ABSTRACT
miR-139-3p exerts tumor-suppressing functions in various cancers. We analyzed and identified that miR-139-3p expression was notably low in gastric cancer (GC) via edgeR differential analysis based on The Cancer Genome Atlas database and quantitative real-time polymerase chain reaction (qRT-PCR) assay. The binding relationship between Kinesin Family Member 18B (KIF18B) and miR-139-3p was predicted by bioinformatics databases, and verified through dual-luciferase assay. Western blot and qRT-PCR results also indicated that miR-139-3p restrained KIF18 expression at mRNA and protein levels. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, wound healing, transwell, flow cytometry assays were introduced to evaluate cell proliferation, migration, invasion, and cell cycle, respectively, where the results indicated that upregulating miR-139-3p inhibited proliferative, migratory, and invasive abilities of GC cells, while caused cell-cycle arrest. Moreover, the results of rescue experiments illustrated that miR-139-3p hampered the progression of GC cells by targeting and suppressing KIF18B. To sum up, we concluded that miR-139-3p suppressed GC progression by targeting KIF18B.
GRAPHICAL ABSTRACT

1. Introduction
Gastric cancer (GC) has high mortality and morbidity worldwide [Citation1,Citation2]. In 2018, it was estimated that 1.03 million new GC cases worldwide ranked sixth in the cancer cases and 780,000 deaths ranked second in cancer deaths [Citation3]. Survival rate of early GC within 5 years can reach more than 95%, but most patients are diagnosed in advanced stage because of low rate of early diagnosis [Citation4]. Combination treatment with neoadjuvant therapy is used for patients with advanced GC, but the survival rate within 5 years is not optimistic of about 20% to 35% [Citation5–7]. Hence, it is vital to further discuss pathogenesis of GC and find potential molecular targets to promote the progress of GC treatment.
Non-coding RNAs are involved in GC progress, where many miRNAs have been reported as tumor promoting or suppressing factors. These miRNAs promote GC in diverse ways. For example, miR-20a-5p facilitates GC cell proliferative ability by targeting X-linked gene and activating phosphatidylinositol 3-kinase/Protein Kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway [Citation8]. Downregulation of miR-1265 in GC causes overexpression of calcium-binding protein 39 and promotes oncogenic autophagy, which helps with tumor growth in GC [Citation9]. miR-106a-3p overexpression suppresses adenomatous polyposis coli expression and induces metastasis by activating Wnt/beta-catenin pathway [Citation10]. Accumulating understandings on the effects of miRNAs on cancer cell phenotype help a lot with developing cancer prognostic prediction and therapy [Citation11,Citation12]. However, it has not been fully understood on functions of miRNAs in GC. miR-139 is considered as a tumor suppressor in several cancers [Citation13]. Both miR-139-3p and miR-139-5p were reported with their tumor-suppressing molecular mechanisms [Citation13,Citation14]. However, it is still valuable to furtherly explore their roles in cancer progression.
Above all, we assumed that miR-139-3p exerted its tumor-suppressing effects on GC by targeting Kinesin Family Member 18B (KIF18B) and blocking its post-transcriptional process. To identify our assumption, we designed a set of in-vitro experiments along with bioinformatics analyses. At last, we concluded that miR-139-3p restrained GC cell process by targeting KIF18B.
2. Materials and methods
2.1. Information of cell lines
Normal gastric mucosa epithelial cell line GES1 (BNCC337969), and human GC cell lines MKN-45 (BNCC337682), AZ521 (BNCC338085), MGC-803 (BNCC100665), NCI-N87 (BNCC338548), SNU-1 (BNCC100338) were all acquired from BeNa Culture Collection (China). Roswell Park Memorial Institute 1640 (Gibco, 11,875,093) medium plus 10% fetal bovine serum (FBS; Gibco, 10099141C) was used for cell incubation at 37°C with 5% CO2.
2.2. Bioinformatics approaches
MiRNA and mRNA expression data of GC were obtained from The Cancer Genome Atlas (TCGA) database. Differentially expressed miRNAs (DEmiRNAs) and mRNAs (DEmRNAs) were acquired through differential analysis using edgeR (|logFC|>2, padj<0.05). miR-139-3p was screened through literature review, and its potential targets were predicted through databases (miRDB, TargetScan and miRWalk), so as to obtain the mRNA with targeted binding sites to the target miRNA. Target mRNA was finally determined by Pearson analysis [Citation15,Citation16].
2.3. Cell transfection
miR-139-3p mimic, the negative control for mimic (mimic NC), miR-139-3p inhibitor, and the negative control for inhibitor (inhibitor NC) were provided by Shanghai GenePharma Pharmaceutical Technology Co., LTD. Small interfering RNA (siRNA) targeting KIF18B was synthesized by Sangon Biotech (Shanghai) Co., Ltd. The pEGFP1-KIF18B (oe-KIF18B) recombinant plasmid was constructed based on pEGFP1 overexpression vector to construct KIF18B overexpression plasmid. Lipofectamine® 3000 (Invitrogen inc., USA) was utilized for transfecting cells [Citation17].
2.4. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Trizol (Invitrogen) was applied to isolate total RNA. Reverse transcription of RNA was conducted using reverse transcription kit (Invitrogen) for obtaining corresponding cDNA. qRT-PCR was completed using miScript SYBR Green PCR Kit (Qiagen, Germany). KIF18B and miR-139-3p were standardized to glyceraldehyde-3phosphate dehydrogenase and U6 Small Nuclear RNA, respectively. 2−ΔΔCt method was introduced for calculating RNA relative expression. Corresponding Primers were exhibited in Supplementary table S1 [Citation18].
2.5. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The SNU-1 cells (5 × 103) were inoculated into 96-well plates. Cell proliferative ability was detected using sterile MTT solution (Beyotime) after cell culture for 12, 24, 48, 72 h. Absorbance at 570 nm was detected by a microplate reader (Sunnyvale, CA, USA) [Citation19].
2.6. Wound healing assay
SNU-1 cells (1 × 106) were inoculated in a 6-well plate. After 80% confluence of cells was reached, the plate was scratched sousing the 200-µl pipette tip. Cell migration status at 0 h and 24 h was photographed [Citation20].
2.7. Transwell assay
2 × 104 cells were inoculated in the upper compartment with 100 μl of serum-free Dulbecco’s Modified Eagle’s Medium (DMEM) and Matrigel. DMEM with 10% FBS (Thermo fisher, USA) was poured into the lower compartment. The migrated cells were stained using 0.5% crystal violet. Then, migration status was observed and photographed using a microscope [Citation20].
2.8. Western blot
After obtaining cell lysate, equal protein amounts were loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis at 100 V, and later were transferred to the nitrocellulose membrane at 100 mA for 120 min. After blocking process, the membrane was reacted with the primary antibodies at 4°C overnight. Next, the secondary antibody goat anti-rabbit IgG labeled with horseradish peroxidase was used to incubate with membrane at room temperature for 120 min. Then luminescence reaction was carried out with the Efficient chemiluminescence (ECL) kit (Solarbio, Beijing, China). Protein imprinting was observed by photographing. Antibodies used were shown in Supplementary table S2. The assay was conducted for three times [Citation18].
2.9. Dual-luciferase assay
KIF18B wild-type (WT) or mutated-type (MUT) 3’-UTR inserted vectors were constructed based on psiCHECK luciferase reporter vector (Sangon Co., LTD, Shanghang, China). GC cells (Thermo fisher, USA) were seeded in a 48-well plate for 24 h of culture. Then miR-139-3p mimic/mimic NC and psiCHECK WT/MUT vectors were transfected into cells together. Luciferase activity was finally detected using luciferase assay reagent (Promega, Fitchburg, WI, USA) [Citation20].
2.10. Flow cytometry (FCM) assay
The cells in each group were fixed in 70% ethanol for 24 h at 4°C. After being washed with PBS, cells were treated with propidium iodide and ribonuclease for 30 min at 4°C. FCM was used to detect cell cycle following the standard process. The number of cells were counted. Cell cycle distribution were analyzed by ModFit with cell cycle fitting software [Citation18].
2.11. Statistical analysis
Prism 8.0 statistical software was utilized for data analysis, and analysis results were presented as mean ± standard deviation. Student’s t-test was conducted for comparisons of two groups, and one-way analysis of variance was used for analyzing comparisons among multiple groups. In the manuscript, * represented P< 0.05, indicating a remarkable difference.
3. Results
We proposed a hypothesis that miR-139-3p suppressed GC cell progress by targeting its downstream mRNA. To verify the hypothesis, cell experiments and bioinformatics prediction were introduced. We found that miR-139-3p targeted KIF18B and blocked tumor progress in GC.
3.1 miR-139-3p expression is low in GC
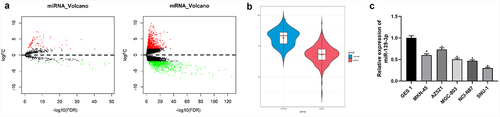
90 DEmiRNAs and 1677 DEmRNAs were acquired through differential expression analysis (). Previous literature illustrated that miR-139-3p, which was contained in the differentially downregulated miRNAs in our analysis, was decreased in many cancers and was closely related to cancer development [Citation21–23]. Decreased miR-139-3p expression status was also showed in GC tissue in this study (). Hence, miR-139-3p was chosen as the study object. miR-139-3p expression status in GC cell lines were assayed by qRT-PCR, in which the result indicated that in comparison with normal gastric mucosal epithelium cell line, miR-139-3p was remarkably less expressed in GC cell lines (). SNU-1 with the lowest miR-139-3p level was utilized for subsequent study.
Figure 1. miR-139-3p is poorly expressed in GC. (a) Volcano plot of DEmRNAs and DEmiRNAs in normal and tumor groups. Red: up-regulated genes, green: down-regulated genes; (b) miR-139-3p expression status in normal and tumor samples; (c) Expression status of miR-139-3p in various cell lines. * P< 0.05.
3.2 miR-139-3p overexpression suppresses cell proliferation, migration, invasion, and arrests cell cycle in GC
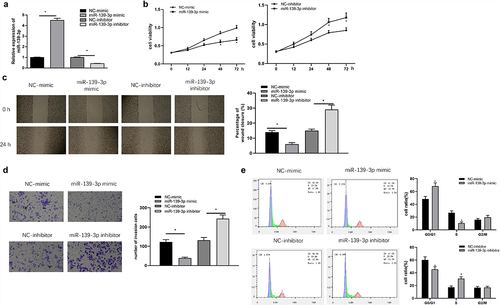
To explore the impacts of miR-139-3p on GC cell behaviors, we transfected miR-139-3p mimic/inhibitor into SNU-1. As shown by qRT-PCR (), the mimic and inhibitors worked well in the cells. Biological function changes of SNU-1 cells after transfection were further detected. The results of MTT (), wound healing (), and transwell () assays presented that proliferative, migratory, and invasive abilities of SNU-1 cells were dramatically suppressed by up-regulating miR-139-3p, while silencing miR-139-3p resulted in opposite results. In addition, result of FCM showed that upregulating miR-139-3p retained SNU-1 cells in G0/G1 phase, while silencing miR-139-3p increased number of SNU-1 cells in the division phase, indicating increased cell proliferation ability (). It could be noted that miR-139-3p restrained progression of GC cells, possibly leading to cell-cycle arrest in G0/G1 phase.
Figure 2. miR-139-3p restrains cell proliferation, migration, invasion, and arrests cell cycle in GC. (a) miR-139-3p level was measured by qRT-PCR; MTT (b), wound healing (c), Tranwell (d) and FCM (e) assays were for cell proliferation, migration and invasion along with cell cycle changes detection, respectively. * P< 0.05.
3.3. miR-139-3p can directly target KIF18B
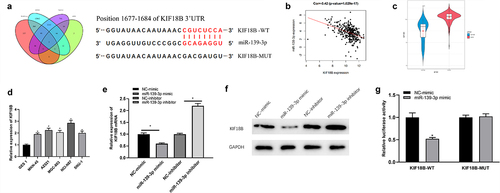
To conduct an in-depth investigation on miR-139-3p modulating biological functions of GC cells, potential target mRNAs of miR-139-3p were predicted based on miRDB, TargetScan, and miRWalk databases. The predicted genes were intersected with 908 upregulated DEmRNAs and then the DEmRNA with targeted binding sites to miR-139-3p was gained (). miR-139-3p negatively modulated KIF18B expression (), and KIF18B level was remarkably high in cancer tissues as TCGA data presented (). Also, we observed the same expression trend of KIF18B in GC cell lines (). qRT-PCR and Western blot indicated that forced miR-139-3p expression decreased KIF18B expression, while silencing miR-139-3p increased KIF18B expression (). Additionally, upregulating miR-139-3p suppressed luciferase activity in KIF18B-WT group, while made no difference on luciferase activity in KIF18B-MUT (). Based on above-mentioned results, miR-139-3p targeted and inhibited KIF18B.
Figure 3. miR-139-3p can target KIF18B directly. (a) Venn diagram of potential targets of miR-139-3p and up-regulated DEmRNAs, and putative binding sites of miR-139-3p on KIF18B 3ʹUTR; (b) Correlation between KIF18B and miR-139-3p; (c) KIF18B expression in normal and tumor samples; (d) KIF18B mRNA level was measured in GC cell lines; E-F: KIF18B mRNA and protein expression upon overexpression or inhibition of miR-139-3p were tested by qRT-PCR (e) and Western blot (f); (g) The binding relationship of miR-139-3p and KIF18B. * P< 0.05.
3.4. KIF18B promotes cell proliferation, migration, invasion, and cell cycle
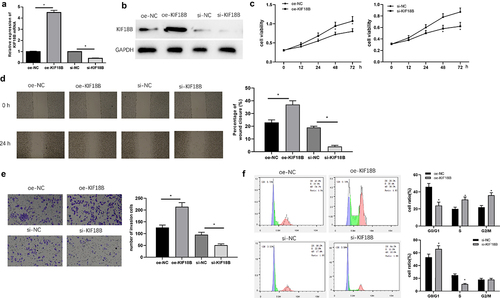
KIF18B has been proved to be overexpressed in tumor tissues and promote malignant process of different tumors [Citation24,Citation25]. However, its effects on GC cells are much less reported. The expression of KIF18B in SNU-1 cell line was dramatically increased after cells were transfected with KIF18B overexpression vector, while that was significantly decreased after cells were transfected with si-KIF18B at mRNA and protein manners (). Further MTT (), wound healing (), and transwell () showed that upregulating KIF18B significantly enhanced SNU-1 cell abilities in proliferation, migration and invasion, while downregulating KIF18B caused the opposite trend. Moreover, the result of FCM exhibited that upregulating KIF18B promoted SNU-1 cell division phase, while silencing KIF18B expression blocked SNU-1 cells in G0/G1 phase (). All above, overexpressed KIF18B promoted cell progression of GC cells.
Figure 4. KIF18B enhances cell proliferation, migration, invasion, and activates cell cycle of GC. After overexpression or inhibition of KIF18B expression, KIF18B mRNA and protein levels were evaluated by qRT-PCR (a) and Western blot (b). Cell proliferative, migratory, invasive abilities and cell cycle changes were tested by MTT (c), wound healing (d), transwell (e) and FCM (f) assays, respectively. * P< 0.05.
3.5. miR-139-3p/KIF18B regulatory axis modulates GC cell behaviors
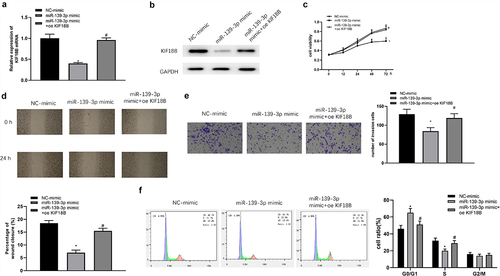
To identify the effects of miR-139-3p/KIF18B axis on GC cells, we co-transfected oe-KIF18B/oe-NC and miR-139-3p mimic/NC mimic into SNU-1 cells. As suggested by qRT-PCR and Western blot, reduced expression of KIF18B caused by overexpressing miR-139-3p could be reversed once the cells were transfected with oe-KIF18B (). MTT (), wound healing (), transwell (), and FCM () results indicated that the suppressive impacts of miR-139-3p overexpression on GC cell functions were restored by overexpression of KIF18B. These results illustrated that miR-139-3p hampered GC cell proliferation, invasion, migration, and arrested cell cycle by suppressing KIF18B.
Figure 5. miR-139-3p restrains proliferation, migration, invasion, and arrests cell cycle in GC by regulating KIF18B. With SNU-1 cells transfected with miR-139-3p mimic and oe-KIF18B, KIF18B mRNA and protein expression was assayed by qRT-PCR (a) and Western blot (b). Cell proliferation, migration, invasion and cell cycle changes were tested by MTT (c), wound healing (d), transwell (e) and FCM (f) assays. * represents the comparison with NC mimic group and # represents comparison with miR-139-3p mimic group.
4. Discussion
The anti-tumor role of miR-139 has been identified in multiple cancers. Ayaka Koma et al. predicted the anti-tumor roles of miR-139-3p and miR-139-5p and their potential targets in neck squamous cell carcinoma, and Cox regression was used for constructing a prognostic model based on the targets [Citation26]. In esophageal squamous cell carcinoma research, Shuo Yan and his colleagues figured out the tumorigenesis role of lncRNA BCAR4, which sponged miR-139-3p to suppress p53/p21 pathway [Citation27]. Compared with the findings above, the trend of our results is consistent with theirs, though we conducted the assays in GC cells. Moreover, we examined cell cycle change by overexpressing miR-139-3p and observed that G0/G1 phase was notably increased while S phase was reduced, indicating that cell cycle was arrested, which was newly reported as the anti-tumor role of miR-139-3p in GC cells.
KIF18B is commonly understood as an oncogene, and several studies reported its tumorigenesis role. For instance, in Bing Hong’s research, KIF18B could upregulate poly ADP-ribose polymerase expression, resulting in enhance of oxaliplatin resistance in colorectal cancer [Citation28]. According to Meng-Jun Qiu’s bioinformatics analysis, Cox regression analysis was processed to identify the prognostic significance of KIF18B in different cancers based on multiple public databases, concluding KIF18B, regarded as a risk factor, could be a promising prognostic biomarker for pan-cancer [Citation29]. Yu-Peng Wu identified that PI3K-AKT-mTOR pathway was activated by KIF18B overexpression, which induced aggressive cell behaviors [Citation30]. Although it was well understood that KIF18B served as an oncogene, no corresponding experimental identifications were processed in GC cells. This study pointed out the molecular mechanism between KIF18B and miR-139-3p, and identified the tumorigenesis role of KIF18B in GC. As can be seen, miR-139-3p/KIF18B axis can provide theoretical supports to develop novel molecular targets in GC.
Some novelties were presented by our study, but limitations remain yet. Lacking in-vivo identification was the most concerned one. To identify miR-139-3p/KIF18B axis in vivo, we are planning to construct mouse models and examine the impacts of miR-139-3p/KIF18B axis on tumor growth in mouse models. For another concern, we have not fully identified the effects of KIF18B on GC cell behaviors. Therefore, we are also planning to furtherly investigate whether miR-139-3p/KIF18B axis can affect angiogenesis, ferroptosis, and lipid drop formation in GC cells.
5. Conclusion
In summary, miR-139-3p was notably less expressed and served as a tumor suppressor in GC. Also, we identified that miR-139-3p hampered GC cell progression by downregulating KIF18B level. This study offers a novel theoretical basis for targeted therapy development of GC.
Authors’ contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Supplemental Material
Download Zip (28.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388(10060):2654–2664.
- Ajani JA, Lee J, Sano T, et al. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3(1):17036.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626.
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713.
- Lazăr DC, Avram MF, Romoșan I, et al. Prognostic significance of tumor immune microenvironment and immunotherapy: novel insights and future perspectives in gastric cancer. World J Gastroenterol. 2018;24(32):3583–3616.
- Yang W, Raufi A, Klempner SJ. Targeted therapy for gastric cancer: molecular pathways and ongoing investigations. Biochim Biophys Acta. 2014;1846(1):232–237.
- Li J, Ye D, Shen P, et al. Mir-20a-5p induced WTX deficiency promotes gastric cancer progressions through regulating PI3K/AKT signaling pathway. J Exp Clin Cancer Res. 2020;39(1):212.
- Xu Z, Li Z, Wang W, et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett. 2019;449:226–236.
- Yang XZ, Cheng TT, He QJ, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17(1):126.
- Li X, Zhang L, Guo X, et al. Self-assembled RNA nanocarrier-mediated chemotherapy combined with molecular targeting in the treatment of esophageal squamous cell carcinoma. J Nanobiotechnology. 2021;19(1):388.
- Cao D, Cao X, Jiang Y, et al. Circulating exosomal microRNAs as diagnostic and prognostic biomarkers in patients with diffuse large B-cell lymphoma. Hematol Oncol. 2021. DOI:10.1002/hon.2956.
- Stavast CJ, van Zuijen I, Karkoulia E, et al. The tumor suppressor MIR139 is silenced by POLR2M to promote AML oncogenesis. Leukemia. 2021. DOI:10.1038/s41375-021-01461-5.
- Zhu X, Jiang S, Wu Z, et al. Long non-coding RNA TTN antisense RNA 1 facilitates hepatocellular carcinoma progression via regulating miR-139-5p/SPOCK1 axis. Bioengineered. 2021;12(1):578–588.
- Wang Y, Zhang J, Liu M, et al. Clinical values and potential pathways of miR-183-5p in gastric cancer: a study based on integrational bioinformatics analysis. J Gastrointest Oncol. 2021;12(5):2123–2131.
- Zhang L, Zou L, Sun P. Relationship between miR-378c and YY1 expression in patients with gastric cancer and the clinicopathological features. Cell Mol Biol Lett. 2021;26(1):12.
- Zou W, Wang Y, Song Q, et al. Ultrasound-targeted microbubble destruction mediated miR-492 inhibitor suppresses the tumorigenesis in non-small cell lung cancer. Ann Med. 2021;53(1):2246–2255.
- Yang Y, Gao M, Li Y, et al. LncRNA CTBP1-AS2 facilitates gastric cancer progression via regulating the miR-139-3p/MMP11 axis. Onco Targets Ther. 2020;13:11537–11547.
- Jiang L, Zhang Y, Guo L, et al. Exosomal microRNA-107 reverses chemotherapeutic drug resistance of gastric cancer cells through HMGA2/mTOR/P-gp pathway. BMC Cancer. 2021;21(1):1290.
- Li Z, Cheng Y, Fu K, et al. Circ-PTPDC1 promotes the progression of gastric cancer through sponging Mir-139-3p by regulating ELK1 and functions as a prognostic biomarker. Int J Biol Sci. 2021;17(15):4285–4304.
- Zhang W, Xu J, Wang K, et al. miR‑139‑3p suppresses the invasion and migration properties of breast cancer cells by targeting RAB1A. Oncol Rep. 2019;42(5):1699–1708.
- Xue F, Li QR, Xu YH, et al. MicroRNA-139-3p inhibits the growth and metastasis of ovarian cancer by inhibiting ELAVL1. Onco Targets Ther. 2019;12:8935–8945.
- Xia Z, Yang X, Wu S, et al. LncRNA TP73-AS1 down-regulates miR-139-3p to promote retinoblastoma cell proliferation. Biosci Rep. 2019;39(5). DOI:10.1042/BSR20190475.
- Wu Y, Wang A, Zhu B, et al. KIF18B promotes tumor progression through activating the Wnt/β-catenin pathway in cervical cancer. Onco Targets Ther. 2018;11:1707–1720.
- Chen FT, Zhong FK. Kinesin family member 18A (KIF18A) contributes to the proliferation, migration, and invasion of lung adenocarcinoma cells in vitro and in vivo. Dis Markers. 2019;2019:6383685.
- Koma A, Asai S, Minemura C, et al. Impact of oncogenic targets by tumor-suppressive miR-139-5p and miR-139-3p regulation in head and neck squamous cell carcinoma. Int J Mol Sci. 2021;22(18):9947.
- Yan S, Xu J, Liu B, et al. Long non-coding RNA BCAR4 aggravated proliferation and migration in esophageal squamous cell carcinoma by negatively regulating p53/p21 signaling pathway. Bioengineered. 2021;12(1):682–696.
- Hong B, Lu R, Lou W, et al. KIF18b-dependent hypomethylation of PARPBP gene promoter enhances oxaliplatin resistance in colorectal cancer. Exp Cell Res. 2021;407(2):112827.
- Qiu MJ, Wang QS, Li QT, et al. KIF18B is a prognostic biomarker and correlates with immune infiltrates in pan-cancer. Front Mol Biosci. 2021;8:559800.
- Wu YP, Ke ZB, Zheng WC, et al. Kinesin family member 18B regulates the proliferation and invasion of human prostate cancer cells. Cell Death Dis. 2021;12(4):302.
