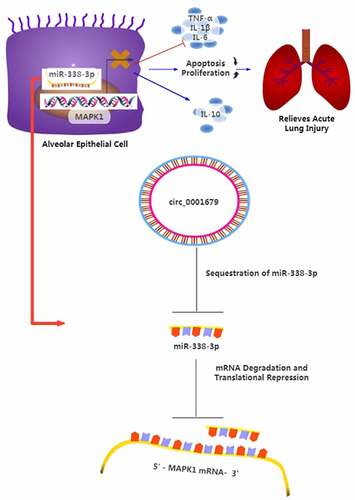
ABSTRACT
The upregulation of circ_0001679 was reported in lipopolysaccharide (LPS)-induced lung injury mouse model, but its functional roles and mechanisms in LPS-induced lung injury remain to be investigated. In this study, we aimed to explore the potential role of circ_0001679 in septic acute lung injury. We initially established an in vitro lung cell injury model using LPS-treated MLE-12 cells. siRNAs targeting circRNA_0001679 were employed to stably knock down circRNA_0001679, followed by functional assays to investigate the effect of circRNA_0001679 silencing. The levels of inflammatory cytokines such as IL-6, IL-β and TNF-α (Tumor necrosis factor-α) were detected by ELISA (Enzyme-linked immunosorbent assay). Meanwhile, protein levels of Bcl-2, cleaved-caspase 3, Bax, and MAPK1 (Mitogen-Activated Protein Kinase 1) proteins expression level were measured by Western blot. We found that Circ_0001679 was upregulated in LPS-induced MLE-12 cells, and silencing circ_0001679 attenuated the growth inhibition and suppressed apoptosis induced by LPS. Circ_0001679 knockdown also lowered levels of IL-6, IL-β and TNF-α, and prevent the activation of cleaved-caspase 3 protein. We further revealed that circ_0001679 functioned as a sponge of miR-338-3p to negatively regulate miR-338-3p activity. miR-338-3p downregulated its downstream target MAPK1, while the upregulation of circ_0001679 maintained a high-level expression of MAPK1 by suppressing miR-338-3p. Collectively, our study indicates that circ_0001679/miR-338-3p/MAPK1 axis may play an important role in the pathogenesis of acute lung injury (ALI).
Graphical abstract
Introduction
A cute respiratory distress syndrome (ARDS)/acute lung injury (ALI) is characterized by non-cardiogenic pulmonary edema, acute respiratory distress, as well as persistent inflammatory response caused by various pulmonary and external factors [Citation1]. ALI/ARDS treatment modalities include general supportive care and protective mechanical ventilation [Citation2]. Although these treatments have improved the survival rate of ALI/ARDS patients, the therapeutic outcome has not met expectations with a mortality rate between 40% and 46% [Citation3]. Therefore, understanding the molecular mechanisms underlying the pathogenesis of acute lung injury is crucial for formulating novel therapeutic approaches.
The main pathophysiological features of ALI/ARDS include the migration of inflammatory cells, fibroproliferation, and activation of apoptosis in alveolar epithelial cells [Citation4]. Inflammatory responses have been recognized as a key factor in ALI pathophysiology and a self-limiting inflammatory response is crucial for regulating the progression of ALI [Citation5]. In the early stage of ALI, local neutrophils and macrophages infiltrate in the lung tissues and secrete various cytokines like TNF-α, IL-6, and IL-1β, which can further activate leukocytes and cause systemic inflammatory response syndrome (SIRS) if inflammation is not controlled [Citation6,Citation7]. Inflammatory responses and cytokines therefore are key contributors to the development and pathogenic progression of ALI/ARDS [Citation8]. During the development of ALI/ARDS, alveolar epithelial cells (AEC) are the main target cells that suffer from cell damage and apoptosis due to inflammations [Citation9]. Apoptosis can be triggered by external pathways or internal pathways originating from the mitochondria. Both pathways can eventually activate cysteine aspartate-specific proteases (caspases), which function as the executioner proteases inducing morphological and biochemical changes during apoptosis [Citation10]. After the onset of diffuse alveolar injury, DNA damage level is increasing and there is an elevated expression of pro-apoptotic protein Bax in Alveolar epithelial cell (AEC) [Citation11]. In the treatment of ALI/ARDS, interventions to control the ‘waterfall’ effect of inflammatory responses and ‘cytokine storms’ in the lung are considered as key measures to prevent alveolar epithelial cell apoptosis [Citation12].
The etiology of ALI is complicated, including sepsis, pneumonia, gastric content aspiration, severe trauma, acute pancreatitis, and blood transfusion [Citation13]. Among these pathogenic factors, sepsis is the most common cause of ALI/ARDS [Citation14]. Lipopolysaccharide (LPS) is the main component of gram-negative bacterial cell wall that stimulates inflammatory cells and activates multiple pathways like JAK/STAT, MAPK, or NF-κB to induce inflammatory cytokines [Citation15–17]. LPS has been recognized as an important factor in ALI-related inflammatory reaction, which is widely used for inducing acute lung injury in animal and cell models [Citation18,Citation19]. Circular RNAs (circRNAs) have been recently implicated in sepsis-related lung injury and inflammatory responses [Citation20,Citation21]. CircRNAs are non-coding RNAs with a covalent closed-loop structure, which are highly stable and show cell-type specific expression. They can not only sponge RNA-binding proteins (RBPs) and microRNAs (miRNAs), but also modulate the translation and transcription of genes involved in a wide range of biological processes [Citation20,Citation21]. MicroRNAs (miRNAs) are small endogenous non-coding RNAs (19–23 nucleotides) which can modulate post-transcriptional gene expression [Citation22]. miRNAs have been found to regulate numerous cellular processes, including cell growth, invasion, apoptosis, differentiation, inflammatory response, immune function and tumorigenesis [Citation23,Citation24]. In sepsis-induced lung injury, circRNAs have been reported to modulate inflammatory responses by sponging specific miRNAs [Citation25,Citation26]. Circ_0001679 was found to be highly expressed in a mice model of ALI, and P2X7R antagonist ameliorates lung injury by regulating the expression of circ_0001212 and circ_0001679 [Citation21]. In human bronchial epithelial cells, it has been reported that circ_0038467 is able to regulate LPS-induced inflammatory injury by sponging miR-338-3p [Citation26].
In our exploratory study, we found that circ_0001679 was upregulated in LPS-induced MLE-12 cells, we therefore hypothesized that circ_0001679 plays a functional role in LPS-induced cell damages in MLE-12 cells. siRNAs targeting circRNA_0001679 were employed to stably knock down circRNA_0001679 and we investigated the cell damages and inflammatory responses after circRNA_0001679 silencing. We further identified miR-338-3p as a downstream miRNA target of circRNA_0001679, and we performed functional assays to demonstrate that circ_0001679/miR-338-3p/ MAPK1 axis may play an important role in the cell model of ALI.
Materials and methods
Cell culture
Mouse lung epithelial MLE-12 cells were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured with 1640 medium (Invitrogen, CA, USA) containing 1% penicillin–streptomycin (Gibco, MD, USA) and 10% fetal bovine serum (FBS, Invitrogen, CA, USA). Cells were maintained in a humified cell incubator at 37°C, 5% CO2. For cell damage induction, LPS was applied at 0.5 μg/ml [Citation27], the induction time was indicated in the figure legend of each experiment. Additionally, to investigate the role of MAPK1 on LPS-induced inflammatory injury, cells were treated with 10 μM of PD98059 (MCE, NH, USA), a MAPK1 inhibitor, for 24 h. Cell line authentication was performed by STR DNA profiling analysis.
Cell transfections
Small interfering RNA (siRNA) targeting circ_0001679 (si-circ_0001679#1, si-circ_0001679#2 and si-circ_0001679#3), circ_0001679 overexpression vector (circ_0001679), the scrambled siRNA negative control (si-NC), miR-338-3p mimic, miR-338-3p inhibitor and miRNA negative control (miR-NC), MAPK1 overexpression vector (MAPK1) and negative control were purchased from GenePharma (Shanghai, China). Cells were harvested by 0.25% trypsin digestion, and seeded into 6-well plate at the density of 3 × 105 cells/well. When cell confluence reached approximately 70%, the cells were transfected with different plasmids, siRNA or miRNA mimic/inhibitor using Lipofectamine 2000 reagent (Invitrogen, MA, USA) [Citation25]. 48 hours post transfection, cells were treated with LPS (0.5 μg/ml) for 24 hours before functional assays. The sequences of synthesized oligonucleotides were displayed as below. Si-circ_0001679#1; 5’-AGACGAGAAUCCUGAGAAACA-3’, Si-circ_0001679#2 5’-CAAAGAGAGUUCGAAUAAAGG-3’; Si-circ_0001679#3 5’-GCAAAGAGAGUUCGAAUAAAG-3’; miR-338-3p 5ʹ-UCCAGCAUCAGUGAUUUUGUUG-3ʹ; miR-338-3p inhibitor 5ʹ-CAACAAAAUCACUGAUGCUGGA-3ʹ;circ_0001679 5ʹ-CTGAGCATCTTCCGTTTG-3ʹ; MAPK1 5ʹ-AGAACATCATTGGCATCA-3ʹ.
Apoptosis detection by flow cytometry
The detection of cell apoptosis was performed using the FITC Annexin V Apoptosis Detection Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. MLE-12 cells in different treatment groups were harvested by 0.25% trypsin digestion and cell concentration was adjusted to 1 × 106/ml. 1 μL Annexin V-FITC and 1 μL PI were added to 1 mL cell resuspension for 30-min incubation in the dark. Stained cells were washed twice with PBS and the percentage of apoptotic cells was detected by BD FACS CantoTM II Flow Cytometer (BD Biosciences) [Citation28].
Cell counting kit-8 (CCK-8) assay
MLE-12 cells in different treatment groups were harvested by 0.25% trypsin digestion and cells were seeded in a 96-well-plate at a density of 1500 cell/well. Cells were cultured in a humidified cell culture incubator for 0, 24, 48, 72 and 96 hours, respectively. 10 μL CCK8 reaction solution (Solarbio, Beijing, China) was added to the cell culture at indicated time point and incubated for 3 h in a humidified cell culture incubator. The light absorption value (OD value) was recorded at 450 nm wavelength on a Synergy H1 microplate reader (Beckman Coulter, CA, USA).
Enzyme-linked immunosorbent assay (ELISA)
The culture supernatants of MLE-12 cells in different treatment groups were collected, followed by 15 min of centrifugation at 1 000 × g and 4°C. The supernatants of each sample were collected and 100 µL supernatant was used for ELISA according to the manufacturer’s instructions of the ELISA assay kit [Citation29]. After the color development, the OD values were detected by a microplate reader (Beckman Coulter, CA, USA). All the ELISA assay kits were provided by Abcam (Cambridge, MA, USA).
Target prediction and dual luciferase assay
Starbase bioinformatics software (https://starbase.sysu.edu.cn/) [Citation30] was used for predicting the biding site between circ_0001679 and miR-338-3p. Then the wild type (WT) and mutated (MUT) binding site was cloned into PmirGLO vector expressing firefly luciferase respectively (Promega). Plasmid containing the Renilla luciferase gene (pRL-TK) was used as a control plasmid. The reporter plasmid and Renilla luciferase control plasmid were co-transfected into cells with either miR-338-3p mimic or miR-NC in a 12-well plate (1 × 10^5 cells/well) using Lipofectamine 3000 reagent (Invitrogen, L3000001). The relative luciferase activity was recorded 48 h after transfection using the Dual-Luciferase Reporter Assay Kit (Promega, Mannheim, Germany) [Citation25].
The predicted binding site between miR-338-3p and MAPK1 3’ untranslated region (3’UTR) was predicted using Starbase bioinformatics software. The wild type (WT) or the mutated (MUT) binding site between miR-338-3p and MAPK1 was cloned into PmirGLO luciferase reporter, respectively (Promega). Plasmid containing the Renilla luciferase gene (pRL-TK) was used as a control plasmid. The reporter plasmid and Renilla luciferase control plasmid were co-transfected into cells with either miR-338-3p mimic or miR-NC in a 12-well plate (1 × 10^5 cells/well) using Lipofectamine 3000 reagent (Invitrogen, L3000001). 48 h post transfection, the relative luciferase activities were measured using Dual-Luciferase Reporter Assay Kit (Promega). The relative firefly luciferase activity was normalized to that of Renilla luciferase in the control plasmid.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The MLE-12 cells in different treatment groups were collected, and Trizol reagent (Invitrogen) was utilized to extract total RNA. Based on manufacturer's instruction, 1 ml of Trizol reagent were added per 1 × 106 cells. TaqMan miRNA reverse transcription kit (Applied Biosystems, CA, USA) or Takara reverse transcription kit (Takara, Dalian, China) was utilized for cDNA synthesis, while TaqMan miRNA quantitative PCR kit (Thermo Fisher Scientific, MA, USA) or real-time quantitative PCR kit (Takara, Dalian, China) was adopted for amplification on ABI 7500 RT-PCR System (Applied Biosystems Inc., CA, USA). β-actin was used as the reference for cicrRNA, and U6 was used as the reference for the moralization of miRNA expression. The 2-ΔΔCt method was applied to calculate the relative expression of circ_0001679 and miR-338-3p [Citation31]. The PCR cycling conditions used: 95°C for 30s for initial denaturation, and then 40 cycles of denaturation at 94°C for 30s, elongation at 60°C for 20s. In this study, all primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China): miR-338-3p sense 5’-GCAGTCCAGCATCAGTG-3’, anti-sense 5ʹ-CAGTGCGTGTCGTGGAGT-3’; U6 sense 5’-TCCGACGCCGCCATCTCTA-3’, antisense 5’-TATCGCACATTAAGCCTCTA-3’ circ_0001679 sense 5ʹ-CTGGACCCTGAGGATGCT-3’, anti-sense 5ʹ-ATGACCCTGCTTTGTGCA-3’; β-actin sense 5’-GGCTGTATTCCCCTCCATCG-3’, anti-sense 5’-CCAGTTGGTAACAATGCCATGT-3’.
RNA pulldown assay
Biotin-labeled miR-NC (negative control) or Biotin-labeled miR-338-3p probe were synthesized by Ruiyuan Biological Company (Nanjing, China). Cells lysates were collected by IP lysis buffer (Beyotime, P0013) and were incubated with biotinylated miR-NC or Biotin-labeled miR-338-3p for 2 h. 10% of total cell lysates was saved as the input. The mixture was further incubated with M-280 streptavidin magnetic beads (Sigma-Aldrich) at 4°C for 4 h. A magnetic bar was used to precipitate the magnetic beads and associated nucleic acids, then the samples were washed 4 times with IP lysis buffer. Both the input and the pull-down samples were purified with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Reverse transcription and qPCR were performed to determine the relative expression of circ_0001679 in the pull-down samples [Citation32].
Western blotting (WB) assay
The procedures of Western blot were adopted from a previous study [Citation33]. RIPA lysate was utilized for extracting total proteins from cells. Cells suspended in RIPA buffer were lyzed on ice for 10 mins and lysed cells were centrifuged at 14,000 rpm for 10 mins. The supernatant containing total protein lysate was quantified by a BCA Protein assay kit (Beyotime Biotechnology P0009; Shanghai, China). Aliquots of proteins (30 µg) were seperated through 10% SDS-PAGE, followed by the transfer of proteins onto PVDF membranes. After blocking with 5% skimmed milk for 1 hour, the membrane was then incubated with primary antibodies overnight at 4°C.: B-cell lymphoma-2 (Bcl-2) (1:500; ab196495, Abcam, Cambridge, UK), BCL2-associated x protein (Bax) (1:1000; ab32053, Abcam), Cleaved caspase 3 (1:1000; ab231289, Abcam), MAPK1 (1:800; ab265600, Abcam) and β-actin (1:5000; ab179467, Abcam). The membrane was washed 3 times with TBST for 5 minutes each. After wash, the membrane was further incubated with goat anti-rabbit IgG H&L (Abcam; ab96899, 1:5000) for additional 1 h under ambient temperature. Enhanced chemiluminescence ECL reagent (Pierce, IL, USA) was employed to visualize protein bands. The protein bands image was captured using Gel Doc 100 system and the Quantity One® imaging software (Bio-Rad, CA, USA).
Statistical methods
SPSS21.0 (Chicago, IL, USA) was adopted for statistical analysis. Measurement data were presented as mean ± SD. The statistical difference between two groups was compared using unpaired Student’s t tests. Comparisons among multiple groups were analyzed using one-way analysis of variance (ANOVA) with Bonferroni post-hoc test. P < 0.05 is considered as statistically significant, each experimental result is the summary of at least three independent experiments.
Results
In the present study, we aim to investigate the role of circ_0001679 in regulating the LPS-induced lung cell injury. We initially established an in vitro lung cell injury model using LPS-treated MLE-12 cells. siRNAs targeting circRNA_0001679 were employed to stably knock down circRNA_0001679, followed by functional assays to investigate the effect of circRNA_0001679 silencing. The levels of inflammatory cytokines such as IL-6, IL-β and TNF-α were detected by ELISA assay. We found that Circ_0001679 was upregulated in LPS-induced MLE-12 cells, and silencing circ_0001679 attenuated the growth inhibition and suppressed apoptosis induced by LPS. We further demonstrated that circ_0001679 functioned as a sponge of miR-338-3p to negatively regulate miR-338-3p activity. The upregulation of circ_0001679 maintained a high-level expression of MAPK1 by suppressing miR-338-3p. These data indicate that circ_0001679/miR-338-3p/MAPK1 axis may play an important role in acute lung injury.
Knockdown of circ_0001679 ameliorates LPS-induced cell injury in MLE-12
A previous study found that circ_0001679 was highly expressed in an LPS-induced ALI mice model, and silencing circ_0001679 could reduce sepsis-induced ALI injury [Citation21]. In our study, we first examined the expression levels of circ_00016791 in MLE-12 upon LPS treatment. Circ_0001679 expression was significantly upregulated in MLE-12 cells after LPS induction (). We next silenced circ_00016791 expression in MLE-12 cell using siRNAs (si-circ_0001679#1, #2, #3). All three siRNAs significantly reduced the level of circ_00016791 in MLE-12 cells (), si-circ_0001679#1 showed a relatively higher knockdown efficiency and was used for the following study.
Figure 1. Knockdown of circ_0001679 inhibits LPS-induced cell injury in MLE-12. (a) Circ_00016791 expression was detected by qRT-PCR in MLE-12 cells after LPS treatment; (b) Circ_00016791 level was measured through qRT-PCR in MLE-12 cells after the transfection with siRNAs (NC, si-circ_0001679#1,#2,#3); (c) The light absorption at 450 nm in different groups of MLE-12 cells at 0 h, 24 h, 48 h and 72 h was detected by CCK-8; (d) The apoptosis levels in different MLE-12 cell groups were measured through flow cytometric analysis; (e) Apoptosis-related proteins Bcl2, cleaved-cas3, and Bax were measured in different groups of MLE-12 cells by Western blot; (f) IL-6, TNF-α, and IL-1β levels in different treatment groups were measured through ELISA. *p < 0.05; **p < 0.01; ***p < 0.001.
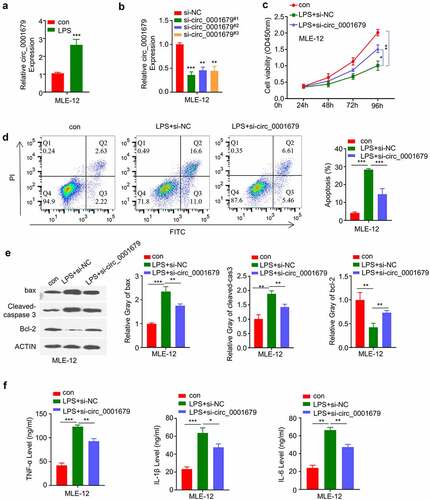
We next performed CCK-8 assay to investigate the effect of circ_00016791 in LPS induced cell proliferation. LPS treatment significantly suppressed cell proliferation in MLE-12 cells, while si-circ_0001679 transfection could partially rescue this effect (). Apoptosis detection by flow cytometry further revealed that LPS induced apoptotic events could also be partially suppressed by circ_0001679 silencing (). Consistently, LPS treatment reduced antiapoptotic protein Bcl-2 level and increased the levels of cleaved-cas3 and Bax. The transfection of si-circ_0001679 attenuated these changes (). We also analyzed the inflammatory cytokines by ELISA and we found that silencing circ_0001679 could mitigate the production of IL-6, IL-1β and TNF-α induced by LPS treatment (). Together, these results indicate that circ_0001679 upregulation is implicated in LPS-induced cell damages.
Circ_0001679 overexpression alone does not cause cell injury in MLE-12
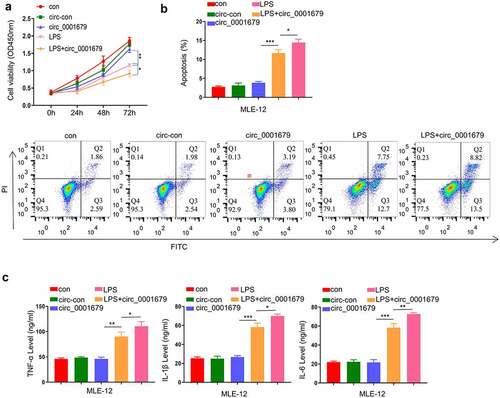
To further confirm the regulatory role of circ_0001679 in LPS-induced cell injury, we constructed circ_0001679 overexpression vector and investigated the effect of circ_0001679 overexpression in cells with or without LPS treatment. Circ_0001679 overexpression alone did not affect cell proliferation, while in LPS-induced cells Circ_0001679 overexpression exacerbated the impairment of cell growth by LPS (). We further examined the apoptosis in different treatment groups. Consistently, circ_0001679 overexpression alone did not induce apoptosis in MLE-12 cells, while the apoptotic events of LPS-induced cells were significantly increased by circ_0001679 overexpression (). The analysis of inflammatory cytokines revealed a similar scenario, in which circ_0001679 overexpression did not affect the levels of TNF-α, IL-1β and IL-6 without LPS induction. However, upon LPS treatment circ_0001679 overexpression promoted the production of TNF-α, IL-1β and IL-6 (). Therefore, circ_0001679 overexpression is implicated in regulating the cell responses only when LPS-induced damages are initiated.
Figure 2. Circ_0001679 overexpression alone does not induce cell injury. (a) The light absorption at 450 nm in different groups of MLE-12 cells at 0 h, 24 h, 48 h and 72 h was detected by CCK-8; (b) The apoptosis levels in different MLE-12 cell groups were measured through flow cytometric analysis; (c) IL-6, TNF-α, and IL-1β levels in different treatment groups were measured through ELISA. *p < 0.05; **p < 0.01; ***p < 0.001.
Circ_0001679 targets miR-338-3p
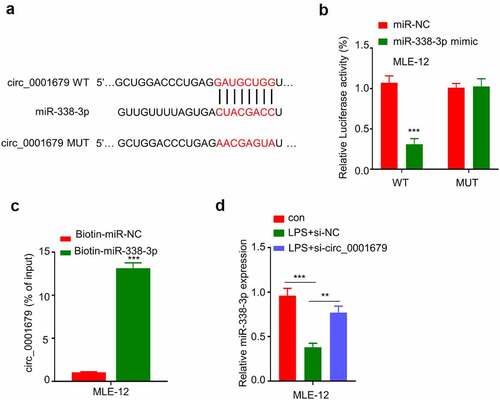
To the downstream mechanism by which circ_0001679 regulates LPS-induced cellular responses, we first predicted the downstream target miRNAs of circ_0001679 by starbase (http://starbase.sysu.edu.cn /index.php). We found that circ_0001679 contain many miRNA binding sites (Figure S1), and miR-338-3p was discovered to harbor a binding site for circ_0001679 (). We applied qRT-PCR to detect the expression of 7 target miRNAs which have been reported to be relevant in lung injury or LPS-induced inflammation response (The list of the literature was in supplementary file). Compared to LPS induced cells, silencing circ_0001679 could greatly upregulate the expression of miR-338-3p while the expression levels of other miRNAs were only mildly increased (Figure S2).Then we performed luciferase reporter assay using WT reporter or reporter containing mutated binding site between circ_00016791 with miR-338-3p. The presence of miR-338-3p mimic significantly inhibited luciferase activity in MLE-12 cells, while this effect was not observed in mutated reporter (). To further show the physical interaction between circ_00016791 with miR-338-3p, we performed RNA pull-down assay using biotin-labeled miR-338-3p probe. As compared to the miR-NC probe, miR-338-3p probe strongly enriched more circ_00016791 (). In addition, LPS treatment caused a downregulation of miR-338-3p, which could be largely reversed by the knockdown of si-circ_0001679 (). Together, these results suggest that circ_0001679 negatively target miR-338-3p.
Figure 3. Circ_0001679 targets miR-338-3p. (a) Starbase was used to predict binding site between circ_00016791 with miR-338-3p; B-C, Functional interaction between circ_00016791 with miR-338-3p was confirmed by RNA pulldown assay (b) and luciferase reporter assay (c); (d) miR-338-3p expression in different treatment groups was measured through qRT-PCR. *p < 0.05; **p < 0.01; ***p < 0.001.
miR-338-3p mediates the effect of circ_0001679 in LPS-induced cell injury
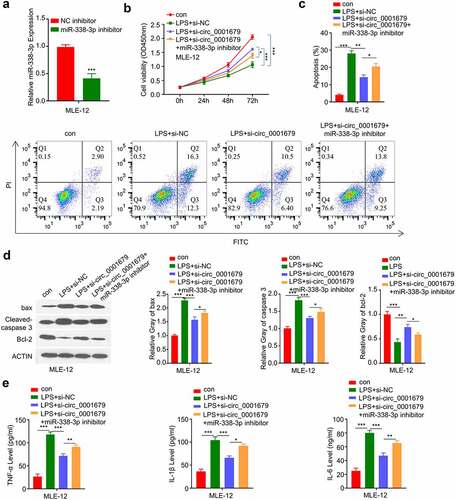
We next sought to investigate whether miR-338-3p mediates the effect of circ_0001679 in LPS-induced cell damages. We applied miR-338-3p inhibitor which could markedly decrease the expression of miR-338-3p (). We found that the rescue effect of circ_00016791 silencing on LPS-induced cell damages were significantly impaired by miR-338-3p inhibitor. (). miR-338-3p inhibitor also impaired the protective effect of circ_00016791 silencing on LPS-induced apoptosis (). Consistently, si-circ_00016791 cause the increased of Bcl2 protein and the decrease of cleaved-cas3 and Bax levels after LPS induction, and miR-338-3p inhibitor partially reversed these effects (). Finally, we demonstrated that the rescue effect of si-circ_00016791 on the proinflammatory cytokine production (IL-6, TNF-α and IL-1β) was dampened by miR-338-3p inhibitor ().
Figure 4. Knockdown of circ_0001679 inhibits LPS-induced cell injury in MLE-12 cells via regulating miR-338-3p. (a) miR-338-3p expression in different treatment groups was measured through qRT-PCR; (b) The light absorption value at 450 nm in different groups of MLE-12 cells at 0 h, 24 h, 48 h, 72 h were detected by CCK-8; (c) The apoptosis levels in different MLE-12 cell groups were measured through flow cytometric analysis; (d) The protein expression levels of Bcl2, cleaved-cas3, Bax and GAPDH in different experimental groups of MLE-12 cells were measured by Western blot; E, IL-6, TNF-α, and IL-1β levels in different treatment groups were measured through ELISA. *p < 0.05; **p < 0.01; ***p < 0.001.
miR-338-3p targets MAPK1
The above results showed that knockdown of circ_0001679 inhibits LPS-induced MLE-12 cell injury through targeting miR-338-3p. We next searched for the downstream target mRNA of miR-338-3p, and found the presence of the miR-338-3p binding site in the 3ʹUTR of MAPK1 mRNA. Dual-luciferase reporter assay confirmed that miR-338-3p mimic was able to inhibit the luciferase activity of WT reporter containing the binding site between MAPK1 mRNA and miR-338-3p, while the activity of the MUT reporter containing the mutated binding site was not affected (). miR-338-3p overexpression markedly decreased MAPK1 protein level (). Moreover, we found that MAPK1 protein level was increased after LPS treatment and the upregulation was attenuated by si-circ_00016791; whereas miR-338-3p inhibitor co-transfection impaired the effect of si-circ_00016791 ().
Figure 5. miR-338-3p targets MAPK1. (a) The presence of miR-338-3p binding site in the 3’ UTR of MAPK1 mRNA was predicted by starbase, and luciferase reporter assay was carried out to validate the functional interaction between miR-338-3p and MAPK1 mRNA; (b) MAPK1 protein expression was measured by Western blot; (c) MAPK1 protein expression in different treatment groups was measured by Western blot; (d) The light absorption at 450 nm in different groups of MLE-12 cells at 0 h, 24 h, 48 h and 72 h was detected by CCK-8; (e) The apoptosis levels in different MLE-12 cell groups were measured through flow cytometric analysis; (f) IL-6, TNF-α, and IL-1β levels in different treatment groups were measured through ELISA. *p < 0.05; **p < 0.01; ***p < 0.001.
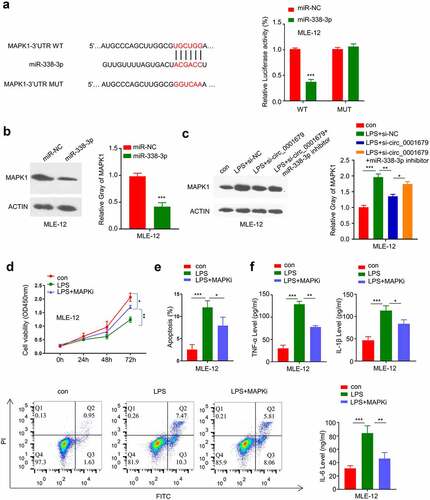
To confirm a functional role of MAPK1 in LPS-induced cell damages, we applied an MAPK1 inhibitor PD98059 to investigate whether MAPK1 activity inhibition could rescue the effect of LPS-induced cell damages. MAPK1 inhibition could at least partially recue the LPS-induced growth inhibition () and as well as decrease the apoptotic events (). Meanwhile, MAPK1 inhibition attenuated the increase of TNF -α, IL-1β and IL-6 levels in LPS-induced MLE-12 cells (). These data suggest that MAPK1 is a downstream effector in LPS-induced cell damages.
MiR-338-3p/MAPK1 axis is implicated in LPS-treated MLE-12 cell injury
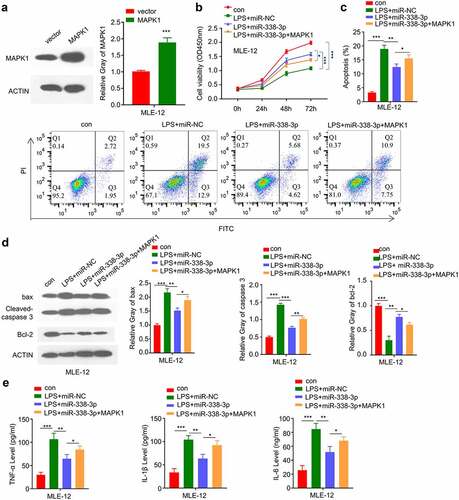
We next attempted to validate whether miR-338-3p/MAPK1 axis is implicated in LPS-treated MLE-12 cell injury. We constructed an MAPK1 overexpression vector that could significantly promote the expression level of MAPK1 (). We then treated the cells with LPS, LPS+miR-338-3p mimic or LPS+miR-338-3p mimic+ MAPK1 overexpression. The cell growth inhibition by LPS was partially released by miR-338-3p mimic, and MAPK1 overexpression further impaired the effect (). MAPK1 overexpression also impaired the recue effect of miR-338-3p mimic on apoptosis (), which was accompanied by the decreased Bcl2 level and increased levels of cleaved-cas3 and Bax after MAPK1 overexpression (). Finally, we demonstrated that the rescue effect of miR-338-3p mimic on the proinflammatory cytokine production (IL-6, TNF-α and IL-1β) was dampened by MAPK1 overexpression (). The above results indicate that MiR-338-3p/MAPK1 axis is implicated in LPS-treated MLE-12 cell injury.
Figure 6. miR-338-3p inhibits LPS-induced cell injury by negatively regulating MAPK1 expression. (a) MAPK1 protein levels in different treatment groups were measured by Western blot; (b) The light absorption value at 450 nm at 0 h, 24 h, 48 h, 72 h in different groups of MLE-12 cells was detected by CCK-8; (c) The apoptosis levels in different MLE-12 cell groups were measured through flow cytometric assay; (d) The protein expression levels of Bcl2, cleaved-cas3, Bax and GAPDH in different groups of MLE-12 cells were measured by Western blot; (e) IL-6, TNF-α, and IL-1β expression levels in different treatment groups were measured through ELISA. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
Although great achievements have been made in the diagnosis and treatment of ALI/ARDS, patients with ALI are still at a high mortality rate [Citation34]. ALI/ARDS is characterized by a severe acute inflammatory response in the lungs, which can result in injuries of alveolar epithelial cells and pulmonary capillary endothelial cells [Citation35]. Alveolar epithelial cells are the major components in the alveolar structure [Citation36], which functions as a mechanical barrier for protecting against pulmonary damage, regulating pulmonary immune response and maintaining pulmonary fluid homeostasis [Citation36,Citation37].
A previous study reported that circ_0001679 was highly expressed in an LPS-induced ALI mice model, and silencing circ_0001679 could reduce sepsis-induced ALI injury [Citation21]. Teng et al. [Citation38] performed whole transcriptome analysis to show that circ_0001679 was highly expressed in lung tissues of the mice model of ALI. These studies strongly suggest that circ_0001679 upregulation is implicated in regulating the inflammatory damages in ALI. However, how circ_0001679 regulates the pathogenesis of ALI remains unclear. In the present study, we used an in vitro ALI model of LPS-induced cell damage in MEL-12 cell to study the functional role down the underlying mechanisms of circ_0001679. We demonstrated that circ_0001679 knockdown could prevent apoptosis and inflammatory response induced by LPS treatment. These data suggest that the knockdown of circ_0001679 can attenuate the inflammatory response in sepsis-induced ALI.
CircRNAs are able to modulate mRNA expression through sponging miRNAs [Citation39]. In sepsis-related studies, it was found that circRNA_MA21 can ameliorate sepsis-induced renal injury by modulating the miR-9-3p/SMG1 axis [Citation40]. circDMNT3B increases intestinal mucosal permeability in septic rats by regulating miR-20b-5p [Citation41]; circC3P1 decreases proinflammatory cytokine production and apoptosis by regulating miR-21 in sepsis-induced ALI [Citation42]. In the present study, we found that circ_0001679 sponges miR-338-3p, and circ00001679 negatively regulate miR-338-3p expression. Importantly, miR-338-3p inhibitor could dampen the effect of circ00001679 silencing in LPS induced cell model. These data altogether suggest miR-338-3p mediates the downstream effect of circ00001679. miR-338-3p is associated with inflammatory response in different disease models [Citation26,Citation43,Citation44]. As demonstrated by Zhang et al., miRNA-338-3p could inhibit the inflammatory response in acetaminophen-induced acute liver injury through CAMK IIα signaling pathway [Citation43]. In a mouse model of allergic encephalomyelitis, circRNA_001076/ miR-338-3p was found to promote immune inflammation via TRIM33/RORγt signaling pathway [Citation44]. MiR-338-3p is reported to be sponged by circ_003867 in LPS-induced inflammatory injury model of human bronchial epithelial cell [Citation26]. However, the expression pattern of miR-338-3p has not been reported in ALI patients. In our study, it was found that LPS treatment reduce the expression of miR-338-3p, which is mediated by the upregulation of circ_0001679. We further demonstrated that suppressing the expression of miR-338-3p could partially reverse the effect of circ_0001679 silencing on cell proliferation, apoptosis and inflammatory responses. Our data suggest that Circ_0001679/miR-338-3p axis is implicated in the regulation of LPS-induced ALI.
CircRNAs are able to regulate gene expression at post-transcriptional or transcriptional levels [Citation45,Citation46]. We further revealed that miR-338-3p directly target MAPK1 mRNA at 3ʹUTR and negatively regulate its expression. Mitogen-activated protein kinases (MAPKs), also known as extracellular signal-regulated kinase (ERK), participate in numerous biological processes, like cell differentiation, growth and cell cycle progression [Citation47]. MAPK1 is a downstream target of MAPK, which is also known as p42 MAPK or ERK2 [Citation48]. It can be activated by different physical or chemical stimuli, which in turn activates the P38 signaling pathway to regulate the immune responses [Citation49]. It was found that in macrophages, the activation of MAPK1 pathway contributes to the enhanced inflammatory cytokine production, like IL-1, IL-6, and TNF-α [Citation50]. In liver cirrhosis and inflammation, the degree of liver fibrosis and inflammation were correlated with MAPK1 activation [Citation51]. In a LPS-induced ALI model, MAPK1 was reported to be regulated by miR-342 [Citation52]. In current study, we found that MAPK1 inhibitor PD08059 was able to significantly ameliorate cell injury caused by LPS induction. In addition, MAPK1 overexpression mitigates effect of miR-338-3p mimic on LPS-induced MEL-12 injury. Overall our data suggest that circ_0001679/miR-338-3p/MAPK1 axis involved in regulating the inflammatory response in ALI.
However, many cellular factors can contribute to the acute lung injury caused by LPS. We found that knockdown of circ_0001697 could attenuate cell injury and inflammatory response induced by LPS, but could not completely inhibit these effects. This may be because that high expression of circ_0001697 is not the only underlying factor leading to cellular injury upon LPS treatment. In addition, targeting the miR-338-3p/MAPK1 axis could only attenuate the observed cellular effects caused by LPS treatment, but not prevent the cellular effects. We therefore conclude that targeting circ_0001697/miR-338-3p/MAPK1 axis may attenuate the pathogenesis of ALI, but could not cure the disease.
Our study provides a possible intervention target for the amelioration of clinical lung injury. However, more evidence is required to validate the role of circ_0001679/miR-338-3p/MAPK1 in human samples. Clinical samples from human ALI patients are required to validate the upregulation circ_0001679 and downregulation of miR-338-3p in human subjects. Meanwhile, the association of expression level of circ_0001679/miR-338-3p and the inflammatory conditions in ALI patients should be evaluated to inform whether circ_0001679/miR-338-3p axis can indicate the inflammatory conditions in patients. The potential protective effect of MAPK1 inhibitor in ameliorating LPS-induced cell damage should be examined in the ALI mouse model.
Conclusion
In summary, our work demonstrated the upregulation of circRNA_0001679 in LPS-induced MLE-12 cells, and its silencing can alleviate LPS-induced cell injury through the miR-338-3p/MAPK1 axis. Our study suggests that circ_0001679 may serve as a potential target in ameliorating ALI, and the functional roles of circ_0001679/miR-338-3p/MAPK1 axis in ALI need to be further validated in human samples and animal models.
Data availability
The data is available from the corresponding author on reasonable request.
Supplemental Material
Download Zip (2.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Butt Y, Kurdowska A, Allen TC. Acute lung injury: a clinical and molecular review. Arch Pathol Lab Med. 2016;140:345–350.
- Krzak A, Pleva M, Napolitano LM. Nutrition Therapy for ALI and ARDS. Critical Care Clinics. 2011;27(3):647–659.
- Zambon M, Vincent JL. Mortality rates for patients with acute lung injury/ARDS have decreased over time. Chest. 2008;133:1120–1127.
- Johnson ER, Matthay MA. Acute lung injury: epidemiology, pathogenesis, and treatment. J Aerosol Med Pulm Drug Deliv. 2010;23:243–252.
- Serhan CN, Krishnamoorthy S, Recchiuti A, et al. Novel anti-inflammatory–pro-resolving mediators and their receptors. Curr Top Med Chem. 2011;11:629–647.
- Yi L, Zhou Z, and Zheng Y, et al. Suppressive effects of GSS on lipopolysaccharide-induced endothelial cell injury and ALI via TNF-α and IL-6. Mediators Inflamm. 2019;30:4251394. doi:10.1155/2019/4251394.
- Fang Y, Xu P, Gu C, et al. Ulinastatin improves pulmonary function in severe burn-induced acute lung injury by attenuating inflammatory response. J Trauma. 2011;71:1297–1304.
- Muralidharan S, Mandrekar P. Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation. J Leukoc Biol. 2013;94:1167–1184.
- Yuan Z, Bedi B, Sadikot RT. Bronchoalveolar lavage exosomes in lipopolysaccharide-induced septic lung injury. J Vis Exp. 2018;21:57737.
- Galani V, Tatsaki E, Bai M, et al. The role of apoptosis in the pathophysiology of acute respiratory distress syndrome (ARDS): an up-to-date cell-specific review. Pathol Res Pract. 2010;206:145–150.
- Bardales RH, Xie SS, Schaefer RF, et al. Apoptosis is a major pathway responsible for the resolution of type II pneumocytes in acute lung injury. Am J Pathol. 1996;149:845–852.
- Baradaran Rahimi V, Rakhshandeh H, and Raucci F, et al. Anti-Inflammatory and anti-oxidant activity of portulaca oleracea extract on LPS-Induced rat lung injury. Molecules. 2019;24:139.
- Derwall M, Martin L, Rossaint R. The acute respiratory distress syndrome: pathophysiology, current clinical practice, and emerging therapies. Expert Rev Respir Med. 2018;12:1021–1029.
- De Freitas Caires N, Gaudet A, Portier L, et al. Endocan, sepsis, pneumonia, and acute respiratory distress syndrome. Crit Care. 2018;22:018–2222.
- Zhu H, Wang Y, Sun J, et al. Tomentosin inhibits lipopolysaccharide-induced acute lung injury and inflammatory response by Suppression of the NF-κB pathway in a mouse model of sepsis. J Environ Pathol Toxicol Oncol. 2020;39:291–298.
- Xie C, Li X, Zhu J, et al. Magnesium isoglycyrrhizinate suppresses LPS-induced inflammation and oxidative stress through inhibiting NF-κB and MAPK pathways in RAW264.7 cells. Bioorg Med Chem. 2019;27:516–524.
- Yu P, Li Y, Wang Y, et al. Lipopolysaccharide restricts murine norovirus infection in macrophages mainly through NF-kB and JAK-STAT signaling pathway. Virology. 2020;546:109–121.
- Liu P, Feng Y, Li H, et al. Ferrostatin-1 alleviates lipopolysaccharide-induced acute lung injury via inhibiting ferroptosis. Cell Mol Biol Lett. 2020;25:020–00205.
- Huang XT, Liu W, Zhou Y, et al. Galectin-1 ameliorates lipopolysaccharide-induced acute lung injury via AMPK-Nrf2 pathway in mice. Free Radic Biol Med. 2020;146:222–233.
- Yang CL, Yang WK, He ZH, et al. Quietness of circular RNA circ_0054633 alleviates the inflammation and proliferation in lipopolysaccharides-induced acute lung injury model through NF-κB signaling pathway. Gene. 2021;766:17.
- Zou Z, Wang Q, Zhou M, et al. Protective effects of P2X7R antagonist in sepsis-induced acute lung injury in mice via regulation of circ_0001679 and circ_0001212 and downstream Pln, Cdh2, and Nprl3 expression. J Gene Med. 2020;22(12):30.
- O’Driscoll L. The emerging world of microRNAs. Anticancer Res. 2006;26:4271–4278.
- Wu G, Zhang J, Fan GG, et al. MiRNA-324-5p inhibits inflammatory response of diabetic vessels by targeting CPT1A. Eur Rev Med Pharmacol Sci. 2020;24:12836–12843.
- Jiang A, Dong C, Li B, et al. MicroRNA-206 regulates cell proliferation by targeting G6PD in skeletal muscle. FASEB J. 2019;33:14083–14094.
- Chen G, Liu T, Yu B, et al. CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle. 2020;19:1696–1705.
- Liu G, Wan Q, Li J, et al. Circ_0038467 regulates lipopolysaccharide-induced inflammatory injury in human bronchial epithelial cells through sponging miR-338-3p. Thorac Cancer. 2020;11:1297–1308.
- Yi X, Wei X, Lv H, et al. Exosomes derived from microRNA-30b-3p-overexpressing mesenchymal stem cells protect against lipopolysaccharide-induced acute lung injury by inhibiting SAA3. Exp Cell Res. 2019;383:111454.
- Crowley LC, Marfell BJ, Scott AP, et al. Quantitation of apoptosis and necrosis by annexin V binding, propidium iodide uptake, and flow cytometry. Cold Spring Harb Protoc. 2016;2016. DOI:10.1101/pdb.prot087288.
- Konstantinou GN. Enzyme-Linked immunosorbent assay (ELISA). Methods Mol Biol. 2017;1592:79–94.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408.
- Zheng Y, Chen CJ, Lin ZY, et al. Circ_KATNAL1 regulates prostate cancer cell growth and invasiveness through the miR-145-3p/WISP1 pathway. Biochem Cell Biol. 2020;98:396–404.
- Kurien BT, Scofield RH. Western blotting: an introduction. Methods Mol Biol. 2015;1312:17–30.
- He YQ, Zhou CC, Yu LY, et al. Natural product derived phytochemicals in managing acute lung injury by multiple mechanisms. Pharmacol Res. 2021;163:29.
- Mokra D, Kosutova P. Biomarkers in acute lung injury. Respir Physiol Neurobiol. 2015;209:52–58.
- Guillot L, Nathan N, Tabary O, et al. Alveolar epithelial cells: master regulators of lung homeostasis. Int J Biochem Cell Biol. 2013;45:2568–2573.
- Zhang H, Cui Y, Zhou Z, et al. Alveolar type 2 epithelial cells as potential therapeutics for acute lung injury/acute respiratory distress syndrome. Curr Pharm Des. 2019;25:4877–4882.
- Teng X, Liao J, Zhao L, et al. Whole transcriptome analysis of the differential RNA profiles and associated competing endogenous RNA networks in LPS-induced acute lung injury (ALI). PloS one. 2021;16:e0251359.
- Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44.
- Shi Y, Sun CF, Ge WH, et al. Circular RNA VMA21 ameliorates sepsis-associated acute kidney injury by regulating miR-9-3p/SMG1/inflammation axis and oxidative stress. J Cell Mol Med. 2020;24:11397–11408.
- Liu J, Liu Y, Zhang L, et al. Down-regulation of circDMNT3B is conducive to intestinal mucosal permeability dysfunction of rats with sepsis via sponging miR-20b-5p. J Cell Mol Med. 2020;24:6731–6740.
- Jiang WY, Ren J, Zhang XH, et al. CircC3P1 attenuated pro-inflammatory cytokine production and cell apoptosis in acute lung injury induced by sepsis through modulating miR-21. J Cell Mol Med. 2020;24:11221–11229.
- Zhang C, Kang L, Zhu H, et al. miRNA-338-3p/CAMK IIα signaling pathway prevents acetaminophen-induced acute liver inflammation in vivo. Ann Hepatol. 2021;21:100191.
- Duan W, Dong Z, Huang Y. TRIM33/RORγt signaling pathway promoted immune inflammation in mice of AE through miR-338-3p/ Circ RNA 001076. Minerva Med. 2020. DOI:10.23736/s0026-4806.20.06647-1
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec(1):R17–29.
- Panni S, Lovering RC, Porras P, et al. Non-coding RNA regulatory networks. Biochimica et biophysica acta. Gene Regulatory Mech. 2020;1863:194417.
- Satoh Y, Endo S, Ikeda T, et al. Extracellular signal-regulated kinase 2 (ERK2) knockdown mice show deficits in long-term memory; ERK2 has a specific function in learning and memory. J Neurosci. 2007;27:10765–10776.
- Lu N, Malemud CJ. Extracellular signal-regulated kinase: a regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated gene expression. Int J Mol Sci. 2019;20. DOI:10.3390/ijms20153792.
- Chen X, Qu Y, Cheng Y, et al. MiR-19b-3p regulates MAPK1 expression in embryonic fibroblasts from the great tit (Parus major) under hypoxic conditions. Cell Physiol Biochem. 2018;46:546–560.
- Braicu C, Buse M, Busuioc C, et al. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers (Basel). 2019;11. DOI:10.3390/cancers11101618.
- Jeng KS, Lu SJ, Wang CH, et al. Liver Fibrosis and Inflammation under the Control of ERK2. Int J Mol Sci. 2020;21. DOI:10.3390/ijms21113796.
- Zhu S, Song W, Sun Y, et al. MiR-342 attenuates lipopolysaccharide-induced acute lung injury via inhibiting MAPK1 expression. Clin Exp Pharmacol Physiol. 2020;47:1448–1454.
