulated the DDP sensitivity of OC cells by negatively regulating miR-634
For confirming that miR-634 participated in circ-LPAR3 regulated OC cell DDP sensitivity, si-circ-LPAR3 and anti-miR-634 were co-transfected into OC cells. Circ-LPAR3 knockdown could remarkably accelerate miR-634 expression, while anti-miR-634 could reverse this effect, confirming that both transfections were successful (). MiR-634 inhibitor reversed the inhibitory function of circ-LPAR3 downregulation on the IC50 of cells to DDP (). Also, we found that the inhibition effect of circ-LPAR3 silencing on cell viabilities, colony numbers, migrated cell numbers and invaded cell numbers, as well as the promotion effect on apoptosis in OC cells were reversed by miR-634 inhibitor (). Besides, miR-634 inhibitor revoked the suppressive of silenced circ-LPAR3 on OC cell glucose consumption and lactate production (). To further confirm the role of miR-634 on the DDP sensitivity of OC cells, we transfected with anti-miR-634 into OC cells. The decreased miR-634 expression confirmed the transfection efficiency of anti-miR-634 was good (). Using MTT assay, we discovered that miR-634 inhibitor could enhance the IC50 of OC cells to DDP, and this effect could be reversed by 2-DG (), indicating that miR-634 could positively regulate the DDP sensitivity of OC cells by regulating cell glycolysis ability.
Figure 5. Circ-LPAR3 silencing and miR-634 inhibitor regulated cell DDP sensitivity. (a-i) SKOV3/DDP and A2780/DDP cells were transfected with si-circ-LPAR3 and anti-miR-634. (a) Transfection efficiency was confirmed by detecting miR-634 expression using qRT-PCR. (b) MTT assay was used to detect the IC50. MTT assay (c), colony formation assay (d), flow cytometry (e), and transwell assay (f-g) were carried out to measure cell viability, colony number, apoptosis and metastasis. (h-i) Cell glycolysis was assessed by detecting glucose consumption and lactate production. (j) The miR-634 expression was tested by qRT-PCR to verify transfection efficiency of anti-miR-634. (k) The IC50 of DDP was evaluated by MTT assay. **P < 0.01, ***P < 0.001, ****P < 0.0001.
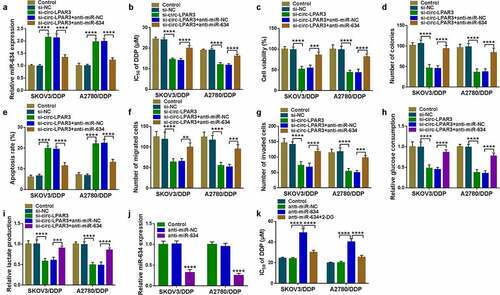
MiR-634 directly interacted with PDK1
Targetscan software was used to predict the targeted mRNAs of miR-634. PDK1 3ʹUTR was discovered to have binding sites with miR-634 (). Besides, miR-634 could suppress the luciferase activity of PDK1 3ʹUTR-WT vector (). PDK1 had an upregulated expression in DDP-resistant OC tissues (). Besides, the protein expression of PDK1 was markedly higher in OC cells, and was highly expressed in DDP-resistant OC cells (). Furthermore, we also found that there had a negative correlation between PDK1 and miR-634 expression in DDP-resistant OC tissues ().
Figure 6. MiR-634 targeted PDK1. (a) The predicted miR-634 binding sites with PDK1 3ʹUTR and the corresponding mutant sequence were presented. (b-c) The interaction between PDK1 and miR-634 was confirmed by dual-luciferase reporter assay. The mRNA and protein expression levels of PDK1 in OC tissues were determined by qRT-PCR (d) and WB analysis (e). (f) The protein expression of PDK1 in cells was assessed by WB analysis. (g) Pearson correlation analysis was used to analyze the correlation between PDK1 and miR-634 in DDP-resistant OC tissues. ***P < 0.001, ****P < 0.0001.
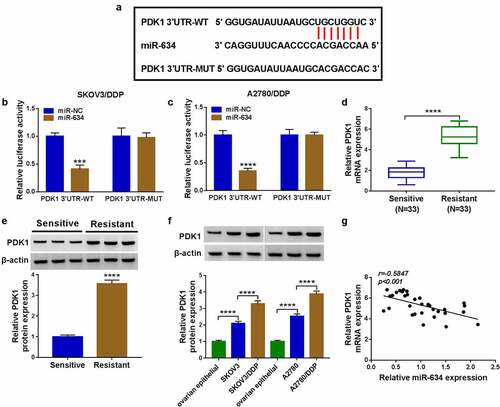
PDK1 reversed the regulation of miR-634 on cell DDP sensitivity
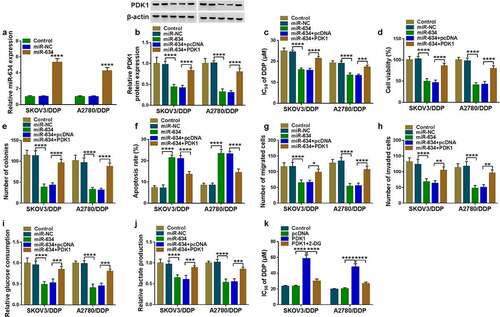
Then, miR-634 mimic was transfected into DDP-resistant OC cells. As shown in , the miR-634 expression was significantly increased, which confirmed the transfection efficiency of miR-634 mimic. To determine the role of PDK1 in the regulation of miR-634 on the DDP sensitivity of OC, miR-634 mimic and PDK1 overexpression vector were co-transfected into OC cells. PDK1 overexpression inverted the suppression effect of miR-634 on PDK1 expression, suggesting that the transfections of both were successful (). Then, the detection results of IC50 revealed that the inhibitory effect of miR-634 overexpression on the IC50 of cells to DDP could be reversed by PDK1 overexpression (). Furthermore, the decreasing function of miR-634 on cell viabilities and colony numbers also were revoked by PDK1 overexpression (). Further detection results revealed that PDK1 also inverted the promotion effect of miR-634 on the apoptosis and the suppressive effect on the migration, invasion, glucose consumption and lactate production in OC cells (). In addition, we also found that 2-DG reversed the promoting effect of PDK1 on the IC50 value, indicating that the promotion of PDK1 overexpression on DDP resistance of OC cells was realized through the glycolysis process ().
Figure 7. Effects of miR-634 and PDK1 overexpression on cell DDP sensitivity. (a) The expression of miR-634 was tested by qRT-PCR. (b-k) SKOV3/DDP and A2780/DDP cells were transfected with miR-634 mimic and PDK1 overexpression vector. (b) PDK1 protein expression was detected by WB analysis. (c) MTT assay was employed to measure IC50. Cell viability, colony number, apoptosis and metastasis were determined by MTT assay (d), colony formation assay (e), flow cytometry (f), and transwell assay (g-h). (i-j) The glucose consumption and lactate production of cells were examined to assess cell glycolysis. (k) The IC50 was determined using MTT assay. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
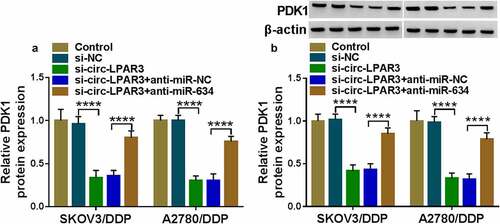
Circ-LPAR3 contributed to cell DDP resistance through elevating PDK1 expression by sponging miR-634
To confirm the regulatory effect of circ-LPAR3 on PDK1, we detected the expression of PDK1 in OC cells co-transfected with si-circ-LPAR3 and anti-miR-634. We found that circ-LPAR3 knockdown significantly inhibited PDK1 mRNA expression, and this inhibition could be effectively reversed by the miR-634 inhibitor (). In addition, at the protein level, we also obtained the results consistent with the mRNA level ().
Discussion
The occurrence of cancer chemoresistance is a complex process with multiple factors, and changes in the cell microenvironment and the expression of multiple genes may lead to chemoresistance [Citation22,Citation23]. Studies indicate that many circRNAs are differentially expressed in the chemoresistant and chemosensitive of tumor tissues, which may be related to cancer chemoresistance development [Citation24]. Our study explored the circ-LPAR3 role in OC DDP resistance. Through detecting the proliferation, apoptosis, metastasis and glycolysis process of DDP-resistant OC cells, we proposed that circ-LPAR3 might contribute to the progression of OC resistance. In addition, we found that the silencing of circ-LPAR3 not only reduced OC tumor growth, but also increased tumor sensitivity to DDP in vivo, suggesting that circ-LPAR3-targeted therapy might be an effective way to inhibit OC chemoresistance.
MiR-634 is underexpressed in many cancers and has been shown to be a tumor suppressor to participate in regulating cancer development, including pancreatic cancer [Citation25], hepatocellular carcinoma [Citation26], and gastric cancer [Citation27]. Peng et al. suggested that miR-634 improved the PTX sensitivity of nasopharyngeal carcinoma [Citation28]. MiR-634 was underexpressed in OC and could repress the proliferation and enhance OC cell DDP sensitivity [Citation29]. In this study, miR-634 was found to be a target for circ-LPAR3. The rescue experiments further illuminated that circ-LPAR3 regulated OC DDP resistance by sponging miR-634. Additionally, consistent with the previous research [Citation29], we also proposed that miR-634 could increase the DDP sensitivity of OC, suggesting that miR-634 also was a meaningful target for the treatment of OC chemoresistance.
Many studies have pointed out that the ability of cellular glycolysis is closely related to cancer malignant progression [Citation30]. As a regulator of pyruvate dehydrogenase, a key rate-limiting enzyme in glycolysis, PDK1 has been found to be overexpressed in many cancers, including OC [Citation31,Citation32]. MiR-4290 could suppress the glycolysis mediated by PDK1 to enhance the DDP sensitivity of gastric cancer [Citation33]. In OC, PDK1 was overexpressed in DDP-resistant OC cells and could accelerate the DDP resistance of OC [Citation34]. Here, we found that PDK1 was targeted by miR-634, and it also was involved in regulating miR-634-mediated OC DDP resistance. Furthermore, the positive regulation of circ-LPAR3 on PDK1 expression also indirectly confirmed the potential mechanism of circ-LPAR3 affecting the glycolysis process of DDP-resistant OC cells.
Conclusion
Overall, our study suggested that circ-LPAR3 promoted the DDP resistance of OC by miR-634/PDK1 pathway. Our findings provided a new biomarker for OC chemoresistance treatment and confirmed a new regulation axis for circ-LPAR3.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Momenimovahed Z, Tiznobaik A, Taheri S, et al. Ovarian cancer in the world: epidemiology and risk factors. Int J Womens Health. 2019;11:287–299.
- La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev. 2017;26(1):55–62.
- Luvero D Plotti F, Aloisia A, et al. Ovarian cancer relapse: from the latest scientific evidence to the best practice. Crit Rev Oncol Hematol. 2019;140:28–38.
- Moufarrij S Dandapani M, Arthofer E, et al. Epigenetic therapy for ovarian cancer: promise and progress. Clin Epigenetics. 2019;11(1):7.
- Glasgow MA Argenta P, Abrahante JE, et al. Biological insights into chemotherapy resistance in ovarian cancer. Int J Mol Sci. 2019;20(9):2131 .
- Ekmann-Gade AW Hogdall CK, Engelholm SA, et al. Neoadjuvant chemotherapy reduces the treatment-free interval after first-line treatment in patients with advanced ovarian cancer. Anticancer Res. 2020;40(5):2765–2770.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Zhang HD Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7.
- Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50.
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521.
- Jeyaraman S, Hanif EAM, Ab Mutalib NS, et al. Circular RNAs: potential regulators of treatment resistance in human cancers. Front Genet. 2019;10:1369.
- Cui C Yang J, Li X, et al. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19(1):58.
- Zheng S, Wang C, Yan H, et al. Blocking hsa_circ_0074027 suppressed non-small cell lung cancer chemoresistance via the miR-379-5p/IGF1 axis. Bioengineered. 2021;12(1):8347–8357.
- Yan J, Xu H. Regulation of transforming growth factor-beta1 by circANKS1B/miR-515-5p affects the metastatic potential and cisplatin resistance in oral squamous cell carcinoma. Bioengineered. 2021;12(2):12420–12430.
- Zhang S, Cheng J, Quan C, et al. circCELSR1 (hsa_circ_0063809) contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids. 2020;19:718–730.
- Zhao Z, Ji M, Wang Q, et al. Circular RNA Cdr1as Upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24–33.
- Xu F, Ni M, Li J, et al. Circ0004390 promotes cell proliferation through sponging miR-198 in ovarian cancer. Biochem Biophys Res Commun. 2020;526(1):14–20.
- Cao Y, Xie X, Li M, et al. CircHIPK2 contributes to DDP resistance and malignant behaviors of DDP-resistant ovarian cancer cells both in vitro and in vivo through circHIPK2/miR-338-3p/CHTOP ceRNA pathway. Onco Targets Ther. 2021;14:3151–3165.
- Zhang Y, Liu Y, Xu X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J Cell Biochem. 2018;119(7):6296–6308.
- Wu J, Liu S, Xiang Y, et al. Bioinformatic analysis of circular RNA-Associated ceRNA network associated with hepatocellular carcinoma. Biomed Res Int. 2019;2019:8308694.
- Song W, Wang W-J, Fu T, et al. Integrated analysis of circular RNA-associated ceRNA network in pancreatic ductal adenocarcinoma. Oncol Lett. 2020;19(3):2175–2184.
- Das M, Law S. Role of tumor microenvironment in cancer stem cell chemoresistance and recurrence. Int J Biochem Cell Biol. 2018;103:115–124.
- Cheng S, Huang Y, Lou C, et al. FSTL1 enhances chemoresistance and maintains stemness in breast cancer cells via integrin beta3/Wnt signaling under miR-137 regulation. Cancer Biol Ther. 2019;20(3):328–337.
- Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71.
- Chen D, Wu X, Zhao J, et al. MicroRNA-634 functions as a tumor suppressor in pancreatic cancer via directly targeting heat shock-related 70-kDa protein 2. Exp Ther Med. 2019;17(5):3949–3956.
- Zhang CZ, Cao Y, Fu J, et al. miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol. 2016;10(10):1532–1541.
- Guo J, Zhang C-D, An J-X, et al. Expression of miR-634 in gastric carcinoma and its effects on proliferation, migration, and invasion of gastric cancer cells. Cancer Med. 2018;7(3):776–787.
- Peng X, Cao P, He D, et al. MiR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel and inhibits cell growth both in vitro and in vivo. Int J Clin Exp Pathol. 2014;7(10):6784–6791.
- van Jaarsveld MT, van Kuijk PF, Boersma AWM, et al. miR-634 restores drug sensitivity in resistant ovarian cancer cells by targeting the Ras-MAPK pathway. Mol Cancer. 2015;14(1):196.
- Abbaszadeh Z, Cesmeli S, Biray Avci C. Crucial players in glycolysis: cancer progress. Gene. 2020;726:144158.
- Wang JJ Siu MK, Jiang YX, et al. Aberrant upregulation of PDK1 in ovarian cancer cells impairs CD8(+) T cell function and survival through elevation of PD-L1. Oncoimmunology. 2019;8(11):e1659092.
- Liu T, Yin H. PDK1 promotes tumor cell proliferation and migration by enhancing the warburg effect in non-small cell lung cancer. Oncol Rep. 2017;37(1):193–200.
- Qian Y Wu X, Wang H, et al. MicroRNA-4290 suppresses PDK1-mediated glycolysis to enhance the sensitivity of gastric cancer cell to cisplatin. Braz J Med Biol Res. 2020;53(5):e9330.
- Zhang MCong Q, Zhang XY, et al. Pyruvate dehydrogenase kinase 1 contributes to cisplatin resistance of ovarian cancer through EGFR activation. J Cell Physiol. 2019;234(5):6361–6370.