ABSTRACT
The critical roles of lncRNAs in drug resistance of malignancies have been widely recognized. This investigation aims to study the function of lncRNA PCAT6 in the resistance of non-small cell lung cancer (NSCLC) to gefitinib. In our study, we demonstrated that prostate cancer-associated transcript 6 (PCAT6) was upregulated in gefitinib-resistant NSCLC. PCAT6 knockdown inhibited gefitinib resistance of NSCLC, as indicated by decreased IC50 value, proliferation, and metastasis, and increased cell apoptosis. Besides, PCAT6 could directly target miR-326 in gefitinib-resistant NSCLC cells and augment NSCLC resistance to gefitinib by serving as ceRNA of miR-326. Furthermore, interferon-alpha receptor 2 (IFNAR2) was validated as a downstream target of miR-326 and miR-326 reduced resistance to gefitinib by inhibiting IFNAR2 expression. Our investigation identified that PCAT6 enhanced gefitinib resistance of NSCLC via miR-326/IFNAR2 axis, which might offer a new therapeutic strategy against gefitinib resistance of NSCLC patients.
Graphical Abstract
Introduction
Lung cancer is one of the most common malignant tumors and non-small cell lung cancer (NSCLC) accounts for approximately 85% of the total lung cancer cases [Citation1,Citation2]. Currently, various treatments, such as surgical resection, chemotherapy, radiotherapy, and targeted therapy have been employed in the management of NSCLC [Citation3,Citation4], but the efficacy of existing therapies is limited and the overall survival rate of NSCLC patients remains only 15% [Citation5]. Increased understanding of carcinogenesis of NSCLC has led to the application of gefitinib, a type of tyrosine kinase inhibitors (TKIs), for the treatment of NSCLC patients [Citation6]. Initial treatment can be effective, but cancer progression can reemerge after 6–15 months of therapy due to the increased gefitinib resistance [Citation6]. Hence, it is important to investigate the underlying mechanisms of gefitinib resistance in NSCLC.
Long non-coding RNA (lncRNA) is a kind of endogenous RNA that is more than 200 nucleotides in length without the protein-coding ability [Citation7]. During the past decade, numerous studies have elucidated the regulatory role of lncRNAs in malignancies, such as bladder, breast, and colorectal cancer [Citation8–10]. Particularly, some lncRNAs were considered to regulate the chemoresistance of NSCLC. For example, silencing of BLACAT1 reduced the afatinib resistance of NSCLC by modulating STAT3 signaling [Citation11]. LncRNA SNHG14 increased NSCLC resistance to cisplatin by sponging miR-133a [Citation12]. LncRNA UCA1 knockout suppressed gefitinib resistance in NSCLC via STAT3 signaling [Citation13]. LncRNA PCAT6, located at 1q32.1, was identified as an oncogene in various cancers, such as breast cancer, bladder cancer, and colon cancer [Citation14–16]. In addition, lncRNA PCAT6 was aberrantly expressed in NSCLC and associated with the NSCLC progression [Citation17]. More importantly, PCAT6 promoted chemoresistance of cervical cancer by absorbing miR-543 to upregulate ZEB1 [Citation18]. PCAT6 suppressed miR-204 to enhance colorectal cancer cell resistance to 5-fluorouracil-based treatment via the HMGA2 pathway [Citation19]. Therefore, a hypothesis can be made that PCAT6 might participate in the gefitinib resistance of NSCLC.
We aimed to explore the precise role and mechanism of PCAT6 in the gefitinib resistance of NSCLC, and we hypothesized that PCAT6 enhanced gefitinib resistance of NSCLC via miR-326/IFNAR2 axis. Our findings might offer novel therapeutic strategies for NSCLC patients to overcome gefitinib resistance.
Patients and methods
Clinical specimens
A total of 78 NSCLC tissues (34 gefitinib resistance and 44 gefitinib sensitivity) were obtained from NSCLC patients harboring EGFR exon 19 deletion (19DEL) or L858R. Sample tissues were stored in liquid nitrogen at −80°C immediately after surgical resection for future use. This research was approved by the Ethics Committee of the First Affiliated Hospital of Jinzhou Medical University and all patients signed written informed consent.
Cell culture
The NSCLC cells (HCC827 and PC9) were purchased from ATCC (USA). Stable gefitinib-resistant cells were generated by continuous treatment with gradually increasing concentrations of gefitinib as previously described [Citation20]. The cells were maintained in DMEM containing 10% FBS (All from Gibco) at 37°C in 5% CO2. Gefitinib (1 µM) was used to maintain the drug resistance of HCC827/GR and PC9/GR cells.
Cell transfection
shRNA targeting PCAT6 (shPCAT6; 5’-UGCAUGCAGCUGCUAGAUACG-3’) with its negative control (shNC; 5’-AAUUCUCCGAACGUGUCACGU-3’), miR-326 mimics (5’-GCAGGGCACGACUGAUCUUGG-3’) and miR-326 inhibitor (5’-UCGCUCGGUCCYGAUCGGGAG-3’) with their controls (NC mimics: 5’-GGAAGUCAUCCAAUGUGCAUU-3’ and NC inhibitor: 5’-UCCCUGGUUGCAGAUCGCGAA-3’), and PCAT6 overexpression plasmid (pcDNA3.1/PCAT6) and IFNAR2 overexpression plasmid (pcDNA3.1/IFNAR2) with their negative control (pcDNA3.1) were obtained from GenePharma (Shanghai). Transfection of aforementioned vectors was conducted using lipofectamine 2000 (Invitrogen).
RT-qPCR
Total RNA was isolated from tissues and cells using TRIzol (Invitrogen). Reverse transcription of total RNA to cDNA was performed using reverse transcription kit (TaKaRa). Thereafter, qPCR was implemented using the SYBR Select Master mix on the ABI7300 system (Applied Biosystems). The gene expressions were calculated with the 2−ΔΔCq method and normalized to U6 or GAPDH. The primer sequences were as follows: PCAT6, forward, 5’-CCCCTCCTTACTCTTGGACAAC-3ʹand reverse, 5’-GACCGAATGAGGATGGAGACAC-3’; IFNAR1, forward, 5’-TTGCGTATAAGAGCAGAAACAGGAAACA-3’ and reverse, 5’-CGAAACTTGAACTATCCATAGCCCACAT-3’; miR-326, forward 5’-ACTGTCCTTCCCTCTGGGC-3’ and reverse 5’-AATGGTTGTTCTCCACTCTCTCTC-3’; GAPDH, forward: 5’-GCACCACCAACTGCTTAGCA-3’ reverse and 5’-GTCTTCTGGGTGGCAGTGATG-3’; U6, forward: 5’-CTCGCTTCGGCAGCACA-3’ and reverse 5’-AACGCTTCACGAATTTGCGT-3’.
CCK-8
The gefitinib-resistant NSCLC cells were plated at a concentration of 1 × 104 cells/well into 96-well plates and cultured for 0, 24,48, and 72 h at 37°C and 5% CO2. Next, 10 µl CCK-8 reagent (Beyotime) was added, followed by an extra 4 h incubation. Absorbance at 450 nm was observed using a microplate reader (Thermo Fisher Scientific, Inc.). To calculate the half-maximal inhibitory concentration (IC50) values, cells were exposed to different concentrations of gefitinib for 24 h [Citation21].
Wound healing assay
The transfected cells were incubated in DMEM without serum and cultured until 70% confluence. Then, a pipette tip was employed to create a single scratch in the cell monolayer. The migratory distance was measured at 0 h and 24 h by a microscope [Citation22].
Transwell assay
Cell invasive ability was determined using Transwell chambers (EMD Millipore) coated with Matrigel (Corning Inc.). Cells were plated in the upper chambers in serum-free culture medium. The lower chambers were filled with a total of 600 µl DMEM with 10% FBS. After 24 h incubation, the total invasive cells on the surface of the lower chamber were fixed and stained and subsequently counted under a light microscope [Citation23].
TUNEL
The cells were fixed with paraformaldehyde solution, permeabilized with 0.25% Triton-X 100, and then incubated in TUNEL reaction mixture. Subsequently, the TUNEL-stained cells were counterstained with DAPI (2 µg/ml). The TUNEL-positive cells were recorded by a fluorescence microscope (Olympus Corporation) [Citation22].
Luciferase assay
The wild-type (WT) and mutant (MUT) of PCAT6 and IFNAR2 were sub-cloned in the pmirGLO vector (Promega). Subsequently, the above vectors were co-transfected with NC mimics/inhibitor or miR-326 mimics/inhibitor into gefitinib-resistant NSCLC cells using lipofectamine 2000 (Invitrogen). After 48 h, the luciferase activities were evaluated by a Dual-Luciferase Reporter System (Promega) [Citation24].
Immunoprecipitation (RIP) assay
RIP assay was conducted by Magna RNA-binding protein RIP kit (EMD Millipore) according to the manufacturer’s protocol [Citation25]. Cells were lysed with RIP lysis buffer and incubated with magnetic beads conjugated with Ago2 or IgG. Then, the RNAs were extracted and measured by RT-qPCR.
Western blot
The proteins were extracted using radioimmunoprecipitation lysis buffer (Beyotime). Then, protein samples were resolved by SDS-PAGE and transferred to PVDF membranes. Afterward, the membranes were incubated with primary antibodies against IFNAR2 or GAPDH at 4°C. After incubation with secondary antibodies, the protein expression was detected using an ECL reagent (GE Healthcare) [Citation26].
Statistical analysis
The data were analyzed using SPSS version 17.0 software and presented as mean ± standard deviation (SD). Student’s t-test or one-way ANOVA was used to analyze the differences between the groups. P < 0.05 was considered statistically significant.
Results
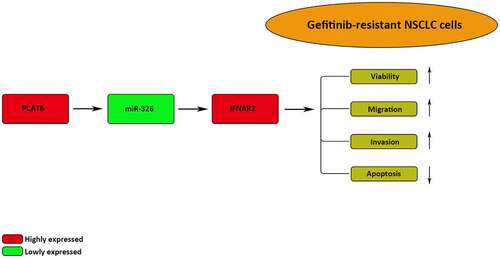
This study aimed to investigate the effect of PCAT6 on the gefitinib resistance of NSCLC and the molecular mechanism involved. Through bioinformatics analysis and functional experiments, it was demonstrated that PCAT6 exhibited an inhibitory effect on the gefitinib sensitivity of NSCLC cells through sponging miR-326 to elevate IFNAR2 expression.
PCAT6 expression is increased in gefitinib-resistance NSCLC
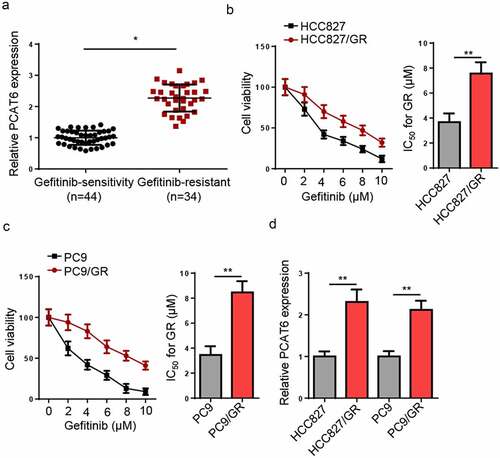
Firstly, RT-qPCR revealed that PCAT6 levels were increased in gefitinib-resistant NSCLC tissues (). Subsequently, gefitinib-resistant NSCLC cells were established, as indicated by increased IC50 values in gefitinib-resistant HCC827 and PC9 cells ( and c), and PCAT6 expression was increased in gefitinib-resistant NSCLC cells compared to corresponding parental cell lines ().
Figure 1. PCAT6 is upregulated in gefitinib-resistance NSCLC. (a) RT-qPCR measured the PCAT6 levels in nonresistant and gefitinib-resistant NSCLC tissues. (b and c) CCK-8 evaluated the IC50 values of HCC827, HCC827/GR, PC9, and PC9/GR cells. (d) RT-qPCR detected the relative expression of PCAT6 in gefitinib-resistant NSCLC cells and the parental cells. **p < 0.01; *p < 0.05.
PCAT6 knockdown reduces gefitinib resistance of NSCLC
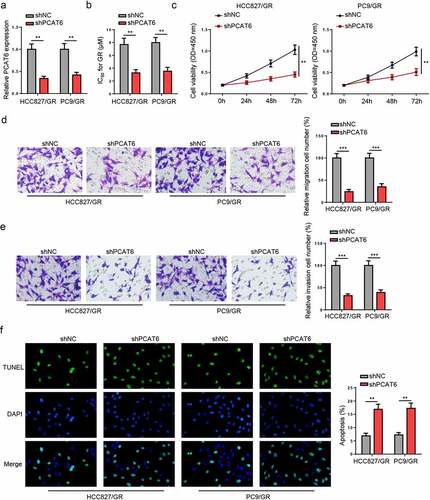
Subsequently, loss-of-function experiments were performed to determine the impact of PCAT6 on gefitinib-resistant NSCLC cellular activities. As presented in , PCAT6 expression was reduced in gefitinib-resistant HCC827 and PC9 cells transfected with shPCAT6. PCAT6 silencing reduced the IC50 value and viability of gefitinib-resistant NSCLC cells ( and c). Moreover, downregulation of PCAT6 suppressed the metastasis ( and d) but promoted apoptosis in gefitinib-resistant NSCLC cells (). These data revealed that PCAT6 silencing decreased the resistance of gefitinib-resistant NSCLC cells to gefitinib.
Figure 2. PCAT6 knockdown reduces gefitinib resistance of NSCLC. (a) PCAT6 levels in gefitinib-resistant HCC827 and PC9 cells in shNC and shPCAT6 groups. (b and c) Changes in IC50 value and cell viability in treated cells after silencing PCAT6. (d and e) Cell migratory and invasive abilities of cells following PCAT6 knockdown. (f) The impact of PCAT6 silence on the apoptosis of gefitinib-resistant cells. ***p < 0.001; **p < 0.01.
PCAT6 targets miR-326 in gefitinib-resistant NSCLC
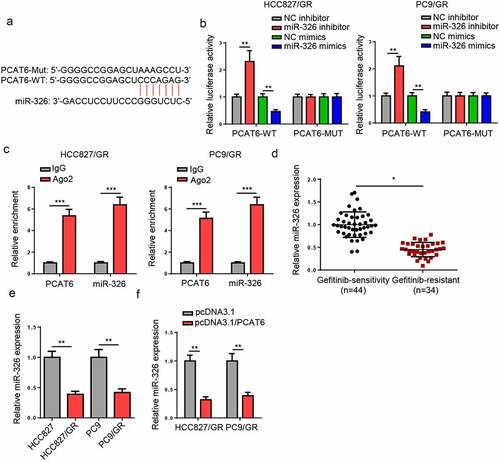
To study the mechanism of PCAT6 in gefitinib-resistant NSCLC, the downstream genes were screened. miR-326 has been reported to act as a tumor suppressor in NSCLC [Citation27–29]. Through using starBase database, PCAT6 was found to contain the binding sequences of miR-326 (). Luciferase activity of PCAT6-WT was increased by the downregulation of miR-326 but reduced by miR-326 overexpression, while no change was observed in PCAT6 mutant group (). Furthermore, RIP assay indicated that miR-326 and PCAT6 were co-immunoprecipitated by anti-AGO2 but not anti-IgG (). In addition, miR-326 was downregulated in gefitinib-resistant NSCLC tissues and cells ( and e). Besides, overexpressing PCAT6 reduced miR-326 levels in gefitinib-resistant NSCLC cells (). Taken together, PCAT6 could suppress miR-326 expression by direct interaction.
Figure 3. PCAT6 knockdown reduces gefitinib resistance of NSCLC. (a) Starbase showed the binding sites between PCAT6 and miR-326. (b) Luciferase reporter analysis for the luciferase activities of PCAT6-WT and PCAT6-MUT in gefitinib-resistant NSCLC cells. (c) RIP assay was used to detect the enrichment of PCAT6 and miR-326 in AGO2 and IgG groups. (d and e) RT-qPCR results of miR-326 expression in NSCLC tissues and cells. (f) Evaluation of miR-326 expression change by RT-qPCR following PCAT6 overexpression. ***p < 0.001; **p < 0.01; *p < 0.05.
PCAT6 augments NSCLC resistance to gefitinib by serving as ceRNA of miR-326
To explore whether PCAT6 regulated NSCLC resistance to gefitinib via miR-326, shPCAT6, or shPCAT6 + miR-326 inhibitor was transfected into gefitinib-resistant HCC827 and PC9 cells. miR-326 expression was enhanced in gefitinib-resistant NSCLC cells transfected with shPCAT6, but was partially decreased by miR-326 inhibitor (). CCK-8 revealed that PCAT6 silencing reduced the IC50 values and cell viability, while these effects were reversed by miR-326 inhibition (Fig, 4B and C). Besides, PCAT6 silence alleviated the metastasis of gefitinib-resistant NSCLC cells, while miR-326 inhibition abrogated the alleviation ( and e). On the contrary, the promotive effect of gefitinib-resistant NSCLC cell apoptosis caused by PCAT6 knockdown was annulled by inhibiting miR-326 (). To summarize, PCAT6 sponged miR-326 to strengthen NSCLC resistance to gefitinib.
Figure 4. PCAT6 augments NSCLC resistance to gefitinib by acting as ceRNA of miR-326. (a) Relative level of miR-326 in gefitinib-resistant cells transfected with shNC, shPCAT6, and shPCAT6+ miR-326 inhibitor. (b and c) CCK-8 showed the cell viability and the IC50 of gefitinib of different groups in vitro. (d and e) The migratory and invasive abilities in gefitinib-resistant NSCLC cells were determined by Transwell assay. (f) The apoptosis of HCC827/GR and PC9/GR cells were detected by TUNEL assay. **p < 0.01; *p < 0.05.
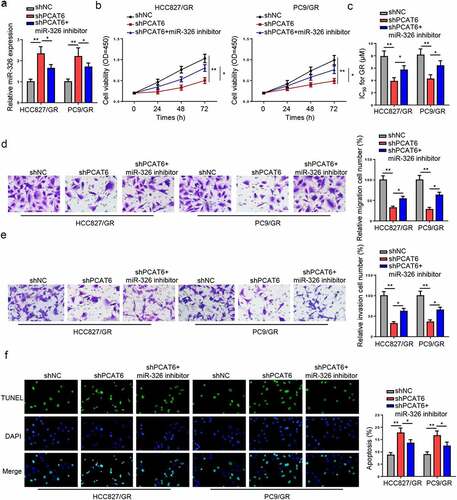
Figure 5. IFNAR2 is a downstream target of miR-326. (a) The complementary sequences between PCAT6 and miR-326. (b) Luciferase activities of IFNAR2-WT and IFNAR2-MUT in gefitinib-resistant HCC827 and PC9 cells. (c and d) RT-qPCR analysis for the IFNAR2 mRNA level in NSCLC tissues and cells. (e) Protein levels of IFNAR2 in NSCLC cells. (f and g) The levels of IFNAR2 in HCC827/GR and PC9/GR cells transfected with pcDNA3.1/PCAT6 or pcDNA3.1/PCAT6+ miR-326 mimics. **p < 0.01; *p < 0.05.
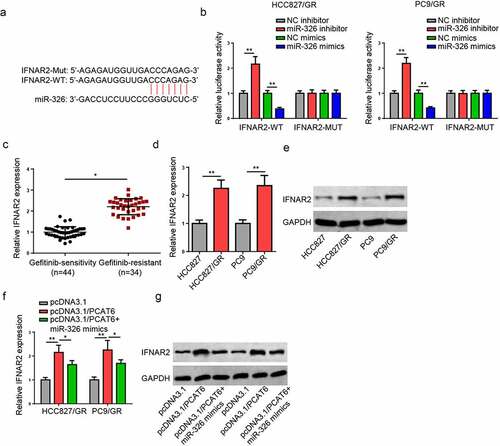
IFNAR2 is a downstream target of miR-326
The PCAT6/miR-326 axis was further investigated by exploring its downstream mechanism. The upregulated IFNAR2 expression was observed in lung cancer, and high IFNAR2 expression contributed to shorter progression-free survival (PFS) and overall survival (OS) [Citation30]. Through StarBase database, IFNAR2 was screened as a putative gene of miR-326 (). Moreover, miR-326 mimics significantly repressed and miR-326 inhibitor increased the luciferase activity of WT of IFNAR2, while no significant difference in MUT IFNAR2 group (). Furthermore, IFNAR2 levels were substantially upregulated in gefitinib-resistant NSCLC tissues and cells (). In addition, the upregulation of PCAT6 increased the IFNAR2 level while miR-326 overexpression mitigated this effect ( and g). The aforementioned results implicated that PCAT6 could regulate IFNAR2 expression by targeting miR-326.
miR-326 reduces NSCLC cell resistance to gefitinib by inhibiting IFNAR2
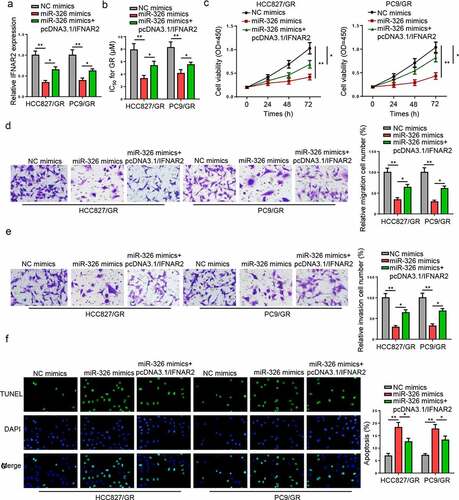
To further evaluate whether IFNAR2 was a downstream modulator of the PCAT6/miR-326 axis in regulating the gefitinib resistance of NSCLC, rescue assays were conducted. RT-qPCR indicated that IFNAR2 overexpression restored the inhibitory effect of miR-326 mimics on IFNAR2 expression (). Functional assays uncovered that miR-326 overexpression suppressed the IC50 values, cell viability, and metastasis but promoted apoptosis in gefitinib-resistant NSCLC cells, while overexpression of IFNAR2 partially reversed these effects (). In sum, miR-326 could increase the gefitinib sensitivity of NSCLC by suppressing IFNAR2.
Figure 6. miR-326 reduces NSCLC cell resistance to gefitinib by inhibiting IFNAR2. (a) The relative expression of IFNAR2 was measured in gefitinib-resistant NSCLC cells transfected with NC mimics, miR-326 mimics, and miR-326 mimics+ pcDNA3.1/PCAT6. (b-f) CCK-8, transwell, and TUNEL assays indicated the cell proliferation, IC50 values, migration, invasion, and apoptosis of gefitinib-resistant NSCLC cells in different groups. **p < 0.01; *p < 0.05.
Discussion
With the development of medical technology, multiple-chemotherapy drugs, such as platinum/pemetrexed, gefitinib, paclitaxel, and cisplatin, have been widely applied to treat human cancers [Citation31–34]. Gefitinib has been used as the first-line therapy for NSCLC with EGFR mutation [Citation35]. Nevertheless, gefitinib resistance is a major obstacle for NSCLC therapy [Citation36]. Accumulating researches have indicated that dysregulated lncRNAs were related to the chemoresistance in NSCLC. Chen et al. disclosed that lncRNA HOST2 suppressed miR-621 to enhance gefitinib resistance of NSCLC [Citation37]. Xu et al. demonstrated that lncRNA UCA1 caused acquired gefitinib resistance by inhibiting CDKN1A in NSCLC [Citation38]. Li et al. indicated that RHPN1-AS1 inhibited gefitinib resistance of NSCLC via miR-299-3p/TNFSF12 axis [Citation39]. PCAT6 has been reported to be dysregulated and function as an oncogenic gene in multiple cancers. For instance, PCAT6 promoted the occurrence and development of triple-negative breast cancer by modulating VEGFR2 [Citation15]. PCAT6 facilitated the tumorigenesis of ovarian cancer through suppressing PTEN [Citation40]. PCAT6 accelerated the viability and inhibited apoptosis in colon cancer cells by modulating ARC level [Citation14]. In this research, it was disclosed that PCAT6 expression was highly expressed in gefitinib-resistant NSCLC, and silencing PCAT6 could reduce NSCLC cell resistance to gefitinib.
Increasing evidence identified that the lncRNA-mediated ceRNA network was involved in the regulation of chemoresistance of various cancers, including NSCLC. For example, UCA1 enhanced the gefitinib resistance of NSCLC by sponging miR-143 to upregulate FOSL2 [Citation41]. LncRNA SNHG1 contributed to cisplatin resistance as a ceRNA to target DCLK1 by sponging miR-330-5p in NSCLC [Citation42]. In addition, several miRNAs, such as miR-34c-3p [Citation43], miR-490-3p [Citation44], miR-449c [Citation45], have been identified as tumor suppressors in NSCLC. In this work, we confirmed that miR-326 was a downstream gene of PCAT6. miR-326 was demonstrated to be related to the chemosensitivity of various tumors. For example, miR-326 downregulation restored the suppressive impact of lncRNA DDX11-AS1 silencing on oxaliplatin resistance of gastric cancer [Citation46]. The promotive effect of circRNA_102272 upregulation on cisplatin resistance of hepatocellular carcinoma was abrogated by miR-326 overexpression [Citation47]. Herein, we demonstrated miR-326 silencing restored the suppressive effect of PCAT6 knockdown on the gefitinib resistance of NSCLC, suggesting that PCAT6 enhanced gefitinib resistance of NSCLC by regulating miR-326.
Interferon-alpha receptor 2 (IFNAR2) is a type of cell surface receptor that mediates the type I interferons (IFNs) to exert various biological functions [Citation30]. The abnormal expression of IFNAR2 was studied in different types of malignancies, including pancreatic cancer, gastrointestinal cancer, chronic myelogenous leukemia, and renal cell carcinoma [Citation48–51]. Importantly, the upregulation of IFNAR2 has been discovered in various histological types of lung cancer [Citation30]. Herein, IFNAR2 was validated as a target gene of miR-326, and PCAT6 upregulated IFNAR2 expression by targeting miR-326. Functional experiments indicated the upregulation of miR-326 enhanced the gefitinib sensitivity of NSCLC cells, while this effect was abolished by IFANR2.
Conclusion
Our study found that PCAT6 enhanced the gefitinib resistance of NSCLC by sponging miR-326 to upregulate IFNAR2. These findings suggested that PCAT6 could be a new therapeutic target to activate NSCLC sensitivity toward gefitinib treatment.
Research highlights
1. PCAT6 expression was elevated in gefitinib-resistant NSCLC
2. PCAT6 enhanced the gefitinib resistance in NSCLC cells
3. PCAT6 upregulated IFNAR2 expression by sponging miR-326
4. PCAT6 increased gefitinib resistance in NSCLC via miR-326/IFNAR2 axis
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- He X, Zhang C, Shi C, et al. Meta-analysis of mRNA expression profiles to identify differentially expressed genes in lung adenocarcinoma tissue from smokers and non-smokers. Oncol Rep. 2018;39:929–938.
- Chen Y, Mao B, Peng X, et al. A comparative study of genetic profiles of key oncogenesis-related genes between primary lesions and matched lymph nodes metastasis in lung cancer. J Cancer. 2019;10:1642–1650.
- Schneider BJ, Ismaila N, Aerts J, et al. Lung cancer surveillance after definitive curative-intent therapy: ASCO guideline. J Clin Oncol. 2020;38:753–766.
- Yuan M, Huang LL, Chen JH, et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. 2019;4:61.
- Ramnath N, Dilling TJ, Harris LJ, et al. Treatment of stage III non-small cell lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e314S–e340S.
- Lei Y, Guo W, Chen B, et al. Tumor‑released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non‑small cell lung cancer. Oncol Rep. 2018;40:3438–3446.
- Jin X, Liu X, Zhang Z, et al. lncRNA CCAT1 acts as a MicroRNA-218 sponge to increase gefitinib resistance in NSCLC by targeting HOXA1. Mol Ther Nucleic Acids. 2020;19:1266–1275.
- Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. Journal of Cellular Physiology. 2019;234:9105–9117.
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond). 2019;39:22.
- Wang L, Cho KB, Li Y, et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer, 20. Int J Mol Sci. 2019.
- Shu D, Xu Y, Chen W. Knockdown of lncRNA BLACAT1 reverses the resistance of afatinib to non-small cell lung cancer via modulating STAT3 signalling. J Drug Target. 2020;28:300–306.
- Xu L, Xu Y, Yang M, et al. LncRNA SNHG14 regulates the DDP-resistance of non-small cell lung cancer cell through miR-133a/HOXB13 pathway. BMC Pulm Med. 2020;20:266.
- Zhang B, Wang H, Wang Q, et al. Knockout of lncRNA UCA1 inhibits drug resistance to gefitinib via targeting STAT3 signaling in NSCLC. Minerva Med. 2019;110:273–275.
- Huang W, Su G, Huang X, et al. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle. 2019;18:69–83.
- Dong F, Ruan S, Wang J, et al. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell Death Dis. 2020;11:728.
- Zhang D, Du D, Yi S, et al. LncRNA PCAT6: a potential biomarker for diagnosis and prognosis of bladder cancer. Ann Diagn Pathol. 2020;49:151642.
- Cui LH, Xu HR, Yang W, et al. lncRNA PCAT6 promotes non-small cell lung cancer cell proliferation, migration and invasion through regulating miR-330-5p. Onco Targets Ther. 2018;11:7715–7724.
- Ma Z, Gu G, Pan W, et al. LncRNA PCAT6 accelerates the progression and chemoresistance of cervical cancer through up-regulating ZEB1 by sponging miR-543. Onco Targets Ther. 2020;13:1159–1170.
- Wu H, Zou Q, He H, et al. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8:2484–2495.
- Cui Z, Liu Z, Zeng J, et al. TRIM59 promotes gefitinib resistance in EGFR mutant lung adenocarcinoma cells. Life Sci. 2019;224:23–32.
- Hong Y, Liang H, Uzair Ur R, et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421.
- Liu B, Wu S, Ma J, et al. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482.
- Yang B, Diao H, Wang P, et al. microRNA-877-5p exerts tumor-suppressive functions in prostate cancer through repressing transcription of forkhead box M1. Bioengineered. 2021;12(1):9094–9102.
- Zhang J, Cheng F, Rong G, et al. Circular RNA hsa_circ_0005567 overexpression promotes M2 type macrophage polarization through miR-492/SOCS2 axis to inhibit osteoarthritis progression. Bioengineered. 2021; 12:8920–8930.
- Li Z, Yu Z, Meng X, et al. LncRNA LINC00968 accelerates the proliferation and fibrosis of diabetic nephropathy by epigenetically repressing p21 via recruiting EZH2. Biochem Biophys Res Commun. 2018;504:499–504.
- Cai L, Ye L, Hu X, et al. MicroRNA miR-330-3p suppresses the progression of ovarian cancer by targeting RIPK4. Bioengineered. 2021;12:440–449.
- Sun C, Huang C, Li S, et al. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7:8341–8359.
- Wu Y, Cheng K, Liang W, et al. lncRNA RPPH1 promotes non-small cell lung cancer progression through the miR-326/WNT2B axis. Oncol Lett. 2020;20:105.
- Tan Q, Liu C, Shen Y, et al. Circular RNA circ_0000517 facilitates the growth and metastasis of non-small cell lung cancer by sponging miR-326/miR-330-5p. Cell J. 2021;23:552–561.
- Tanaka S, Hattori N, Ishikawa N, et al. Interferon (alpha, beta and omega) receptor 2 is a prognostic biomarker for lung cancer. Pathobiology. 2012;79:24–33.
- Lin JJ, Schoenfeld AJ, Zhu VW, et al. Efficacy of platinum/pemetrexed combination chemotherapy in ALK-positive NSCLC refractory to second-generation ALK inhibitors. J Thorac Oncol. 2020;15:258–265.
- Hida T, Ogawa S, Park JC, et al. Gefitinib for the treatment of non-small-cell lung cancer. Expert Rev Anticancer Ther. 2009;9:17–35.
- Weaver BA. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25:2677–2681.
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378.
- Liu X, Lu X, Zhen F, et al. LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol Ther Nucleic Acids. 2019;16:155–161.
- Wang L, Dong X, Ren Y, et al. Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC by epigenetically regulating the PTEN/AKT signaling pathway. Cell Death Dis. 2018;9:129.
- Chen ZY, Liu HY, Jiang N, et al. LncRNA HOST2 enhances gefitinib-resistance in non-small cell lung cancer by down-regulating miRNA-621. Eur Rev Med Pharmacol Sci. 2019;23:9939–9946.
- Xu T, Yan S, Wang M, et al. LncRNA UCA1 induces acquired resistance to gefitinib by epigenetically silencing CDKN1A expression in non-small-cell lung cancer. Front Oncol. 2020;10:656.
- Li X, Zhang X, Yang C, et al. The lncRNA RHPN1-AS1 downregulation promotes gefitinib resistance by targeting miR-299-3p/TNFSF12 pathway in NSCLC. Cell Cycle. 2018;17:1772–1783.
- Kong FR, Lv YH, Yao HM, et al. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2019;23:8230–8238.
- Chen X, Wang Z, Tong F, et al. lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Mol Ther Nucleic Acids. 2020;19:643–653.
- Ge P, Cao L, Zheng M, et al. LncRNA SNHG1 contributes to the cisplatin resistance and progression of NSCLC via miR-330-5p/DCLK1 axis. Exp Mol Pathol. 2021;120:104633.
- Huang W, Yan Y, Liu Y, et al. Exosomes with low miR-34c-3p expression promote invasion and migration of non-small cell lung cancer by upregulating integrin α2β1. Signal Transduct Target Ther. 2020;5:39.
- Zhang ZY, Gao XH, Ma MY, et al. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep. 2020;10:9024.
- Miao LJ, Huang SF, Sun ZT, et al. MiR-449c targets c-Myc and inhibits NSCLC cell progression. FEBS Lett. 2013;587:1359–1365.
- Song W, Qian Y, Zhang MH, et al. The long non-coding RNA DDX11-AS1 facilitates cell progression and oxaliplatin resistance via regulating miR-326/IRS1 axis in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24:3049–3061.
- Guan Y, Zhang Y, Hao L, et al. CircRNA_102272 promotes cisplatin-resistance in hepatocellular carcinoma by decreasing MiR-326 targeting of RUNX2. Cancer Manag Res. 2020;12:12527–12534.
- Booy S, Hofland LJ, Waaijers AM, et al. Type I interferon receptor expression in human pancreatic and periampullary cancer tissue. Pancreas. 2015;44:99–105.
- Ota H, Nagano H, Doki Y, et al. Expression of type I interferon receptor as a predictor of clinical response to interferon-alpha therapy of gastrointestinal cancers. Oncol Rep. 2006;16:249–255.
- Ito K, Tanaka H, Ito T, et al. Initial expression of interferon alpha receptor 2 (IFNAR2) on CD34-positive cells and its down-regulation correlate with clinical response to interferon therapy in chronic myelogenous leukemia. Eur J Haematol. 2004;73:191–205.
- Ariyasu T, Fujioka N, Yamamoto S, et al. Correlation between interferon alpha receptor protein expression and sensitivity to interferon alpha subtypes in human renal carcinoma cell lines. Cancer Genomics Proteomics. 2004;1:87–94.
