ABSTRACT
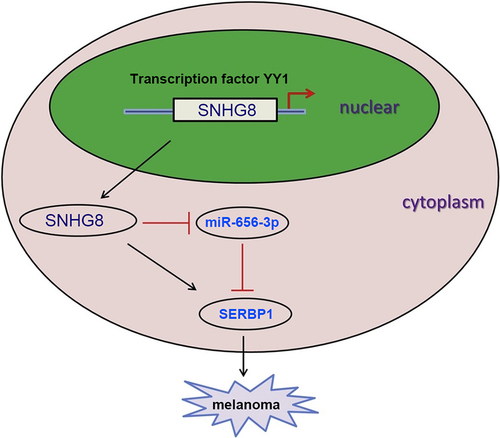
Long non-coding (lnc) RNA serves a vital role in the cellular processes of carcinoma. This study aimed to explore the accurate mechanism underlying lncRNA small nucleolar RNA host gene 8 (SNHG8) in melanoma. In this study, lncRNA SNHG8 expression were upregulated in melanoma tissues and cells, and lncRNA SNHG8 knockdown reduced melanoma cell viability, migration and invasion. Moreover, lncRNA SNHG8 expression could be induced by transcription factor YY1. In addition, we found that miR-656 could directly bind to lncRNA SNHG8 and SERPINE1 mRNA binding protein 1 (SERBP1). Rescue assays indicated that miR-656 overexpression inhibited the aforementioned cellular activities in melanoma cells, which were reversed by SERBP1 overexpression. In conclusion, this work elucidated that YY1-induced upregulation of lncRNA SNHG8 boosted the development of melanoma via the miR-656-3p/SERBP1 axis, providing a novel therapeutic strategy for melanoma treatment.
Graphical Abstract
KEYWORDS:
Introduction
Melanoma is a common malignant skin tumor with an imbalanced transformation of melanocytes [Citation1], which display enhanced proliferation and metastasis [Citation2]. Despite the low incidence rate of melanoma, in recent years, the incidence rate has increased by 6.1% between 2005 and 2017 in China [Citation3]. Several treatment strategies are used to alleviate melanoma, such as chemotherapy, chemoimmunotherapy, and radiotherapy; however, their effects are limited in patients with melanoma [Citation4]. Patients with metastatic melanoma are difficult to treat, hence the high and unsatisfactory recurrence and prognosis of these patients [Citation5], and the relatively high mortality rate [Citation6]. Therefore, understanding the pathogenesis of melanoma is important for the future treatment of patients.
Long non-coding (lnc) RNAs are a type of non-coding (nc) RNAs, that are >200 nucleotides in length, but display no protein-coding ability [Citation7]. LncRNAs exert different functions in human cancer. For example, E2F transcription factor 6-downregulated lncRNA cancer susceptibility 2 (CASC2) contributed to a poor prognosis and enhanced the progression of gastric cancer [Citation8]. LncRNA CDKN2B antisense RNA 1 indicated a poor prognosis in patients with intrahepatic cholangiocarcinoma [Citation9]. LncRNA HOX transcript antisense RNA was associated with chemoresistance in an ovarian cancer cell line via homeobox A7 [Citation10]. Previous studies showed that lncRNAs, such as HSD11B1-AS1 [Citation11], FGD5-AS1 [Citation12], and PVT1 [Citation13], could be used as potential diagnostic or prognostic biomarkers in melanoma. LncRNA small nucleolar RNA host gene 8 (SNHG8) has been demonstrated to be a tumor promoter in various cancers, such as gastric cancer [Citation14], hepatocellular carcinoma [Citation15], and lung cancer [Citation16]. Nevertheless, the role of lncRNA SNHG8 in melanoma has not been elucidated.
MicroRNAs (miRNAs/miRs), which are 18–25 nucleotides in length, are another subtype of ncRNA, with functional roles in carcinoma. For example, miR-377 targeted zinc finger E-box binding homeobox 2 to inhibit cervical cancer cell proliferation and invasion [Citation17]. miR-148b served as a tumor inhibitor by targeting endoplasmic reticulum metallopeptidase 1 in endometrial carcinoma cell lines [Citation18]. miR-135a upregulated Rac family small GTPase 1 to confer gefitinib-resistance in non-small cell lung cancer (NSCLC) [Citation19]. In addition, lncRNAs can act as competing endogenous RNAs (ceRNAs) for miRNAs to regulate miRNA and miRNA-targeted mRNAs [Citation20], which influence the initiation and progression of human cancers [Citation21,Citation22]. Among the miRNAs, miR-656 was demonstrated to interact with lncRNA SNHG8 [Citation23]. miR-656 has been reported to be sponged by lncRNA nc RNA activated by DNA damage (NORAD), a tumor booster for the initiation and development of non-small cell lung cancer [Citation24]. Prior to the present study, the role of miR-656 in melanoma had not been investigated.
In our research, we hypothesized that lncRNA SNHG8 could act as an oncogene in melanoma by regulating the miR-656/SERBP1 axis. Therefore, the aim of the present study was to investigate the biological function and molecular mechanisms of lncRNA SNHG8 in melanoma, which might provide a novel therapeutic target for melanoma treatment.
Materials and Methods
Tissue samples
A total of 32 paired melanoma and adjacent non-tumor tissues were obtained from patients with melanoma at the Second Hospital of Jilin University. The 32 patients included 17 women and 15 men, with a mean age of 51 years (range, 23–68 years). The inclusion criteria were as follows: patients were diagnosed as melanoma with no history of other tumors; patients did not receive any preoperative treatment, such as chemotherapy or radiotherapy; patients provided written informed consents. The following were the exclusion criteria: patients with other skin diseases or other malignancies; patients who had been treated prior to admission. The diagnoses of these samples were verified, in a blinded manner, by experienced pathologists at the Department of Pathology based on the American Society of Clinical Oncology guidelines [Citation25]. This study was approved by the Ethics Committee of the Second Hospital of Jilin University (Aug 2020, Changchun, China).
Cell culture
Cutaneous melanoma cells (A375, A875, M14, and SK-MEL-5) were purchased from the American Type Culture Collection. Human normal epidermal melanocytes (HEMa-LP) were purchased from Invitrogen (Thermo Fisher Scientific, Inc.). The melanoma cells were cultured in DMEM supplemented with 10% FBS (both from Gibco) at 37°C in a humified incubator with 5% CO2, and the HEMa-LP cells were cultured in medium 254 supplemented with human melanocyte growth supplement-2 (HMGS-2) (both from Gibco).
Cell transfection
SNHG8-targeting short hairpin RNA (sh-SNHG8; 5’-GCAGCACCAUCUCUGGUGUGU-3’) and a non-targeting shRNA [sh-negative control (NC); 5-AAUGCUCCGCACGUGUCACAU-3’] were designed by Guangzhou RiboBio Co., Ltd. miR-656 overexpression and knockdown were performed using miR-656 mimic (5’-UGCAAUGUUAGGACGUGUGAGG-3’) and miR-656 inhibitor (5’-UUCACCGAAGCUGUCAGCUAU-3’), respectively (Guangzhou RiboBio Co., Ltd.). SERBP1 overexpression vectors (pcDNA3.1/SERBP1) and the pcDNA3.1 vector were also synthesized by Guangzhou RiboBio Co., Ltd. Cell transfection was conducted with Lipofectamine® 2000 reagent (Invitrogen).
RT-qPCR
Total RNA was extracted from melanoma tissues and cells (HEMa-LP, A375, A875, M14, and SK-MEL-5) using TRIzol® (Invitrogen). RNA was reversely transcribed into cDNA using the cDNA Reverse Transcription kit (Invitrogen). Then, qPCR was performed on an Applied Biosystems 7500 system and with the SYBR Select Master Mix (both from Applied Biosystems). The following primers were used for qPCR: lncRNA SNHG8 forward, 5’-CCCGAGAACCGTCAGTTTGA-3’ and reverse, 5’-ACACCCGTTTCCCCAACTAC-3’; miR-656 forward, 5’-ACACTCCAGCTGGGAATATTATACAGTCA-3’ and reverse, 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGAGGUUG-3’; SERBP1 forward, 5’-CCACCTCGTGAACGAAGATT-3’ and reverse, 5’-ACCACCACGACCTCGAATAG-3’; GAPDH forward, 5’-TGGTCACCAGGGCTGCTT-3’ and reverse, 5’-AGCTTCCCGTTCTCAGCC-3’; U6 forward, 5’-CTTCGGCAGCACATATAC-3’ and reverse, 5’-GAACGCTTCACGAATTTGC-3’.
ChIP assay
The EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore) was applied for ChIP assay. Crosslinked chromatin DNA was obtained and sonicated into 200-bp to 1000-bp fragments, and then immunoprecipitated with anti-YY1 (Abcam) or anti-IgG antibody (Abcam) overnight at 4 °C. Finally, the immunoprecipitated DNA was analyzed by RT-qPCR [Citation26].
CCK-8 assay
Briefly, the transfected cells were seeded into 96-well plates (1x104 cells/well) and cultured for 0, 24, 48, and 72 h. Then,10 μl CCK-8 solution (Dojindo) was added to each well and incubated at 37°C for 2 h. The optical density was measured at 450 nm using a microplate reader [Citation27].
Colony formation assay
The transfected melanoma cells were seeded in 6-well plates (4x102 cells/well) and incubated for 2 weeks, following which, each well was washed with PBS. Subsequently, the cells were fixed with paraformaldehyde (Sigma-Aldrich), stained with crystal violet, and counted using a light microscope.
Wound healing assay
To investigate cell migration, a wound healing assay was performed. Briefly, at 90% confluence, sterile tips were employed to make a single scratch in the cell monolayer. Images of the migrated cells were acquired at 0 and 24 h using a light microscope [Citation28].
Matrigel assay
Cell invasive ability was determined using Transwell chambers (8-μm; Costar; Corning, Inc.) [Citation29]. The cells (2x104) with serum-free medium were plated into the upper chambers. In the lower chambers, culture medium containing 10% FBS was added. After incubation for 2 days. The invading cells were fixed with 4% paraformaldehyde, stained with 0.5% crystal violet, and observed using a light microscope.
Luciferase reporter assay
LncRNA SNHG8 or SERBP1 sequences with wild-type (WT) or mutant (Mut) miR-656-interacting sites were cloned into the pmirGLO reporter vector (Promega). The A375 and A875 cells were co-transfected with Renilla plasmid, either the four reporter plasmids and miR-656 mimics or NC mimics. Following incubation for 48 h at 37°C, luciferase activity was evaluated using the dual-luciferase reporter assay system (Promega) [Citation30].
Western blot analysis
Proteins were extracted using radioimmunoprecipitation lysis buffer (Beyotime), separated using 10% SDS-PAGE, and transferred to nitrocellulose membranes (EMD Millipore). Then, the membranes were incubated with anti-SERBP1 (no. ab55993; Abcam) or anti-GAPDH (no. ab70699; Abcam) overnight at 4°C. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Abcam) for 1 h at 37°C. The protein bands were visualized using an ECL reagent and an Image Quant LAS 4000 Mini Biomolecular Imager (both from GE Healthcare).
Statistical analysis
Statistical analyses were conducted using SPSS software (version 19.0; IBM Corp.). The data are presented as the mean ± SD. The comparison between the experimental and control groups was estimated using Student’s t-test or one-way ANOVA. Kaplan-Meier method and the log-rank test were used to determine the overall survival rate. The median expression of lncRNA SNHG8 was used as the cutoff value. P < 0.05 was considered statistically significant.
Results
In the present study, lncRNA SNHG8 levels in melanoma tissues and cells were investigated, as well as the potential regulatory mechanisms of lncRNA SNHG8 in melanoma cells.
LncRNA SNHG8 level is upregulated in melanoma tissues and cells
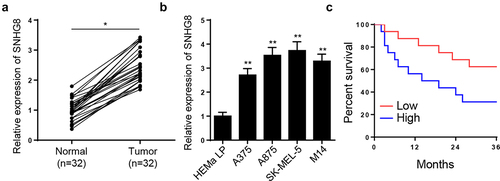
To explore the possible role of lncRNA SNHG8 in melanoma, lncRNA SNHG8 levels in melanoma tissues and cells were compared to that in adjacent normal tissues and human normal epidermal melanocytes (HEMa-LP). In melanoma tissues, the mRNA expression level of lncRNA SNHG8 was significantly upregulated (). In addition, in the four melanoma cell lines (A375, A875, M14, and SK-MEL-5), the mRNA expression level of lncRNA SNHG8 was significantly increased (). In addition, Kaplan–Meier curves determined that patients with melanoma expressing a high level of lncRNA SNHG8 suffered shorter OS ().
Figure 1. LncRNA SNHG8 level is enhanced in melanoma tissues and cells
LncRNA SNHG8 is activated by YY1
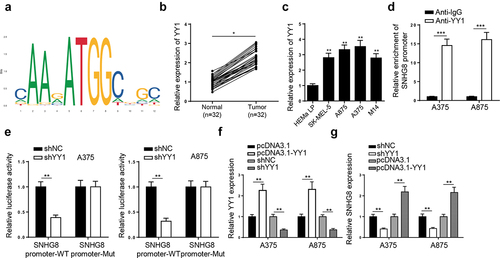
Accumulating studies have revealed that transcription factor YY1 can transcriptionally activate various lncRNAs [Citation19]. Hence, we wondered whether YY1 could regulate lncRNA SNHG8 expression at a transcription level. By use of JASPAR and UCSC website, the binding sequence of YY1 to lncRNA SNHG8 promoter (−829 ~ −818) was obtained (). Moreover, the expression of YY1 was obviously upregulated in melanoma tissues and cell lines (SK-MEL-5, A875, A375, and M14) compared to non-tumor tissues and human normal epidermal melanocytes (HEMa-LP), respectively ( and C). Subsequently, ChIP assay further validated the interaction of YY1 with SNHG8 promoter (). Moreover, a luciferase reporter assay demonstrated that YY1 knockdown greatly reduced the luciferase activity of lncRNA SNHG8 promoter-WT but not lncRNA SNHG8 promoter-Mut (). Furthermore, YY1 was overexpressed or depleted in A375 and A875 cells to evaluate the impact on the expression of SNHG8 (). RT-qPCR results indicated that YY1 deletion reduced lncRNA SNHG8 level in melanoma cells, whereas its addition showed an opposite effect (). In summary, YY1 transcriptionally activated lncRNA SNHG8 expression in melanoma cells.
Figure 2. LncRNA SNHG8 is activated by YY1
LncRNA SNHG8 knockdown suppresses melanoma cell viability and metastasis
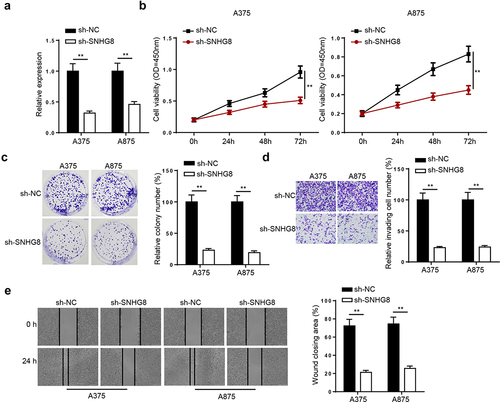
The function of lncRNA SNHG8 in melanoma was investigated by transfecting the A375 and A875 cells with sh-NC or sh-SNHG8. Knockdown efficiency was confirmed using RT-qPCR (). CCK-8 assay indicated that lncRNA SNHG8 knockdown inhibited A375 and A875 cell viability (). The colony formation assay demonstrated that colony formation was reduced by lncRNA SNHG8 knockdown in the A375 and A875 cells (). The Matrigel assay indicated that cell invasion was inhibited in sh-SNHG8-transfected melanoma cells (). The wound healing assay indicated that cell migration was decreased following lncRNA SNHG8 knockdown in A375 and A875 cells (). The data indicated that lncRNA SNHG8 silencing suppressed melanoma cell proliferation, migration, and invasion.
Figure 3. LncRNA SNHG8 knockdown suppresses melanoma cell viability and metastasis
LncRNA SNHG8 binds to miR-656-3p
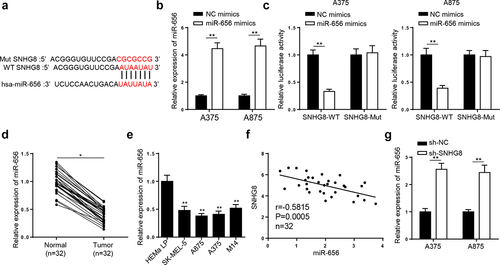
Subsequently, the mechanism underlying lncRNA SNHG8 in melanoma was investigated. StarBase website was applied to predict probable targets of lncRNA SNHG8. Under the condition of CLIP-Data: strict stringency (≥5) and Pan-Cancer = 6, miR-656-3p were screened. The binding sites between lncRNA SNHG8 and miR-656-3p were exhibited in . Then, RT-qPCR analysis revealed that miR-656-3p enhancement markedly lifted miR-656-3p level in A375 and A875 cells (). Later, it was shown that miR-656-3p supplementation repressed the luciferase activity of SNHG8-WT in the A375 and A875 cells, while it had no significant effect on reporter gene with SNHG8-Mut (). Moreover, miR-656-3p was downregulated in melanoma tissues and cells ( and E). Furthermore, an inverse correlation between lncRNA SNHG8 and miR-656-3p level was found in melanoma tissues (). Besides, lncRNA SNHG8 interference upregulated miR-656-3p in A375 and A875 cells (). In a word, lncRNA SNHG8 sponged miR-656-3p in melanoma.
Figure 4. LncRNA SNHG8 binds to miR-656-3p
miR-656-3p directly targeted SERBP1
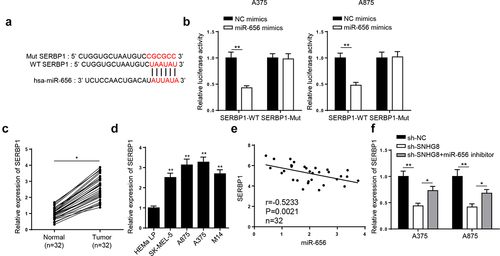
Firstly, SERBP1 was forecasted to bind with miR-656-3p by 4 databases (StarBase, PITA,miRmap, and microT) (). Then it was demonstrated that miR-656-3p mimics restrained SERBP1-WT luciferase activities in A375 and A875 cells, but not those of SERBP1-Mut (). Then, we demonstrated that SERBP1 expression was lifted in melanoma tissues and cells ( and D). In addition, it was uncovered that SERBP1 level was negatively related to miR-656-3p in melanoma tissues (). Finally, RT-qPCR results indicated that SERBP1 level was remarkably reduced by lncRNA SNHG8 silence, which could be reversed following miR-656-3p inhibition (). Altogether, the results elucidated that lncRNA SNHG8 bound to miR-656-3p to enhance SERBP1 level.
Figure 5. miR-656-3p directly targeted SERBP1
miR-656 inhibits melanoma cell proliferation, migration, and invasion via SERBP1
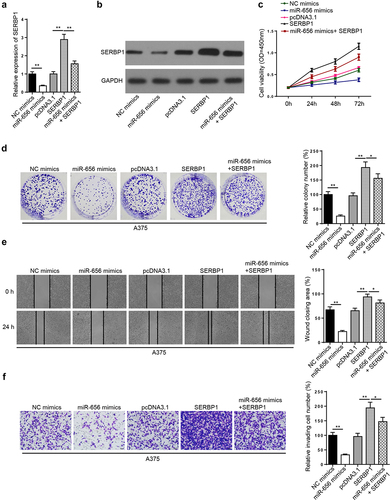
To investigate whether the role of lncRNA SNHG8 in melanoma was involved in the miR-656/SERBP1 axis, the A375 cells were transfected with either NC mimics, miR-656 mimics, pcDNA3.1, SERBP1, and miR-656 mimics + SERBP1. As evidenced by RT-qPCR and Western blot analysis, SERBP1 mRNA and protein expression levels were markedly reduced by miR-656 mimics, but notably increased by SERBP1 overexpression, respectively. miR-656 overexpression-mediated downregulation of SERBP1 was reversed by co-transfection with miR-656 mimics and SERBP1 overexpression plasmid ( and B). The CCK-8 and colony formation assays indicated that cell proliferation was decreased after miR-656 overexpression, but was enhanced after SERBP1 overexpression. Furthermore, SERBP1 overexpression partially abolished the suppressive effect of miR-656 overexpression on A375 cell proliferation ( and D). The results of the wound healing assay indicated that miR-656 and SERBP1 overexpression inhibited and enhanced cell migration, respectively. miR-656 overexpression-induced inhibition of migration was rescued by SERBP1 overexpression (). The Matrigel assay revealed that cell invasion was inhibited by miR-656 overexpression, but was enhanced by SERBP1 overexpression. Furthermore, SERBP1 upregulation restored the inhibitory effect of miR-656 overexpression on cell invasion (). Collectively, the results indicated that SERBP1 overexpression abolished miR-656 mimics-mediated inhibition of A375 cell proliferation, migration, and invasion.
Figure 6. miR-656-3p impedes melanoma cell viability, migration, and invasion via SERBP1
Discussion
The present study indicated that lncRNA SNHG8 expression was significantly increased in melanoma tissues and cells, and that lncRNA SNHG8 knockdown decreased melanoma cell proliferation, migration, and invasion by sponging miR-656; therefore, increasing the expression level of SERBP1. The present study not only identified a novel regulatory mechanism underlying lncRNA SNHG8/miR-656/SERBP1 in melanoma but also suggested a novel therapeutic target for patients with melanoma.
Increasing evidence has revealed the role of lncRNAs in melanoma, such as RMEL3 [Citation31], nuclear paraspeckle assembly transcript 1 [Citation32], CASC2 [Citation33], MIR31 host gene [Citation34], and colon cancer associated transcript 1 [Citation35]. LncRNA SNHG8 was found to be associated with the development of a number of carcinomas. For example, lncRNA SNHG8 was involved in endometrial cancer development by enhancing c-MET expression by competitive binding to miR-152 [Citation36]. LncRNA SNHG8 was a pivotal regulator in the progression of non-small-cell lung cancer cells and sponged miR-542-3p by targeting cyclin D1/cyclin dependent kinase 6 [Citation16]. LncRNA SNHG8 increased the progression and decreased the chemosensitivity of pancreatic adenocarcinoma cells [Citation37]. Consistent with the aforementioned reports, we found that lncRNA SNHG8 expression was increased in melanoma tissues and cells. Furthermore, lncRNA SNHG8 silencing inhibited melanoma cell proliferation, migration, and invasion. In addition, we found that YY1 directly binds to lncRNA SNHG8 promoter and facilitated lncRNA SNHG8 expression. These data indicated that YY1-induced lncRNA SNHG8 upregulation promoted the tumorigenesis of melanoma.
Subsequently, the present study investigated the mechanism underlying lncRNA SNHG8 in melanoma. An increasing number of studies have focused on the competitive endogenous (ce) RNA networks that lncRNAs can interact with miRNAs in competition with miRNA-targeted mRNA [Citation38]. For example, lncRNA CASC21 exerted carcinogenic functions in colorectal cancer by mediating the miR-7-5p/Yes1 associated transcriptional regulator axis [Citation39]. LncRNA taurine up-regulated 1 was associated with cerebral ischemia/reperfusion injury by targeting miR-145 to increase aquaporin 4 mRNA expression level [Citation40]. LncRNA TMPO antisense RNA 1 served as a ceRNA in promoting osteosarcoma initiation by regulating the miR-199a-5p/Wnt family member 7B axis [Citation41]. Therefore, this study aimed to explore the potential ceRNA mechanism underlying lncRNA SNHG8 in melanoma.
The bioinformatic databases predicted the binding sites between lncRNA SNHG8 and miR-656, and miR-656 and SERBP1. A previous study indicated that miR-656 could be sponged by NORAD, resulting in the occurrence and development of non-small cell lung cancer [Citation24]. By contrast, SERBP1 exerted an oncogenic role in human cancer. Wang et al [Citation42] reported that miR-218 suppressed hepatocellular carcinoma cell metastasis and epithelial-mesenchymal transition by targeting SERBP1. Herein, luciferase reporter assays confirmed the interactions between lncRNA SNHG8 or SERBP1 and miR-656. RT-qPCR analysis was performed to investigate the negative and positive effects of lncRNA SNHG8 on miR-656 and SERBP1, respectively, and the negative effect of miR-656 on SERBP1 in melanoma cells. The results revealed that lncRNA SNHG8 competitively interacted with miR-656 to increase SERBP1 expression levels in melanoma cells. In addition, rescue experiments indicated that miR-656 overexpression inhibited melanoma cell proliferation, migration and invasion, whereas SERBP1 overexpression reversed miR-656 overexpression-mediated effects.
In Conclusion
Our results demonstrated that lncRNA SNHG8 promoted melanoma cell proliferation, migration, and invasion via a ceRNA network, in which lncRNA SNHG8 targeted miR-656 to enhance SERBP1 expression. Our study might provide a novel therapeutic target for melanoma treatment. However, the expression of lncRNA SNHG8, miR-656, and SERBP1 should be determined in a larger cohort, and in vivo assays are required to confirm our conclusion in the future study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Mo H, Guan J, Yuan ZC, et al. Expression and predictive value of miR-489 and miR-21 in melanoma metastasis. World J Clin Cases. 2019;7:2930–2941.
- Zhang C, Li H, Wang J, et al. MicroRNA-338-3p suppresses cell proliferation, migration and invasion in human malignant melanoma by targeting MACC1. Exp Ther Med. 2019;18:997–1004.
- Wu Y, Wang Y, Wang L, et al. Burden of melanoma in China, 1990-2017: findings from the 2017 global burden of disease study. Int J Cancer. 2020;147:692–701.
- Stadler S, Weina K, Gebhardt C, et al. New therapeutic options for advanced non-resectable malignant melanoma. Adv Med Sci. 2015;60:83–88.
- Holbrook K, Lutzky J, Davies MA, et al. Intracranial antitumor activity with encorafenib plus binimetinib in patients with melanoma brain metastases: a case series. Cancer. 2020;126:523–530.
- Santos CAD, Souza DLB. Melanoma mortality in Brazil: trends and projections (1998-2032). Cien Saude Colet. 2019;24:1551–1561.
- Zhang XY, Zheng LW, Li CJ, et al. Dysregulated Expression of Long Noncoding RNAs in Endometriosis. Crit Rev Eukaryot Gene Expr. 2019;29:113–121.
- Li Y, Jiang L, Lv S, et al. E2F6-mediated lncRNA CASC2 down-regulation predicts poor prognosis and promotes progression in gastric carcinoma. Life Sci. 2019;232:116649.
- Angenard G, Merdrignac A, Louis C, et al. Expression of long non-coding RNA ANRIL predicts a poor prognosis in intrahepatic cholangiocarcinoma. Dig Liver Dis. 2019;51:1337–1343.
- Liu S, Lei H, Luo F, et al. The effect of lncRNA HOTAIR on chemoresistance of ovarian cancer through regulation of HOXA7. Biol Chem. 2018;399:485–497.
- Liu K, Zhang L, Li X, et al. High expression of lncRNA HSD11B1-AS1 indicates favorable prognosis and is associated with immune infiltration in cutaneous melanoma. Oncol Lett. 2022;23:54.
- Gao Y, Zhu H, Mao Q. Expression of lncRNA FGD5-AS1 correlates with poor prognosis in melanoma patients. J Gene Med. 2020;22:e3278.
- Xu H, Gong J, Liu H. High expression of lncRNA PVT1 independently predicts poor overall survival in patients with primary uveal melanoma, 12 (2017) e0189675.
- Zhang P, Li S, Chen Z, et al. LncRNA SNHG8 promotes proliferation and invasion of gastric cancer cells by targeting the miR-491/PDGFRA axis. Hum Cell. 2020;33:123–130.
- Dong J, Teng F, Guo W, et al. lncRNA SNHG8 Promotes the Tumorigenesis and Metastasis by Sponging miR-149-5p and Predicts Tumor Recurrence in Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;51:2262–2274.
- Chen C, Zhang Z, Li J, et al. SNHG8 is identified as a key regulator in non-small-cell lung cancer progression sponging to miR-542-3p by targeting CCND1/CDK6. Onco Targets Ther. 2018;11:6081–6090.
- Ye C, Hu Y, Wang J. MicroRNA-377 Targets Zinc Finger E-box-Binding Homeobox 2 to Inhibit Cell Proliferation and Invasion of Cervical Cancer. Oncol Res. 2019;27:183–192.
- Qu J, Zhang L, Li L, et al. miR-148b Functions as a Tumor Suppressor by Targeting Endoplasmic Reticulum Metallo Protease 1 in Human Endometrial Cancer Cells. Oncol Res. 2018;27:81–88.
- Zhang T, Wang N. miR-135a Confers Resistance to Gefitinib in Non-Small Cell Lung Cancer Cells by Upregulation of RAC1. Oncol Res. 2018;26:1191–1200.
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038.
- Conte F, Fiscon G, Sibilio P, et al. An Overview of the Computational Models Dealing with the Regulatory ceRNA Mechanism and ceRNA Deregulation in Cancer. Methods Mol Biol. 2021;2324:149–164.
- Fiscon G, Paci P, Iannello G. MONSTER v1.1: a tool to extract and search for RNA non-branching structures. BMC Genomics. 2015;16:S1.
- Tian X, Liu Y, Wang Z, et al. lncRNA SNHG8 promotes aggressive behaviors of nasopharyngeal carcinoma via regulating miR-656-3p/SATB1 axis. Biomed Pharmacother. 2020;131:110564.
- Chen T, Qin S, Gu Y, et al. Long non-coding RNA NORAD promotes the occurrence and development of non-small cell lung cancer by adsorbing MiR-656-3p. Mol Genet Genomic Med. 2019;7:e757.
- Schnipper LE, Davidson NE, Wollins DS, et al. American Society of Clinical, American Society of Clinical Oncology Statement: a Conceptual Framework to Assess the Value of Cancer Treatment Options. J Clin Oncol. 2015;33:2563–2577.
- Zhang M, Xu Y, Yin S, et al. YY1-induced long non-coding RNA PSMA3 antisense RNA 1 functions as a competing endogenous RNA for microRNA 214-5p to expedite the viability and restrict the apoptosis of bladder cancer cells via regulating programmed cell death-ligand 1. Bioengineered. 2021;12:9150–9161.
- Yao Y, Li X, Cheng L, et al. Circular RNA FAT atypical cadherin 1 (circFAT1)/microRNA-525-5p/spindle and kinetochore-associated complex subunit 1 (SKA1) axis regulates oxaliplatin resistance in breast cancer by activating the notch and Wnt signaling pathway. Bioengineered. 2021;12:4032–4043.
- Feng LL, Shen FR, Zhou JH, et al. Expression of the lncRNA ZFAS1 in cervical cancer and its correlation with prognosis and chemosensitivity. Gene. 2019;696:105–112.
- Ji L, Fan X, Zhou F, et al., lncRNA RPL34-AS1 inhibits cell proliferation and invasion while promoting apoptosis by competitively binding miR-3663-3p/RGS4 in papillary thyroid cancer, 235 (2020) 3669–3678.
- Zhang Y, Zhang Y, Wang S, et al. SP1-induced lncRNA ZFPM2 antisense RNA 1 (ZFPM2-AS1) aggravates glioma progression via the miR-515-5p/Superoxide dismutase 2 (SOD2) axis. Bioengineered. 2021;12:2299–2310.
- Cardoso C, Serafim RB, Kawakami A, et al. The lncRNA RMEL3 protects immortalized cells from serum withdrawal-induced growth arrest and promotes melanoma cell proliferation and tumor growth. Pigment Cell Melanoma Res. 2019;32:303–314.
- Xia Y, Zhou Y, Han H, et al. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J Cell Physiol. 2019;234:19592–19601.
- Zhang Y, Qian W, Feng F, et al. Upregulated lncRNA CASC2 May Inhibit Malignant Melanoma Development Through Regulating miR-18a-5p/RUNX1. Oncol Res. 2019;27:371–377.
- Montes M, Nielsen MM, Maglieri G, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015;6:6967.
- Lv L, Jia JQ, Chen J. The lncRNA CCAT1 Upregulates Proliferation and Invasion in Melanoma Cells via Suppressing miR-33a. Oncol Res. 2018;26:201–208.
- Yang CH, Zhang XY, Zhou LN, et al. LncRNA SNHG8 participates in the development of endometrial carcinoma through regulating c-MET expression by miR-152. Eur Rev Med Pharmacol Sci. 2018;22:1629–1637.
- Song Y, Zou L, Li J, et al. LncRNA SNHG8 promotes the development and chemo-resistance of pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci. 2018;22:8161–8168.
- Wu M, Li W, Huang F, et al. Comprehensive Analysis of the Expression Profiles of Long Non-Coding RNAs with Associated ceRNA Network Involved in the Colon Cancer Staging and Progression. Sci Rep. 2019;9:16910.
- Zheng Y, Nie P, Xu S. Long noncoding RNA CASC21 exerts an oncogenic role in colorectal cancer through regulating miR-7-5p/YAP1 axis. Biomed Pharmacother. 2020;121:109628.
- Shan W, Chen W, Zhao X, et al. Long noncoding RNA TUG1 contributes to cerebral ischaemia/reperfusion injury by sponging mir-145 to up-regulate AQP4 expression. J Cell Mol Med. 2020;24:250–259.
- Cui H, Zhao J. LncRNA TMPO-AS1 serves as a ceRNA to promote osteosarcoma tumorigenesis by regulating miR-199a-5p/WNT7B axis. J Cell Biochem. 2020;121:2284–2293.
- Wang T, Xu L, Jia R, et al. MiR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin (Shanghai). 2017;49:383–391.
