ABSTRACT
This study aimed to investigate the roles of microRNA-886 (miR-886) and long non-coding RNA (lncRNA) OXCT1-AS1 in osteosarcoma (OS). We predicted that they might interact with each other. The expression of OXCT1-AS1 and miR-886 (mature and premature) in osteosarcoma and paired non-tumor tissues from 66 OS patients was negatively correlated. Overexpression and silencing assays showed that OXCT1-AS1 suppresses miR-886 maturation. RNA–RNA pulldown and subcellular fractionation assays demonstrated the direct interaction between OXCT1-AS1 and miR-886. BrdU proliferation assays revealed that OXCT1-AS1 promoted OS cell proliferation, and miR-886 reduced the enhancing effects of OXCT1-AS1 on OS cell proliferation. Western blot showed that OXCT1-AS1 had no effects on the levels of epithelial–mesenchymal transition biomarkers. Overall, OXCT1-AS1 suppresses miR-886 maturation to promote OS cell proliferation.
Graphical Abstract
Introduction
As the most common malignant tumor detected in bone tissues, osteosarcoma (OS) is a type of tumor formed by bone-forming cells [Citation1,Citation2]. OS is relatively rare and only affects about 1000 patients in the US [Citation3]. However, OS is more common in teens and children, accounting for about 3% of total cancers in teens between 15 and 19 years old [Citation4]. With the development of anti-tumor approaches, more than 74% of OS patients with localized tumors can survive 5 years [Citation5,Citation6]. However, more than 20% of OS patients are diagnosed with tumor spreading by the time of diagnosis [Citation7]. Even after adequate treatment, more than 70% of metastatic OS patients will die within 5 years after diagnosis [Citation5,Citation6].
At present, most OS patients are treated with surgical resections combined with chemotherapy applied before and after surgeries [Citation8,Citation9]. However, tumor recurrence is common, and the prognosis is poor in many cases [Citation8]. Molecularly targeted therapies are emerging novel anti-tumor approaches focusing on controlling tumor progression by regulating cancer-related gene expression [Citation10,Citation11]. The main challenge for the development of molecular targeted therapy is the lack of effective targets [Citation10,Citation11]. Previous studies have characterized a considerable number of molecular players with critical functions in OS. Some of these molecular players are proven to be potential targets to treat OS [Citation12]. Non-coding RNAs, such as miRNAs and lncRNAs, regulate gene expression to affect cancer progression. Therefore, they are potential targets to treat OS [Citation13]. MiR-886 was recently reported to downregulate CXCL12 (SDF1) expression in human marrow stromal cells [Citation14]. In another study, miR-886 was regarded as a postoperative outcome signature of patients with the metastatic bone disease [Citation15]. Our pilot study using IntaRNA 2.0 prediction has revealed potential binding between miR-886 and lncRNA OXCT1-AS1. Both miR-886 and lncRNA OXCT1-AS1 play critical roles in cancer biology, while their roles in OS are unclear [Citation16,Citation17]. LncRNA OXCT1-AS1 is a novel lncRNA with an oncogenic role in bladder and lung cancers [Citation16,Citation18]. We predicted that they might interact with each other. This study aimed to explore the relationship between miR-886 and OXCT1-AS1 in OS and their potential function for tumorigenesis.
Materials and methods
Tissue preparation
A total of 66 OS patients received surgical resection of the primary tumors at the First People’s Hospital of Shangqiu City between May 2019 and May 2021. Tumors were dissected by experienced histopathologists to prepare both OS and paired non-tumor tissue samples and stored in a liquid nitrogen tank prior to RNA extraction. Recurrent OS patients were excluded from this study. All patients signed informed consent. This study was approved by the Ethics Committee of the First People’s Hospital of Shangqiu City. shows all patients’ clinical data.
Table 1. Chi-squared test analysis of the correlations between patients’ clinical data and OXCT1-AS1 expression
Cell and Cell Culture Method
Two human OS cell lines Saos2 and MG63 and normal human osteoblasts cell line hFOB 1.19 from Chinese Type Culture Collection were used for in vitro cell assays. These cells were cultured in DMEM (Gibco) containing FBS (10%) at 37°C in an incubator with 95% humidity and 5% CO2.
Cell transfections
OXCT1-AS1 expression vector (pcDNA3.1) and miR-886 mimic were prepared by Sangon Biotech (Shanghai, China). OXCT1-AS1 si-RNA and its corresponding scrambled siRNA (si-NC) were from ThermoFisher Scientific (Waltham, MA, USA). To overexpress/inhibit OXCT1-AS1 and miR-886 expression, 10 mM OXCT1-AS1 vector or 100 mM si-OXCT1-AS1 or miR-886 mimic were transfected into 107 cells using Neon Electroporation Transfection system (ThermoFisher Scientific). After transfection, cells were cultured in fresh media prior to the subsequent assays.
RNA preparations
Total RNAs were isolated from tissue and cell samples using RNAiso Plus (Takara). In brief, samples were homogenized in RNAiso Plus solution for 20 min at 4°C. After centrifugation, RNAs were extracted using chloroform at 1/2 volume of RNAiso Plus and precipitated using isopropanol. After washing with 70% ethanol, RNA samples were dissolved in nuclease-free water and treated with DNase I digestion to remove genomic DNAs. RNA concentration and integrity were determined using Bioanalyzer. RNA isolations were repeated if RIN values were lower than 8.0.
Quantitative reverse transcription PCR (RT-qPCRs)
cDNA samples were prepared from about 2000 ng total RNAs as a template using PrimeScript RT Master Mix (Takara) through reverse transcriptions. After that, 1 μl cDNA sample was used for qRT-PCRs on LightCycler (Roche) with 18S rRNA and U6 as internal controls to determine the expression of OXCT1-AS1 and miR-886 (mature and premature). The primer sequences were lncRNA OXCT1-AS1 forward 5′-CTGGACTGCGTTCACGTTTC-3′ and reverse 5′-CTGGACTGCGTTCACGTTTC-3′, hsa-mir-886-mature forward 5′-GAACGCGGGTGCTTACTG-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′, hsa-mir-886-precursor forward 5′-CGGGTCGGAGTTAGCTCAAGCGG-3′ and reverse 5′-AAGGGTCAGTAAGCACCCGCG-3′, 18S rRNA forward 5′-TCAACACGGGAAACCTCACCA-3′ and reverse 5′-CACCACCCACCGAATCAAGAA-3′, and U6 forward 5′-CGCTTCGGCAGCACATATACTAAAATTGGAAC-3′ and reverse 5′-GCTTCACGAATTTGCGTGTCATCCTTGC-3′. All qPCRs were performed at 95°C for 1 min followed by 40 cycles of 10 s at 95°C and 48s at 58°C. Each experiment included three technical replicates. Ct values of target genes were normalized to internal controls using the 2−ΔΔCt method [Citation19].
Subcellular fractionation assay
CST Cell Fractionation Kit (9038S) was applied to prepare cytoplasmic and nuclear fractions of cells [Citation20]. Cells were harvested, washed with ice-cold PBS three times, and incubated on ice with cell fractionation buffer for 20 min. After that, cell lysates were centrifuged for 10 min at 1500 g to separate cytoplasmic fraction, which was the supernatant. Cytoplasmic fraction was transferred to a RNase-free tube, and the nuclear fraction, which was the cell pellet, was further incubated with nuclear lysis buffer for 60 min at 4°C. After that, the nuclear lysate was centrifuged for 10 min at 14,000 g to harvest nuclear extract. After that, RNAs were extracted from both fractions and subjected to RT-qPCRs to detect OXCT1-AS1.
RNA-RNA pulldown assay
RNA–RNA pulldown assay was performed as previously studied [Citation20]. OXCT1-AS1 was synthesized through in vitro transcriptions using T7 RNA polymerase (NEB) and purified using GeneJET RNA Purification Kit. Negative control (NC) RNAs was from ThermoFisher (Cat# AM7155, ThermoFisher). OXCT1-AS1 and NC were biotin-labeled using Biotin RNA Labeling Mix (Roche) and purified using GeneJET RNA Purification Kit. The biotin-NC and biotin-OXCT1-AS1 were then transfected into cells as mentioned above. 48 h after transfection, cells were harvested and incubated with cell lysis buffer on ice for 20 min. After that, cell lysates were incubated with streptavidin magnetic beads to pull down the RNA complex. The RNA complexes were purified and subjected to RT-qPCRs to determine premature miR-886 levels.
5-Bromodeoxyuridine incorporation proliferation assay
5-Bromodeoxyuridine (BrdU) incorporation, which directly reflects DNA synthesis, was determined to analyze cell proliferation. Cells were harvested at 48 h after transfections and transferred to a 96-well plate. After that, cells were cultured in fresh medium for another 48 h and incubated with 10 μM BrdU for 2 h at room temperature. Cells were then incubated with peroxidase-coupled BrdU antibodies (Sigma-Aldrich) for 1 h. After washed with PBS three times, cells were incubated with peroxidase substrate, and the OD values at 450 nm were determined. Cells without incubation with BrdU antibody were used as the blank controls.
Western blot analysis
Total proteins were extracted from the transfected cells. After quantification with BCA Protein Assay Kit, about 30 μg proteins from each sample were separated by SDS-PAGE and transferred onto membranes. The proteins on the membranes were first coupled with antibodies against caspase-3 (1:1000; sc-65497), E-cadherin (1:1000; sc-8426), N-cadherin (1:1000; sc-8424), and cyclinD1 (1:1000; sc-8396) and then with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:1000; sc-542741) from Santa Cruz Biotechnology (Santa Cruz, CA). In addition, proteins were also coupled with rabbit anti-GAPDH antibody (TA-09, Yatai hengxin, Beijing, China) and incubated with HRP-labeled anti-rabbit secondary antibody (1:500) for 1 h. The signals were visualized with ECL substrate and imaged.
Statistical analysis
Data were analyzed using GraphPad Prism 6 (GraphPad). Differences between two groups were compared using Student’s t-tests. Correlations were explored with Pearson’s correlation coefficient. The 66 OS patients were divided into high and low OXCT1-AS1 level groups, and the correlations between patients’ clinical data and OXCT1-AS1 expression were analyzed with Chi-squared test. Differences with a p < 0.01 were considered statistically significant.
Results
Differential expression of OXCT1-AS1 and mRNA-886 in osteosarcoma
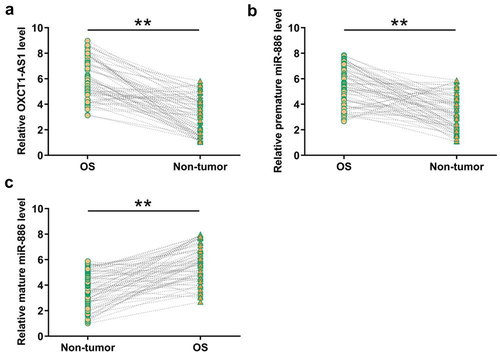
To identify the expression pattern of OXCT1-AS1 and miR-886 in osteosarcoma clinical samples, RT-qPCRs were performed to determine the levels of OXCT1-AS1, premature miR-886, and mature miR-886 in paired OS and non-tumor tissue samples from 66 osteosarcoma patients. It was observed that OS tissues had increased levels OXCT1-AS1 (, p < 0.01) and premature miR-886 (, p < 0.01), but decreased mature miR-886 level (, p < 0.01) compared to non-tumor tissues. Chi-squared test analysis showed that OXCT1-AS1 was not closely correlated with patients’ age, gender tumor site, Enneking stage, and lymph node metastasis but was closely correlated with tumor size ().
Correlations of OXCT1-AS1 with premature or mature miR-886
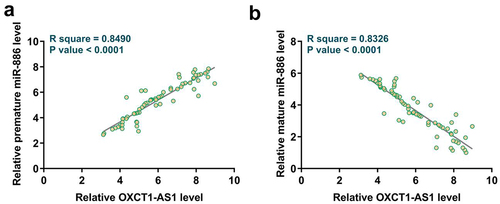
To explore the coordination or antagonism relationship of OXCT1-AS1 with premature or mature miR-886 in OS tissues, correlations between expression levels of OXCT1-AS1 and premature or mature miR-886 in OS tissues were analyzed with Pearson’s correlation coefficient. It was observed that OXCT1-AS1 was positively correlated with premature miR-886 () but was inversely correlated with mature miR-886 ().
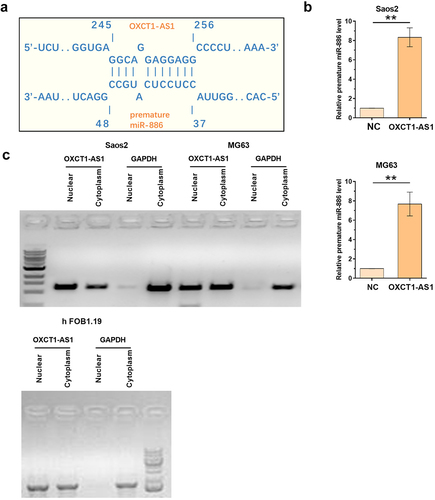
The direct interaction of OXCT1-AS1 with premature miR-886 and the subcellular location of OXCT1-AS1 in osteosarcoma cells
Given the possible inverse correlation between OXCT1-AS1 and miR-886, RNA pulldown assay and subcellular fractionation assay were performed to identify their potential target relationship. The potential interaction between OXCT1-AS1 and premature miR-886 was predicted by IntaRNA 2.0. It was observed that OXCT1-AS1 and premature miR-886 could form strong base pairing (). RNA–RNA pulldown assay was applied to confirm the direct interaction between OXCT1-AS1 and premature miR-886. The premature miR-886 level was much higher in bio-OXCT1-AS1 group compared to bio-NC group (, p < 0.01), confirming their direct interaction. Subcellular fractionation assay was applied to detect OXCT1-AS1 in both nuclear and cytoplasm of Saos2, MG63, and hFOB 1.19 cells. OXCT1-AS1 was detected in both nuclear and cytoplasm fractions (). The basic normal OXCT1-AS1 expression in SaOS2, MG-63, and hFOB 1.19 cells were shown in Supplement data 1.
Figure 3. The direct interaction of OXCT1-AS1 with premature miR-886 and the subcellular location of OXCT1-AS1 in OS cells The potential interaction between OXCT1-AS1 and premature miR-886 was predicted by IntaRNA 2.0 (a) RNA-RNA pulldown assay was applied to confirm the direct interaction between OXCT1-AS1 and premature miR-886 (b). Subcellular fractionation assay was applied to detect OXCT1-AS1 in both nuclear and cytoplasm of both Saos2, MG63 and hFOB 1.19 cells (c). ** p < 0.01.
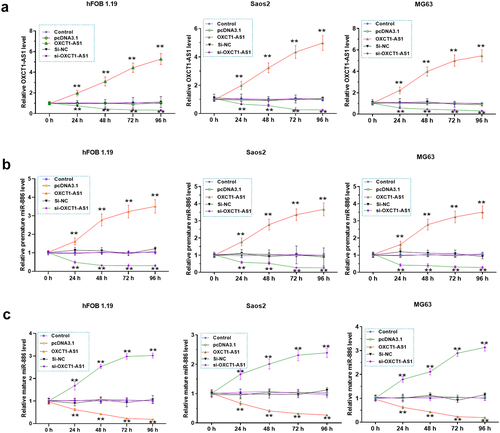
The effects of OXCT1-AS1 on premature and mature miR-886 levels
We then examined the effects of OXCT1-AS1 overexpression/silencing on premature and mature miR-886 expression in vitro. SaOS2 and MG-63 cells were transfected with OXCT1-AS1 expression vector or si-OXCT1-AS1, and OXCT1-AS1 levels were confirmed from 24 h to 96 h (, p < 0.01). In addition, we showed that OXCT1-AS1 overexpression increased premature miR-886 level (, p < 0.01) but decreased mature miR-886 level (, p < 0.01). By contrast, OXCT1-AS1 silencing decreased premature miR-886 level (, p < 0.01) but increased mature miR-886 level (, p < 0.01).
Figure 4. The effects of OXCT1-AS1 on premature and mature miR-886 expression Saos2, MG63 and hFOB 1.19 cells were transfected by OXCT1-AS1 expression vector or si-OXCT1-AS1 (a). The effects of OXCT1-AS1 on the expression of premature (b) and mature (c) miR-886 were analyzed with RT-qPCRs. ** p < 0.01.
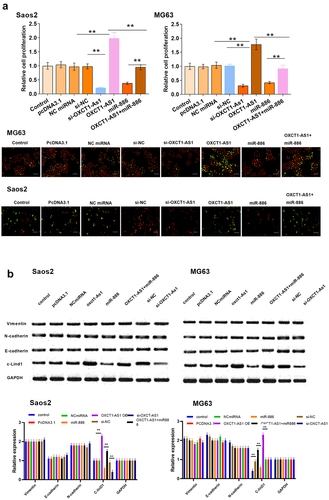
The involvement of OXCT1-AS1 and microRNA-886 in the proliferation of Saos2 and MG63 cells
We further investigated the involvement of OXCT1-AS1 and miR-886 in the malignant behaviors of OS cells. Saos2 and MG63 cell proliferation after the overexpression of OXCT1-AS1 and/or miR-886 was analyzed using BrdU assay. OXCT1-AS1 overexpression promoted OS cell proliferation and miR-886 overexpression inhibited cell proliferation. Moreover, miR-886 reduced the enhancing effects of OXCT1-AS1 on cell proliferation (, p < 0.01). Moreover, we also detected the epithelial–mesenchymal transition markers (Vimentin, E-cadherin, N-cadherin) and cyclin D1 expression after transfecting OXCT1-AS1 and/or miR-886 (, p < 0.01). Except for cyclinD1, no change was observed for these markers.
Figure 5. The involvement of OXCT1-AS1 and miR-886 in the proliferation of Saos2 and MG63 cells (a) Saos2 and MG63 cell proliferation after the overexpression/silencing of OXCT1-AS1 and/or miR-886 was analyzed with BrdU assay. Red signal indicates the nuclei. Green signal indicates the Brdu ** p < 0.01. (b) Western blot of vimentin, E-cadherin, N-cadherin, cyclin D1 and GAPDH. ** p < 0.01.
Discussion
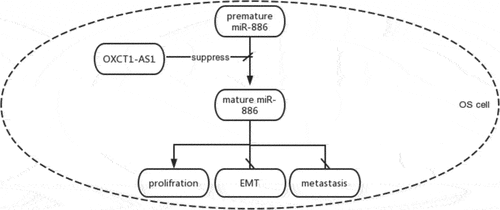
The study explored the roles of OXCT1-AS1 and miR-886 in OS. We demonstrated upregulated OXCT1-AS1 level and downregulated miR-886 level (both mature and premature) in OS tissue samples compared to non-tumor tissue samples and revealed that OXCT1-AS1 might participate in OS cells’ malignant behavior by affecting miR-886 maturation in the OS cell model.
OXCT1-AS1 is a newly identified lncRNA with characterized functions in bladder cancer and glioblastoma [Citation16,Citation17]. As revealed by microarray analysis, OXCT1-AS1 is overexpressed in bladder cancer and interacts with the axis of miR-455-5p/JAK1 to promote cancer progression [Citation16]. OXCT1-AS1 is also overexpressed in glioblastoma [Citation17]. OXCT1-AS1 overexpression in glioblastoma sponges miR-195 to upregulate CDC25A, thereby increasing tumorigenesis [Citation17]. However, the role of OXCT1-AS1 in osteosarcoma has not been explored. This study showed that OXCT1-AS1 is overexpressed in OS, and OXCT1-AS1 overexpression is only correlated with tumor size but not tumor metastasis, suggesting the role of OXCT1-AS1 in OS tumor growth. These findings were confirmed by the observation that OXCT1-AS1 overexpression enhances OS cell proliferation. Therefore, OXCT1-AS1 may participate in OS progression by promoting tumor growth.
Mature miR-886 was reported to be an oncogenic miRNA in cervical cancer, in which miR-886 is overexpressed to suppress tumor cell apoptosis via downregulating Bax [Citation21]. Our study showed that miR-886 maturation is inhibited in OS patients and miR-886 plays an inhibitory role in OS cell proliferation. Therefore, miR-886 may play opposite roles in different types of cancers and miR-886 precursor and mature form may have different functions in cancers. Our study also showed that altered miR-886 maturation, but not expression, participates in cancer biology. Interestingly, OXCT1-AS1 directly interacted with premature miR-886 and suppressed its maturation in OS cells. Together with the fact that OXCT1-AS1 could be detected in both nuclear and cytoplasm of OS cells, we speculated that OXCT1-AS1 could sponge premature miR-886 in nuclear to suppress its maturation process. All these revealed a new OXCT1-AS1/miR-886 signaling axis in OS cells. However, the detailed molecular function of the downstream target gene of the axis remains unclear. The switch role of mature/premature miR-886 and its relationship with OXCT1-AS1 still need to be further explored in more cancer types. OXCT1-AS1 overexpression together with ratio of mature/premature miR-886 may provide prognosis signatures for OS.
Conclusions
OXCT1-AS1 expression is increased, and miR-886 maturation is decreased in OS. In addition, OXCT1-AS1 may sponge premature miR-886 in the nuclear to suppress its maturation, thereby promoting OS cell proliferation.
Authors’ contributions
Wen Dai, Han Liu: study concepts, literature research, clinical studies, data analysis, experimental studies, manuscript writing and review; Wen Dai: study design, literature research, experimental studies and manuscript editing; Han Liu: definition of intellectual content, clinical studies, data acquisition, and statistical analysis.
All authors have read and approve the submission of the manuscript.
Availability of supporting data
The data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but are available on request from the corresponding author.
Ethical approval and consent to participate
All patients signed the written informed consent. All procedures were approved by the Ethics Committee of the First People’s Hospital of Shangqiu City and completed in keeping with the standards set out in the Announcement of Helsinki and laboratory guidelines of research in China.
Supplemental Material
Download Zip (194.6 KB)Supplementary material
Supplemental data for this article can be accessed here.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Czarnecka AM, Synoradzki K, Firlej W, et al. Molecular Biology of Osteosarcoma. Cancers (Basel). 2020;12(8):2130.
- Gorlick R, Khanna C. Osteosarcoma. Journal of Bone and Mineral Research: The Official Journal of the American Society for Bone and Mineral Research. 2010;25(4):683–691.
- Levy M, Leclerc BS. Fluoride in drinking water and osteosarcoma incidence rates in the continental United States among children and adolescents. Cancer Epidemiol. 2012;36(2):e83–8.
- Nie Z, Peng H. Osteosarcoma in patients below 25 years of age: an observational study of incidence, metastasis, treatment and outcomes. Oncol Lett. 2018;16(5):6502–6514.
- Anderson ME. Update on Survival in Osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292.
- Picci P, Mercuri M, Ferrari S, et al. Survival in high-grade osteosarcoma: improvement over 21 years at a single institution. Ann Oncol. 2010;21(6):1366–1373.
- Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma Overview. Rheumatol Ther. 2017;4(1):25–43.
- Zhang Y, Yang J, Zhao N, et al. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol Lett. 2018;16(5):6228–6237.
- Spraker-Perlman HL, Barkauskas DA, Krailo MD, et al. Factors influencing survival after recurrence in osteosarcoma: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2019;66(1):e27444.
- Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol. 2018;834:188–196.
- Sayles LC, Breese MR, Koehne AL, et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019;9(1):46–63.
- Zhou W, Hao M, Du X, et al. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17(96):301–307.
- Nicolas FE. Role of ncRNAs in Development, Diagnosis and Treatment of Human Cancer. Recent Patents on Anti-cancer Drug Discovery. 2017;12(2):128–135.
- Pillai MM, Yang X, Balakrishnan I, et al. MiR-886-3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS One. 2010;5(12):e14304.
- Xu SB, Fan RH, Qin X, et al. microRNA Prognostic Signature for Postoperative Success of Metastatic Orthopedic Cancers: implications for Precision Microsurgery. Front Cell Dev Biol. 2021;9:704505.
- Chen JB, Zhu YW, Guo X, et al. Microarray expression profiles analysis revealed lncRNA OXCT1-AS1 promoted bladder cancer cell aggressiveness via miR-455-5p/JAK1 signaling. J Cell Physiol. 2019;234(8):13592–13601.
- Zhong C, Yu Q, Peng Y, et al. Novel LncRNA OXCT1-AS1 indicates poor prognosis and contributes to tumorigenesis by regulating miR-195/CDC25A axis in glioblastoma. J Exp Clin Cancer Res. 2021;40(1):123.
- Li B, Zhu L, Li L, et al. lncRNA OXCT1-AS1 Promotes Metastasis in Non-Small-Cell Lung Cancer by Stabilizing LEF1, In Vitro and In Vivo. Biomed Res Int. 2021;2021:4959381.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408.
- Schuck PL, Ball LE, Stewart JA. The DNA-binding protein CST associates with the cohesin complex and promotes chromosome cohesion. J Biol Chem. 2021;297(3):101026.
- Li JH, Xiao X, Zhang YN, et al. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol Oncol. 2011;120(1):145–151.