ABSTRACT
Circular RNA circ_0000285 is differentially expressed in several malignancies; however, its role in gliomas is under investigation. Reverse transcription quantitative polymerase chain reaction was conducted to evaluate the expression of circ_0000285, miR-197-3p, and CDC28 protein kinase regulatory subunit 1B (CKS1B) in glioma tissues and cells. Cell Counting Kit-8 and Transwell invasion assays coupled with Western blotting analysis using anti-Bax and anti-Bcl-2 antibodies were performed to evaluate cell proliferation, invasion, and apoptosis. Luciferase reporter and AGO2 RNA immunoprecipitation assays were conducted to verify the interaction between miR-197-3p and circ_0000285 or CKS1B. Xenograft tumor growth was evaluated in mice. We noted that circ_0000285 was highly expressed in glioma tissues and cells and that circ_0000285-silencing retarded tumor growth both in vitro and in vivo. This effect was mediated by the binding of circ_0000285 to miR-197-3p, which silenced CKS1B, an essential driver of glioma cell proliferation and invasion. Thus, circ_0000285 boosted glioma progression by regulating the miR-197-3p/CKS1B axis, highlighting a novel competing endogenous RNA circuit of glioma progression.
Abstract
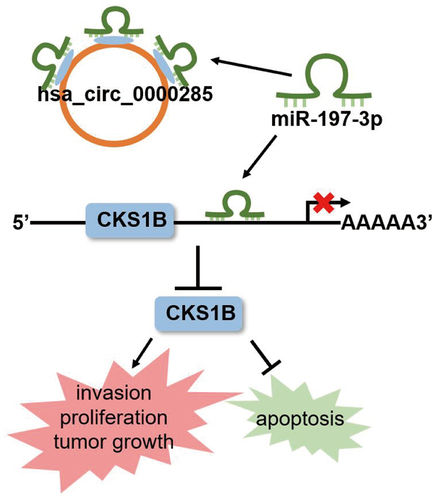
Introduction
Glioma is a frequently diagnosed intracranial cancer that constitutes approximately 30% of intracranial cancers [Citation1]. In contrast to other common malignancies, the incidence of gliomas is gradually increasing worldwide [Citation2]. Despite notable improvements in modalities against gliomas, the mortality and recurrence rates of gliomas remain high [Citation3]. Consequently, elucidating how glioma progresses at the molecular level is very important for exploring diagnostic and therapeutic targets against glioma.
Circular RNAs (circRNAs) are non-coding RNAs characterized by their circular configuration. Although circRNAs do not possess coding ability, their critical role in gene regulation is being recognized [Citation4]. Through gene regulation, circRNAs are implicated in various biological processes, including cell proliferation, energy metabolism, tumorigenesis, and embryonic development [Citation5]. In glioma, increasing number of circRNAs were described to have pro- or anti-tumoral functions [Citation6,Citation7]. For example, circ-0014359 stimulates the oncogenicity of PI3K signaling events and drives glioma tumorigenesis [Citation8]. Circ-ATXN1 exhibits a proangiogenic function, thereby favoring the malignant behavior of glioma cells [Citation9]. Circ_0005198 v silencing represses glioma cell sensitivity to temozolomide (TMZ) and favors glioma tumorigenesis [Citation10]. Circ_0001162 promotes glioma progression by promoting the malignant phenotypes of cancer cells in vitro and in vivo [Citation11]. Circ_0000285 has recently been identified as an oncogenic circRNA in hepatocellular carcinoma [Citation12], osteosarcoma [Citation13], and cervical cancer [Citation14]. For example, ectopic expression of circ_0000285 drives the malignant behavior of osteosarcoma by enhancing the oncogenic activity of transforming growth factor-beta [Citation15]. Conversely, circ_0000285 expression is downregulated in bladder cancer and is associated with the clinical pathological hallmarks of bladder cancer, including cisplatin resistance [Citation16], suggesting its tumor-suppressive role in bladder cancer. Thus, the function of circ_0000285 in malignancies may be context dependent. However, little is known about the role of circ_0000285 in glioma.
In this study, we investigated the expression of circ_0000285 in gliomas and its role in vitro and in vivo. Furthermore, the competing endogenous RNA (ceRNA) network involved in circ_0000285 functioning was determined using a panel of cell function assays. We hypothesized that circ_0000285 affects glioma cell proliferation, migration, and apoptosis by targeting the miR-197-3p/CKS1B axis as a carcinogenic circRNA. Our findings highlight the role of circ_0000285 in glioma progression, suggesting that targeting circ_0000285 may be a promising strategy against glioma.
Methods
Tissue samples
In total, 16 paired glioma and matching noncancerous tissues were surgically resected from patients when first diagnosed with glioma before receiving any treatment at Suizhou Hospital, Hubei University of Medicine. The Ethics Committee of Suizhou Hospital, Hubei University of Medicine approved all the investigations. Consent was obtained from each enrolled patient.
Cell maintenance
Glioblastoma cell lines SHG-44 and A172 were purchased from the American Type Culture Collection (ATCC, USA), glioblastoma cell line U251 was purchased from Millipore Sigma, USA, and normal human astrocyte cells (NHA) were purchased from Lonza (Basel, Switzerland). NHA cells were cultured using the AGMTM Astrocyte Growth Medium BulletKit (Lonza, Switzerland) as the per manufacturer’s instructions. SHG-44, U251, and A172 cells were maintained in Eagle’s Minimum Essential Medium. All cells were grown in a 5% CO2 atmosphere at 37°C.
Oligonucleotides and transfection
MiR-197-3p mimic/inhibitor, mimic/inhibitor NC, si-circ_0000285 (si-circ), si-NC, CKS1B overexpression vector (OE-CKS1B), and empty vector were synthesized by Origene (China). For cell transfection, 2.5 × 103 U251 and SHG-44 cells were seeded in 96-well plates and grown until 80% confluence. The synthesized oligonucleotides were introduced into U251 and SHG-44 cells using Lipofectamine 2000 (Thermo Fisher Scientific). Lentiviral particles containing the short hairpin RNA (shRNA) sh-circ_0000285 or sh-NC were purchased from Origene, China. The 80% confluent SHG-44 and U251 cells were infected with the lentiviral particles for 48 h. Following puromycin maintenance for 4 weeks, cells were subjected to reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis.
Subcellular fractionation assay
Nucleoplasm and cytoplasmic fractionation were performed using a PARIS nuclear/cytoplasmic separation kit (Life Technologies, USA). Briefly, U251 and SHG-44 cells were split and centrifuged, the cytoplasm in the supernatant was placed in separate tubes, and the remaining precipitate was cultured with a cell disruption buffer to lyse the nucleus and centrifuged again to separate the lysate components. The endogenous controls of the nucleus and cytoplasm were Uracil6 (U6) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively [Citation17].
RNase R treatment
The RNA obtained from U251 and SHG-44 cells was digested with RNase R (Epicenter Technologies, USA). Two micrograms of total RNA was incubated with 4 U/μg RNase R for 30 min at 37°C, and an RNeasy MinElute Cleanup Kit (Qiagen, USA) was used to purify the incubated RNA. The levels of circ_0000285 and its linear transcripts (linear circ_0000285) were measured using RT-qPCR [Citation18].
RT-qPCR
Total RNA was isolated from tissues or cells using TRIzol reagent. Complementary DNA (cDNA) was synthesized using a cDNA Reverse Transcription Kit (Takara, Japan). RT-PCR was conducted using a Venor GeM Classic kit (Minerva Biolabs, Germany) on an ABI Prism 7500 Fast Real-Time PCR system. Gene expression normalized to GAPDH was analyzed using the 2−ΔΔCt method [Citation19]. Primers used are listed in .
Table 1. Sequences of quantitative RT-PCR primers
Cell proliferation and invasion assay
To assess cell proliferation, 2 × 104 U251 and SHG-44 cells were seeded in 96-well plates. A 10-μL cell counting kit-8 (CCK-8) solution was added to the maintenance medium after incubation for 24 h, 28 h, and 72 h. After 3 h, the plates were read at OD450 on an ELISA plate reader (Bio-Rad, USA) [Citation20].
Cell invasion was determined using Transwell assays. Transwell inserts pre-coated with matrigen were placed into a 24-well companion plate. After medium with FBS was added to the lower chamber, 2.0 × 105 cells in 200 μL medium were added to the upper chambers and the invasion chamber was assembled. After 36 h of cell culture, the inserts were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet solution for another 15 min. Invasive cells were quantified under a microscope, and images were captured [Citation21].
Western blot
Cells were lysed using radioimmunoprecipitation assay buffer (Beyotime, China) and then quantitatively assessed using a bicinchoninic acid protein assay kit (Pierce, USA) and NanoDrop ND 2000 c Spectrophotometer (Thermo Scientific, USA) as per the manufacturer’s instructions. Equal amounts of denatured proteins were loaded onto 10% sulfate-polyacrylamide gel and separated via electrophoresis run at 75 to 175 V. The proteins in the gels were transferred onto polyvinylidene fluoride membranes using a wet transfer system and blocked with 5% milk in TBS at 25°C for 1 h. Membranes were continuously exposed to TBST containing anti-CKS1B antibodies (Cat#:C0748, Sigma-Aldrich, USA), anti-Bax antibodies (Cat #33-6400, 1:1000, Thermo Fisher Scientific, USA), anti-Bcl-2 antibodies (Cat #MA5-11,757, 1:1000, Thermo Fisher Scientific, USA), or anti-GAPDH antibodies (Cat #MA1-16,757,1:1000, Thermo Fisher Scientific, USA) at 4°C. Antibody-bound proteins were probed using an anti-mouse/goat HRP secondary antibody (Cat #31,430 or Cat #31,402, 1:1000, Invitrogen, USA) at room temperature for 1 h. Chemiluminescent detection (Beyotime, China) was used to visualize the target proteins, and the intensity of protein signals was estimated using Quantity One 4.1 (Bio-Rad Laboratories, USA) [Citation22].
AGO2 RNA immunoprecipitation (RIP) assays
A RIP-Assay Kit (MEDICAL & BIOLOGICAL LABORATORIES CO., LTD., Japan) was used to detect interactions between circ_0000285 and miR-197-3p. Briefly, the prepared beads immunized with anti-Ig and AGO2 antibodies were mixed with the lysed SHG-44 and U251 cells at 4°C. After 3 h, the bead-antibody RIP complexes were collected for RT-qPCR [Citation23].
Luciferase reporter assays
PmiR-RB-REPORT™ vectors (RiboBio, China) were used to construct the wild-type (WT) and mutant type (MUT) luciferase by fusing WT or MUT binding fragment of circ_0000285 or CKS1B 3ʹUTR to miR-197-3p. The recombinant vectors, named circ_0000285-WT, circ_0000285-MUT, CKS1B-MUT, and CKS1B-WT, were co-introduced into SHG-44 and U251 cells with miR-197-3p mimic or NC. At 48 h post-transfection, fluorescence was measured using the Promega Luciferase Assay System [Citation24].
In vivo tumor growth assays
The procedures were approved by the Ethics Committee of Suizhou Hospital, Hubei University of Medicine. Athymic nude mice (4–5 weeks old, 20–25 g) were purchased from the Experimental Animal Center of Wuhan University (Wuhan, China). SHG-44 cells (1 × 106 cells) transfected with si-circ_0000285 or si-NC were subcutaneously injected into athymic nude mice (n = 5) in a pathogen-free environment. Tumor size was monitored weekly. After 5 weeks, the mice were sacrificed, and the tumors were dissected for tumor weight [Citation11].
Statistical tests
Data were analyzed using GraphPad Prism 9.0. Student’s t-test was used to compare two variables. One-way analysis of variance was used to evaluate the differences among multiple groups. Pearson’s correlation analysis was used to determine the correlation between gene expression levels. Statistical significance was set at p < 0.05.
Results
Through bioinformatics analysis, circ_0000285, miR-197-3p, and CKS1B were identified as genes of interest in this study. Tissue sample analysis and in vitro loss-of-function experiments revealed that circ _0000285 affects glioma cell proliferation, migration, and apoptosis by targeting the miR-197-3p/CKS1B axis. These results suggest that targeting circ_0000285 is a promising strategy for the treatment of gliomas.
Cytoplasmic circ_0000285 is robustly expressed in glioma
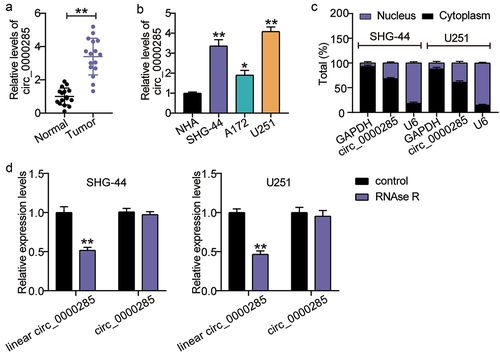
The differential expression of circ_0000285 has been described in several cancers [Citation14,Citation15]; hence, we evaluated its expression in gliomas. As shown in ), the relative expression of circ_0000285 was enhanced in glioma tissues compared with that in normal tissues. We have analyzed the association between circ_0000285 expression and clinicopathological characteristics of glioma cancer patients. Glioma patients were classified into circ_0000285 high expression group (n = 8) and low expression group (n = 8). The results showed that circ_0000285 high expression was correlated with pathological type and tumor size. However, there were no significant associations between circ_0000285 expression and gender, age, and KPS (Supplementary Table S1). For further verification, we performed RT-qPCR analysis in several glioma cells (SHG-44, A172, and U251) and NAH cells. Compared with NAH cells, circ_0000285 RNA levels were elevated in glioma cells ()). An increase in circ_0000285 expression was detected in SHG-44 and U251 cells. Thus, we chose both glioma cell lines for subsequent assays. RT-qPCR was also performed to determine the intracellular localization of circ_0000285, and the results showed its predominant enrichment in the cytoplasm of SHG-44 and U251 cells ()). Additionally, circ_0000285 was enriched upon RNAse R treatment ()), suggesting its circular characteristics.
Figure 1. Cytoplasmic circ_0000285 is robustly expressed in glioma. A. Verification of the relative expression level of upregulated circ_0000285 by RT-qPCR in glioma tissues (N = 16), **P < 0.001. B. RT-qPCR analysis of the expression of circ_0000285 in glioma cells, vs.NHA, *P < 0.05, **P < 0.001; C. Intracellular localization of circ_0000285 in SHG-44 and U251 cells. D. RNase R was used to treat circ_0000285 in SHG-44 and U251 cells to determine the circularity of the RNA, vs.Control,**P < 0.001. N = 3, repetition = 3.
Circ_0000285 silencing decreases carcinogenic potency both in vitro and in vivo
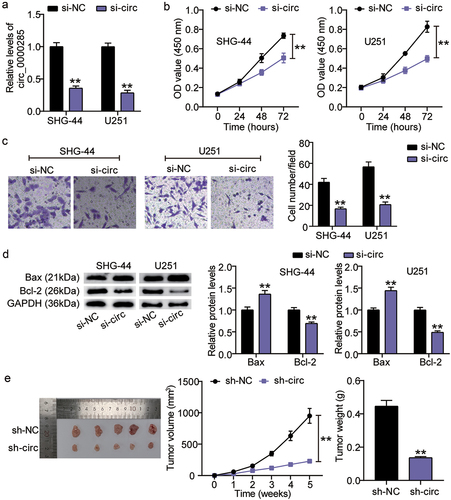
To determine the function of circ_0000285 in gliomas, we established circ_0000285-silenced U251 and SHG-44 cells. RT-qPCR analysis demonstrated a 3-fold reduction in circ_0000285 expression in both cell lines ()). Assessment of cell proliferation showed that circ_0000285 knockdown significantly weakened the proliferative capacity of U251 and SHG-44 cells ()). A similar inhibitory effect was observed in the invasion of U215 and SHG-44 cells upon circ_0000285 knockdown ()). Additionally, apoptosis induction was confirmed by elevated Bax and decreased Bcl-2 expression levels ()). Importantly, suppressed tumorigenesis was observed in mice bearing circ_0000285-silenced SHG-44 cells ()). Collectively, loss-of-function assays demonstrated that circ_0000285 knockdown mitigated glioma growth in vitro and in vivo.
Figure 2. Circ_0000285 silence lessens tumorigenic potency both in vitro and in vivo. A. RT-qPCR analysis of circ_0000285 expression in SHG-44 and U251 cells transfected with si-circ_0000285 and si-NC, vs. si-NC, **P < 0.001. B. CCK8 assays assessing the proliferation of SHG-44 and U251 cells upon circ_0000285 silence or not, vs. si-NC, **P < 0.001. C. The quantification of the invasion assay is done by counting 3 different random fields and plotted average cell number per field. D. Western blotting analysis of Bax and Bcl-2 expression in SHG-44 and U251 cells transfected with si-circ_0000285 and si-NC, vs. si-NC, **P < 0.001. E. Subcutaneous tumor growth, xenograft tumor images and tumor weights of xenografts from SHG-44 cells with or without circ_0000285 silence in nude mice, vs. si-NC, **P < 0.001. si-circ, circ_0000285 siRNA; si-NC, si-circ negative control. N = 3, repetition = 3.
miR-197-3p and CKS1B might be the downstream effectors of circ_0000285 in glioma
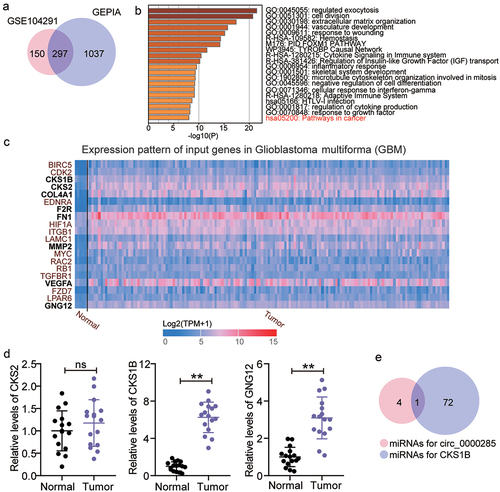
GSE104291, including glioma samples and normal brain samples, is an mRNA microarray dataset used to screen the upregulated genes in glioma samples; GEPIA was also used to screen the upregulated genes in glioma samples. With adj. P < 0.05 and logFC>2, 297 upregulated genes were screened from GSE104291 and GEPIA ()). Uploading the 297 upregulated genes to Metascape for biological process enrichment, 20 genes were found to be enriched in cancer pathways ()). Among the 20 genes, CKS1B, CDC28 protein kinase regulatory subunit 2 (CKS2), collagen type IV alpha 1 chain (COL4A1), coagulation factor II thrombin receptor (F2R), fibronectin 1 (FN1), matrix metallopeptidase 2 (MMP2), vascular endothelial growth factor A (VEGFA), and G protein subunit gamma 12 (GNG12) were found to be overexpressed in glioma samples according to TCGA data ()). Because COL4A1, F2R, FN1, MMP2, and VEGFA have previously been investigated in glioma [Citation25–29], the genes of interest were selected from CKS1B, CKS2, and GNG12. After performing RT-qPCR, CKS1B, presenting with the highest expression in glioma tissues compared with CKS2 and GNG12, was selected as our gene of interest ()). Next, we used starBase to predict the miRNAs that could bind to circ_0000285 and CKS1B and found that only miR-197-3p overlapped ()).
Figure 3. Circ_0000285 might regulate miR-197-3p/CKS1B axis in glioma. A. 297 upregulated genes were overlapped from GSE104291 and GEPIA. GSE104291 and GEPIA including the mRNA expression profile in glioma samples were used to screen the upregulated genes with adj.P < 0.05 and logFC>2. B. Pathways in cancer was enriched by Metascape. Metascape was used to enrich the key biological processes for 297 upregulated genes. C. The expression pattern of 20 genes in glioblastoma multiforma according to the data from TCGA. D. The expression of CKS1B, CKS2 and GNG12 in glioma tissues (N = 16) and adjacent normal tissues (N = 16) was detected by qRT-PCR. (E) miR-197-3p was only miRNA that could bind to circ_0000285 and CKS1B based on starBase prediction. Repetition = 3.
Verification of circ_0000285 targeting miR-197-3p
Considering the possible involvement of miR-197-3p in circ_0000285-modulated glioma progression, we further verified their direct regulatory relationship. Using starBase (http://starbase.sysu.edu.cn/), the circ_0000285 sequence was found to contain a putative binding seed with miR-197-3p ()). The circ_0000285-MUT and circ_0000285-WT luciferase reporter assays were used to determine whether miR-197-3p affected the luciferase activity mediated by circ_0000285-MUT and circ_0000285-WT. The constructed circ_0000285-MUT and circ_0000285-WT luciferase vectors were co-introduced into SHG-44 and U251 cells, in addition to the miR-197-3p mimic or NC. MiR-197-3p mimic had no impact on circ_0000285-MUT-mediated luciferase activity but significantly reduced circ_0000285-WT-mediated luciferase activity ()). A simultaneous increase in circ_0000285 and miR-197-3p levels was also observed in SHG-44 and U251 cells ()). However, miR-197-3p expression levels decreased in glioma tissues and cells () and (e)). Additionally, the binding interaction between circ_0000285 and miR-197-3p was further validated by Pearson correlation analysis, demonstrating a negative correlation between circ_0000285 and miR-197-3p expression in glioma tissues ()). Collectively, these results demonstrate the specificity of circ_0000285 and miR-197-3p interactions.
Figure 4. Circ_0000285 targets miR-197-3p. A. miR-197-3p binding sites prediction of circ_0000285 in Starbase. B. Luciferase reporter experiments were performed with Firefly-luciferase-circ_0000285, control Renilla-luciferase and miR-197-3p mimic or mimic NC, vs.miR-NC,**P < 0.001. C. AGO2-dependent RNA immunoprecipitation (RIP) assay in SHG-44 and U251 cells, vs.Anti-lgG,**P < 0.001. D. Verification of the relative expression level of downregulated miR-197-3p by RT-qPCR in glioma tissues. E. RT-qPCR analysis of the expression of miR-197-3p in glioma cells, vs.NHA,**P < 0.001. F. The Pearson correlation between miR-197-3p mRNA expression and circ_0000285 expression in glioma tissues. N = 3, repetition = 3.
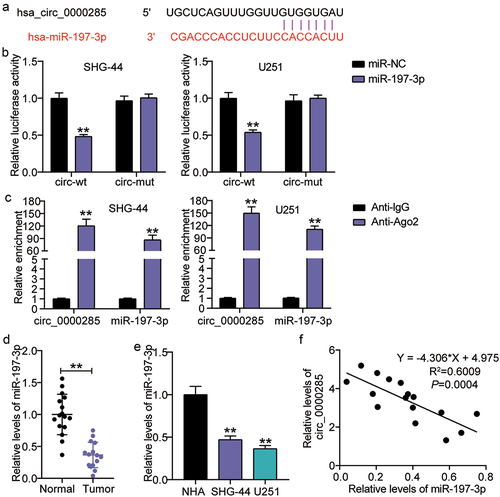
Malignant cell phenotypes suppressed by circ_0000285 silencing are restored upon miR-197-3p inhibition
To determine whether miR-197-3p is required for circ_0000285-mediated glioma cell behavior, we co-transfected an miR-197-3p inhibitor and si-circ_0000285 into SHG-44 and U251 cells. RT-qPCR analysis revealed that circ_0000285 knockdown enhanced endogenous miR-197-3p expression, while the miR-197-3p inhibitor abrogated this effect ()). Increased proliferation occurred in SHG-44 and U251 cells upon miR-197-3p inhibition but was decreased by circ_0000285 silencing ()). Quantification of invasive cells showed that miR-197-3p silencing diminished the invasive behavior of glioma cells, and the promoting effect was offset by si-circ_0000285 ()). A reduction in apoptosis of SHG-44 and U251 cells by the miR-197-3p inhibitor was mitigated by additional transfection with si-circ_0000285 ()). Collectively, our results show that circ_0000285 acts as an oncogene by sponging miR-197-3p.
Figure 5. Cell malignant phenotypes suppressed by circ_0000285 silence is restored upon miR-197-3p inhibitor. SHG-44 and U251 cells were transfected with si-circ_0000285, si-NC, miR-197-3p inhibitor, inhibitor NC, si-circ_0000285+ miR-197-3p inhibitor. 48 h posttransfection. A. RT-qPCR analysis of miR-197-3p expression. B. Cell viability was measured with CCK8. C. Cell invasion was detected by Transwell invasion assay. D. Western blotting analysis of Bax and Bcl-2 expression. Vs.si-NC,**P < 0.001; vs.inhibitor-NC, ++P < 0.001; vs,si-circ+inhibitor, ##P < 0.001. si-circ, circ_0000285 siRNA; si-NC, si-circ negative control; inhibitor, miR-197-3p inhibitor; inhibitor-NC, inhibitor negative control. N = 3, repetition = 3.
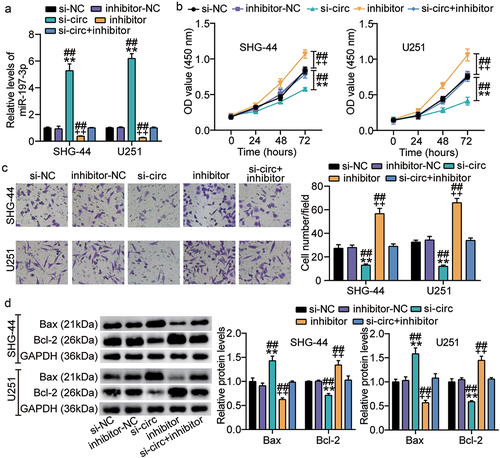
miR-197-3p targets CKS1B
Using starBase analysis, we further verified that miR-197-3p contains sequences complementary to the CKS1B 3ʹUTR ()). To determine the interplay between CKS1B and miR-197-3p, we established CKS1B-3ʹUTR and CKS1B-MUT luciferase constructs and co-transfected the SHG-44 and U251 cells with miR-197-3p mimic or mimic NC. Monitoring luciferase activities showed a reduction in luciferase activity in glioma cells co-transfected with miR-197-3p mimic and CKS1B-MUT luciferase constructs, while no change was observed in other groups ()). These data are consistent with the above-mentioned bioinformatics prediction that miR-197-3p might bind to the 3ʹUTR of CKS1B and edit CKS1B expression in glioma cells. As shown in ), SHG-44 and U251 cells exhibited enhanced expression. The relative levels of CKS1B were negatively correlated with miR-197-3p expression in the glioma tissues ()). In summary, these findings indicate that CKS1B is a direct target of miR-197-3p in glioma cells.
Figure 6. miR-197-3p targets CKS1B. A. Binding sequence of miR-197-3p in CKS1B 3′UTR predicted by starBase. B. SHG-44 and U251 cells were transfected with CKS1B-3ʹUTR-WT or CKS1B-3ʹUTR-MUT luciferase vectors. After 48 h, the luciferase activity was monitored, vs.miR-NC,**P < 0.001 C. RT-qPCR analysis of CKS1B expression in MHA, SHG-44 and U251 cells, vs.NHA,**P < 0.001. D. Pearson Correlation analysis was used in miR-197-3p and CKS1B expression in glioma tissues. N = 3, repetition = 3.
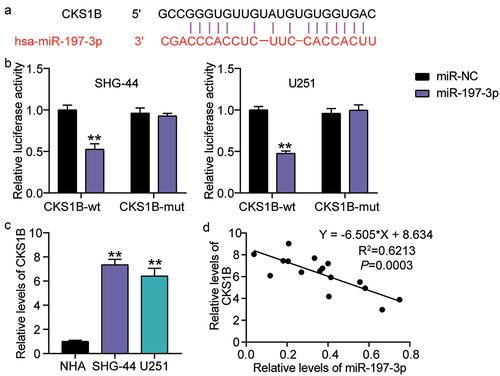
Silencing CKS1B also compromises the malignant behaviors of glioma cells induced by miR-197-3p inhibitor
To confirm that CKS1B is implicated in miR-197-3p-manipulated cell malignant behaviors, we co-introduced si-CKS1B and miR-197-3p inhibitors into SHG-44 and U251 cells. Western blot analysis demonstrated that the miR-197-3p inhibitor mitigated the loss of CKS1B expression caused by si-CKS1B ()). CKS1B silencing arrested the proliferation and invasion of glioma cells, while these effects were nullified by transfection with the miR-197-3p inhibitor () and (c)). In addition, apoptotic stimulation was detected upon CKS1B silencing, while co-introduction of the miR-197-3p inhibitor and si-CKS1B abrogated the activation ()). These data show that miR-197-3p suppresses proliferation and invasion and induces apoptosis activation by downregulating the oncongenicity of CKS1B.
Figure 7. Silencing CKS1B also compromises the malignant behaviors of glioma cells induced by miR-197-3p inhibitor. SHG-44 and U251 cells were cotransfected with si-CKS1B, si-NC, miR-197-3p inhibitor, inhibitor NC, si-CKS1B+ inhibitor. A. Western blotting analysis of CKS1B expression. B. Cell viability was measured with CCK8. C. Cell invasion was detected by Transwell invasion assay. D. Western blotting analysis of Bax and Bcl-2 expression, vs. si-NC, **P < 0.001; vs. inhibitor-NC, ++P < 0.001; vs. si-CKS1B+inhibitor, #P < 0.05, ##P < 0.001. si-CKS1B, CKS1B siRNA; si-NC, si-CKS1B negative control; inhibitor, miR-197-3p inhibitor; inhibitor-NC, inhibitor negative control. N = 3, repetition = 3.
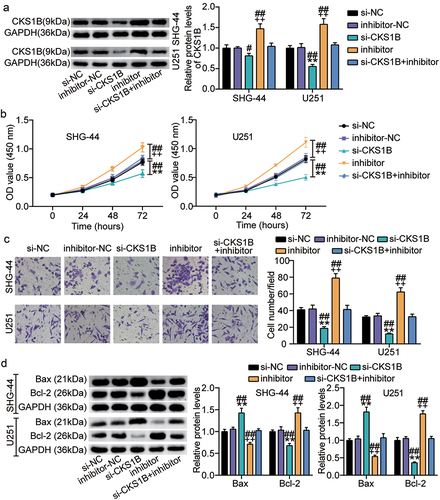
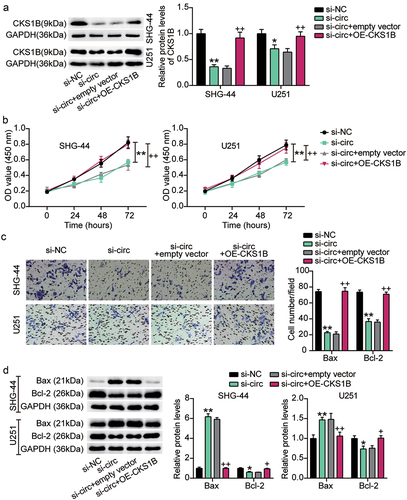
CKS1B overexpression reversed the malignant behavior of glioma cells inhibited by circ_0000285 knockdown
Next, we introduced both OE-CKS1B and si-circ into SHG-44 and U251 cells to assess the effects of CKS1B and circ_0000285 on glioma biological functions. Western blotting analysis showed that si-circ-induced CKS1B deletion was reversed by OE-CKS1B ()). Furthermore, CCK-8 and Transwell assays showed that CKS1B overexpression almost completely alleviated the proliferation and invasion of SHG-44 and U251 cells inhibited by circ_0000285 knockdown (). In addition, compared with the si-circ+empty vector group, Bax protein levels decreased and Bcl-2 levels increased in the si-circ+OE-CKS1B group ()). These data suggest that CKS1B is positively regulated by circ_0000285 and that its overexpression promotes the proliferation and invasion of glioma cells and inhibits apoptosis.
Figure 8. CKS1B overexpression reversed the malignant behavior of glioma cells inhibited by circ_0000285 knockdown. SHG-44 and U251 cells were cotransfected with si-circ, si-NC, si-circ+empty vector, si-circ+OE-CKS1B. A. Western blotting analysis of CKS1B expression. B. Cell viability was measured with CCK8. C. Cell invasion was detected by Transwell invasion assay. D. Western blotting analysis of Bax and Bcl-2 expression, vs. si-NC, *P < 0.05, **P < 0.001; vs. si-circ+empty vector, #P < 0.05, ##P < 0.001. si-circ, circ_0000285 siRNA; si-NC, si-circ negative control; OE-CKS1B, CKS1B overexoression. N = 3, repetition = 3.
Discussion
Previously, circ_0000285 was defined as an oncogenic or anti-oncogenic circRNA in cancer [Citation16,Citation30,Citation31]. However, its function in gliomas remains poorly understood. In the current study, we found that circ_0000285 displayed robust expression in glioma tissues, and circ_0000285 silencing mitigated uncontrolled proliferation and invasion and activated apoptosis. The tumor-suppressing effect of circ_0000285 silencing was also observed in vivo. Luciferase and RNA RIP assays revealed that miR-197-3p is the target of circ_0000285 and CKS1B. Rescue in vitro assays revealed that the inhibitory effect of circ_0000285 silencing or CKS1B knockdown on glioma was attenuated by the miR-197-3p inhibitor. Taken together, our findings indicate that circ_0000285 functions as an oncogenic circRNA by targeting the miR-197-3p/CKS1B axis, thereby affecting glioma cell proliferation, migration, and apoptosis.
Generally, the diverse circRNA-manipulated mechanisms for regulating gene expression depend on their subcellular localization [Citation32]. Cytoplasmic circRNAs function as molecular decoys or ceRNAs to modulate mRNA or protein expression [Citation33]. The ceRNA activity of circRNAs has been reported in various cancers [Citation34–36]. Moreover, circ_0000285 exerts its oncogenic potential via its ceRNA activity toward different miRNAs, such as miR-409-3p [Citation13], mi-654-3p [Citation37], and miR-599 [Citation15]. Moreover, miR-197-3p has been reported to be involved in various cancers. For example, the attenuation of β-catenin signaling by miR-197-3p results in a reduction in the proliferation of prostate cancer cells, leading to tumor suppression [Citation38]. Similarly, miR-197-3p inhibits proliferation, migration, and invasion of cervical cancer cells [Citation39]. Conversely, miR-197-3p overexpression drives bladder cancer cell proliferation and inactivates apoptosis [Citation40]. Therefore, miR-197-3p plays different roles in various cancers. Our cell function assays showed that miR-197-3p blockade increased proliferation and migration and inactivated apoptosis in glioma cells, suggesting its tumor-suppressing role in glioma. Furthermore, the interaction between circ_0000285 and miR-197-3p was verified using luciferase reporter and RIP assays. The decrease in the malignant behavior of glioma cells observed upon circ_0000285 silencing could be explained by the increased expression of miR-197-3p. This interaction was further confirmed by the antagonism of circ_0000285 and miR-197-3p in the glioma tissues.
The CKS1B gene is located on chromosome 1q21.3, and consists of three exons. CKS1B binds to the catalytic subunit of cyclin-dependent kinases and is essential for their biological functions. During the past decades, increasing evidence has demonstrated that CKS1B is overexpressed and plays a pro-tumor role in various cancers [Citation41,Citation42]. For example, CKS1B overexpression reprograms cell metabolism and then favors the colorectal cancer progression in vitro [Citation43]. A meta-analysis including 2,224 cancer participants showed that high CKS1B expression is associated with advanced T stage and lymph node metastasis [Citation44]. In the present study, we elucidated that CKS1B was highly expressed in glioma tissues, and CKS1B silencing blunted glioma cell proliferation and invasion and activated apoptosis. Therefore, CKS1B expression is essential for glioma progression, with respect to promotion of cell proliferation and migration. In contrast, according to bioinformatic analysis, CKS1B is the target of miR-197-3p in gliomas. This prediction was validated using a luciferase reporter assay. Notably, the in vitro growth-suppressing effects of CKS1B-silencing were reverted by miR-197-3p inhibitor treatment. Therefore, miR-197-3p exerts its anti-tumorigenic role in gliomas by downregulating CKS1B expression.
This study has several limitations. First, the number of clinical samples was not sufficient to avoid selection bias. Second, as circRNAs can have multiple targets, the regulatory network controlled by circ_0000285 might be complex and requires further characterization. In addition, CKS1B has been reported to regulate oncogenic signaling circuits in different cancers [Citation42,Citation45,Citation46]; therefore, the downstream signaling events in glioma require further investigation.
Conclusions
In summary, we identified circ_0000285, a novel oncogenic circRNA in glioma. circ_0000285 promotes glioma progression by targeting miR-197-3p and upregulating CKS1B expression. These findings provide insight on novel players and networks involved in glioma tumorigenesis, as well as highlight the clinical potential of blocking circ_0000285 as therapeutic intervention for glioma.
Authors’ contributions
TD and YL conducted the study, collected and analyzed the data. YLY and LYY designed the study and methods. FFL and XBW analyzed and interpreted the data. QYZ and MSX conducted literature analysis and prepared the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Suizhou Hospital, Hubei University of Medicine (Suizhou, China). The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki. All patients signed written informed consent.
Consent for publication
Consent for publication was obtained from the participants.
Supplemental Material
Download MS Word (15 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40(1):1–14.
- Braunstein S, Raleigh D, Bindra R, et al. Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J Neurooncol. 2017;134(3):541–549.
- Angelopoulou E, Piperi C. Emerging role of plexins signaling in glioma progression and therapy. Cancer Lett. 2018;414:81–87.
- Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. 2020;22(12):2162–2169.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Ding C, Yi X, Wu X, et al. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12.
- Jiang Y, Zhou J, Zhao J, et al. The U2AF2 /circRNA ARF1/miR-342-3p/ISL2 feedback loop regulates angiogenesis in glioma stem cells. J Exp Clin Cancer Res. 2020;39(1):182.
- Shi F, Shi Z, Zhao Y, et al. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun. 2019;510(4):614–620.
- Liu X, Shen S, Zhu L, et al. SRSF10 inhibits biogenesis of circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2 pathway. J Exp Clin Cancer Res. 2020;39(1):121.
- Deng Y, Zhu H, Xiao L, et al. Circ_0005198 enhances temozolomide resistance of glioma cells through miR-198/TRIM14 axis. Aging (Albany NY). 2020;13(2):2198–2211.
- Zhou D, Lin X, Wang P, et al. Circular RNA circ_0001162 promotes cell proliferation and invasion of glioma via the miR-936/ERBB4 axis. Bioengineered. 2021;12(1):2106–2118.
- Zhang XJ, Cao G, Fu J, et al. The role of hsa_circ_0000285 in metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2020;24(13):7204.
- Long Z, Gong F, Li Y, et al. Circ_0000285 regulates proliferation, migration, invasion and apoptosis of osteosarcoma by miR-409-3p/IGFBP3 axis. Cancer Cell Int. 2020;20:481.
- Zhang W, Zhang S. Downregulation of circRNA_0000285 suppresses cervical cancer development by regulating miR197-3p-ELK1 axis. Cancer Manag Res. 2020;12:8663–8674.
- Zhang Z, Pu F, Wang B, et al. Hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene Ther. 2020;27(5):186–195.
- Chi BJ, Zhao DM, Liu L, et al. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66(2):197–202.
- Chen L, Shi J, Wu Y, et al. CircRNA CDR1as promotes hepatoblastoma proliferation and stemness by acting as a miR-7-5p sponge to upregulate KLF4 expression. Aging (Albany NY). 2020;12(19):19233–19253.
- Jia Q, Ye L, Xu S, et al. Circular RNA 0007255 regulates the progression of breast cancer through miR-335-5p/SIX2 axis. Thorac Cancer. 2020;11(3):619–630.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Lu J, Lin J, Zhou Y, et al. MiR-328-3p inhibits lung adenocarcinoma-genesis by downregulation PYCR1. Biochem Biophys Res Commun. 2021;550:99–106.
- Bi YN, Guan JP, Wang L, et al. Clinical significance of microRNA-125b and its contribution to ovarian carcinogenesis. Bioengineered. 2020;11(1):939–948.
- Liu H, Chi Z, Jin H, et al. MicroRNA miR-188-5p as a mediator of long non-coding RNA MALAT1 regulates cell proliferation and apoptosis in multiple myeloma. Bioengineered. 2021;12(1):1611–1626.
- Khan AA, Agarwal H, and Reddy SS, et al. MicroRNA 27a is a key modulator of cholesterol biosynthesis. Mol Cell Biol. 2020;40(9):e00470–19.
- Song AF, Kang L, Wang YF, et al. MiR-34a-5p inhibits fibroblast‑like synoviocytes proliferation via XBP1. Eur Rev Med Pharmacol Sci. 2020;24(22):11675–11682.
- Jiang Y, He J, Guo Y, et al. Identification of genes related to low-grade glioma progression and prognosis based on integrated transcriptome analysis. J Cell Biochem. 2020;121(5–6):3099–3111.
- Gao G, Yang M, Wang F, et al. Coagulation factor 2 thrombin receptor promotes malignancy in glioma under SOX2 regulation. Aging (Albany NY). 2020;12(11):10594–10613.
- Liao YX, Zhang ZP, Zhao J, et al. Effects of fibronectin 1 on cell proliferation, senescence and apoptosis of human glioma cells through the PI3K/AKT signaling pathway. Cell Physiol Biochem. 2018;48(3):1382–1396.
- Sincevičiūtė R, Vaitkienė P, Urbanavičiūtė R, et al. MMP2 is associated with glioma malignancy and patient outcome. Int J Clin Exp Pathol. 2018;11(6):3010–3018.
- Plate KH, Breier G, Weich HA, et al. Vascular endothelial growth factor and glioma angiogenesis: coordinate induction of VEGF receptors, distribution of VEGF protein and possible in vivo regulatory mechanisms. Int J Cancer. 1994;59(4):520–529.
- Yang D, Jin Y, Cheng S, et al. The interaction between circular RNA hsa_circ_0000285 and miR-599 in thyroid cancer. Eur Rev Med Pharmacol Sci. 2020;24(13):7219.
- Qin JB, Chang W, Yuan GH, et al. Circular RNA hsa_circ_0000285 acts as an oncogene in laryngocarcinoma by inducing Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(19):9773.
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141.
- Zhu J, Zhang X, Gao W, et al. lncRNA/circRNA‑miRNA‑mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20(4):3160–3174.
- Meinecke-Tillmann S, Evers P, Meinecke B, et al. [The problems of PMSG superovulation treatment of Merino sheep in relation to an embryo transfer program: relation between PMSG plasma concentration and superovulation reaction]. DTW Deutsche Tierarztliche Wochenschrift. 1988;95(4):167–174.
- Wang J, Zhao X, Wang Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32.
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20:173.
- Yao T, Zha D, Hu C, et al. Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene. 2020;747:144661.
- Huang Q, Ma B, Su Y, et al. miR-197-3p represses the proliferation of prostate cancer by regulating the VDAC1/AKT/β-catenin signaling axis. Int J Biol Sci. 2020;16(8):1417–1426.
- Gu Q, Hou W, Shi L, et al. Circular RNA ZNF609 functions as a competing endogenous RNA in regulating E2F transcription factor 6 through competitively binding to microRNA-197-3p to promote the progression of cervical cancer progression. Bioengineered. 2021;12(1):927–936.
- Jiang Y, Wei T, Li W, et al. Circular RNA hsa_circ_0002024 suppresses cell proliferation, migration, and invasion in bladder cancer by sponging miR-197-3p. Am J Transl Res. 2019;11(3):1644–1652.
- Shi W, Huang Q, Xie J, et al. CKS1B as drug resistance-inducing gene-A potential target to improve cancer therapy. Front Oncol. 2020;10:582451.
- Zeng Z, Gao ZL, Zhang ZP, et al. Downregulation of CKS1B restrains the proliferation, migration, invasion and angiogenesis of retinoblastoma cells through the MEK/ERK signaling pathway. Int J Mol Med. 2019;44(1):103–114.
- Wang X, Tao G, Huang D, et al. Circular RNA NOX4 promotes the development of colorectal cancer via the microRNA‑485‑5p/CKS1B axis. Oncol Rep. 2020;44(5):2009–2020.
- Zhang Y, Chen Y, You F, et al. Prognostic and clinicopathological significance of Cks1 in cancer: evidence from a meta-analysis. J Cell Physiol. 2019;234(8):13423–13430.
- Wang H, Zhang Z, Yan Z, et al. CKS1B promotes cell proliferation and invasion by activating STAT3/PD-L1 and phosphorylation of Akt signaling in papillary thyroid carcinoma. J Clin Lab Anal. 2021;35(1):e23565.
- Shi L, Wang S, Zangari M, et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget. 2010;1(1):22–33.
