ABSTRACT
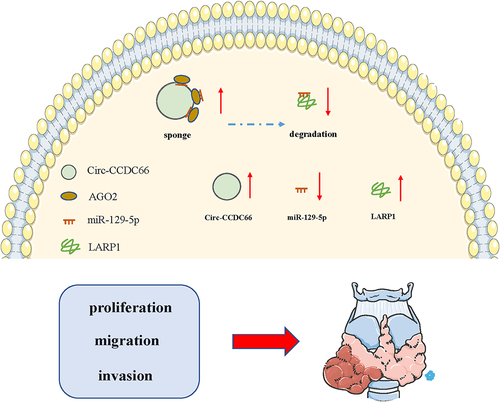
Circular RNAs (circRNAs) play vital roles in the development and progression of various diseases. CircRNA coiled-coil domain containing 66 (circ-CCDC66) has been reported to be involved in several cancers, but its biological function and underlying mechanism in papillary thyroid carcinoma (PTC) remain unclear. We detected the relative expression level of circ-CCDC66 in PTC specimens and cell lines using real-time reverse transcription PCR. In addition, EdU assay, transwell assay, and xenograft analysis were performed to measure the effect of circ-CCDC66 on the proliferative, migratory, and invasive capacities of PTC cells. We also investigated the potential mechanism of circ-CCDC66 by bioinformatics analysis, RNA immunoprecipitation, and dual-luciferase reporter assay. We observed that circ-CCDC66 expression was upregulated in PTC specimens and cell lines and was correlated with poor clinical characteristics of PTC patients. Moreover, in vitro experiments demonstrated that knockdown of circ-CCDC66 markedly suppressed the proliferative, migratory, and invasive capacities of PTC cells. Mechanistically, miR-129-5p was a target gene of circ-CCDC66 and was downregulated in PTC tissues. LARP1, a downstream target of miR-129-5p, was upregulated in PTC tissues. In addition, we confirmed that inhibition of circ-CCDC66 could repress xenograft tumor growth. Circ-CCDC66 promoted PTC proliferation, migration, invasion, and tumor growth by sponging miR-129-5p and promoting LARP1 expression.
Graphical Abstract
1. Introduction
Thyroid carcinoma (TC) is the most common malignant tumor of the endocrine system, with an incidence increasing by approximately 3% every year [Citation1,Citation2]. TC mortality rates are generally low; however, TC mortality rates have increased by approximately 1.1% per year over the past 40 years [Citation3,Citation4]. Papillary TC (PTC) accounts for 85% of all TC cases, affecting all age groups [Citation5]. It is a commonly diagnosed TC with relatively low malignancy rates [Citation6,Citation7]. In recent years, the incidence of PTC has increased annually due to environmental, genetic, and hormonal factors. While most PTC cases have good prognosis, some PTC cases are invasive, leading to the involvement of adjacent organs, extraglandular invasion, or lymph node metastasis at the early stage. As a result, surgical resection rate is markedly reduced, resulting in poor prognosis [Citation8,Citation9]. Therefore, an in-depth understanding of the molecular mechanisms underlying invasiveness and metastasis of PTC is urgently needed for better assessment of invasive risk, and clinical guidance. Many studies have shown that certain proteins are responsible for the biological behaviors of PTC cells and can be utilized as therapeutic targets [Citation10,Citation11]. It is generally believed that invasiveness, local recurrence, and distant metastasis are the main factors affecting cancer prognosis. However, the precise pathogenesis attributed to the invasiveness and metastasis of PTC is not fully understood.
Circular RNAs (circRNAs) are a novel type of endogenous non-coding RNA (ncRNA) characterized by a closed covalent loop in the absence of a 5ʹ cap and 3ʹ poly (A) tail. They are abundantly, robustly, and conservatively expressed [Citation12–14]. According to their sources, circRNAs are classified as ecRNAs, elciRNAs, and ciRNAs, and the former accounts for the majority of circRNA [Citation15–17]. CircRNAs have diverse biological functions in cancer development, such as serving as competing endogenous RNAs (ceRNAs), which competitively bind microRNAs (miRNAs) to further affect the biological functions of their target genes, which is the most reported mechanism of circRNAs in cancer development [Citation18–20]. In brief, miRNAs are single-chain small-molecular RNAs that can inhibit translation or induce degradation of target genes by recognizing and binding to the 3ʹ UTR. Moreover, miRNAs have been reported to be involved in various cancers [Citation21]. miR-335 can inhibit the aggressive phenotypes of ovarian cancer cells by inhibiting COL11A1 expression [Citation22]. miR-210-3p can inhibit pancreatic tumor growth and proliferation by targeting MUC4 [Citation23]. miR-129-5p has been reported to act as a tumor suppressor in various cancers, including gastric cancer [Citation24], colon cancer [Citation25], hepatocellular cancer [Citation26], prostate cancer [Citation27], and breast cancer [Citation28]. CircRNAs also exert a ‘sponge’ effect on proteins to alter their subcellular distribution, mediate transcription of parent genes, and promote protein–protein interaction [Citation29,Citation30]. In addition, circRNAs have translational potential, as some circRNAs contain an internal ribosome entry site (IRES) and open reading frame (ORF), and are capable of intracellularly translating proteins [Citation31,Citation32]. It has also been reported that circRNAs (elciRNAs and ciRNAs) can regulate the expression of parent genes by mediating the transcription activities of RNA Polymerase II and other transcription factors [Citation33,Citation34]. Collectively, circRNAs are functional mediators involved in cancer cell behavior through multiple mechanisms.
CircRNA coiled-coil domain containing 66 (circ-CCDC66) has been identified as a potential therapeutic target in cancer. It has been reported that circ-CCDC66 is upregulated in colorectal cancer (CRC) cases and affects cancer development by mediating certain oncogenes. Knockdown of circ-CCDC66 inhibits the growth and invasiveness of CRC in xenograft and in situ mouse models [Citation35]. Circ-CCDC66 has been reported to be upregulated not only in renal cancer cell lines but also in tumor stem cell spheres, and enhances the enrichment of tumor stem cells [Citation36]. Circ-CCDC66 can promote proliferation, invasion, and migration of glioma cells by suppressing miR-320a and promoting FOXM1 expression [Citation37]. Circ-CCDC66 can upregulate REXO1 expression to aggravate cervical cancer progression by binding to miR-452-5p [Citation38]. Moreover, many studies have confirmed that corresponding preclinical research can be carried out for some key genes, providing a new perspective for tumor prevention and treatment [Citation39–43]. However, the exact function and clinical potential of circ-CCDC66 in PTC remain to be explored.
In the present study, circ-CCDC66 expression was found to be upregulated in PTC specimens and cell lines. Knockdown of circ-CCDC66 markedly suppressed the proliferative, migratory, and invasive capacities of PTC cells. Furthermore, functional experiments demonstrated that circ-CCDC66 promoted the development of PTC by upregulating La-related protein 1 (LARP1) by exerting a ‘sponge’ effect on miR-129-5p.
2. Materials and methods
2.1. Participants and specimens
This study was approved by the Ethics Committee of the Fourth Affiliated Hospital of Anhui Medical University (Hefei, China) and conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent. A total of 60 PTC and paired paracancerous specimens were collected, frozen in liquid nitrogen, and stored at −80°C. Tumor staging was assessed based on the guidelines proposed by the Union for International Cancer Control. Clinical data was recorded, and none of the recruited patients underwent preoperative radiotherapy or chemotherapy, or had other types of malignancies.
2.2. Cell culture
The human thyroid cell line Nttry-ori-3-1 and PTC cell lines IHH4, BCPAP, K1 and TPC-1 were obtained from ATCC (Manassas, VA, USA). All cells were cultured in RPMI-1640 (Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 100 U/mL penicillin, and 0.1 mg/mL streptomycin in a humidified 5% CO2 incubator at 37°C and passaged at 80–90% confluence. The identity of the cells used in the experiment was confirmed using short tandem repeat analysis. All cell lines were tested for Mycoplasma every 3 months.
2.3. Real-time reverse transcription PCR (qRT-PCR)
Total cellular RNA was extracted using TRIzol (Invitrogen) and stored at −80°C until further analysis. The Prime Script RT Reagent Kit (Takara, Dalian, China) was used to obtain cDNA from the reverse transcription of RNAs (500 ng). qRT-PCR was performed using 2 µL cDNA as the template, 1 µL each of forward and reverse primers and the SYBR Green qPCR Mix kit. The PCR reaction conditions were as follows initial denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30s, and extension at 72°C for 30 s; final extension at 72°C for 10 min. qRT-PCR was performed on the ABI7900 fluorescent PCR instrument (Applied Biosystems, Waltham, MA, USA). U6 and GAPDH were used as the internal reference genes. The relative mRNA levels were calculated using the 2−ΔΔCT method [Citation44]. Sequences of primers used for qRT-PCR are listed in .
Table 1. Sequences of primers for qRT-PCR
2.4. Actinomycin D assay
The actinomycin D assay was performed as described previously [Citation45]. K1 and TPC-1 cells were exposed to 2 μg/mL actinomycin D (Sigma-Aldrich, St. Louis, MO, USA). qRT-PCR was then performed as described above to detect relative levels of circ-CCDC66 and mRNA levels of CCDC66.
2.5. RNase R assay
The RNase R assay was performed as described previously [Citation45]. Briefly, 2 mg RNA with or without 5 U/μg RNase R (Epicenter Technologies, Madison, WI, USA) was incubated for 30 min in a water bath at 37°C. Then, RNeasy MinElute kit (Qiagen, Hilden, Germany) was used to purify the sample, and qRT-PCR was performed as described above.
2.6. Cell transfection
Small interfering (si) RNAs targeting circ-CCDC66 (si-circ-CCDC66#1 and #2), LARP1 siRNA, and corresponding negative controls were synthesized by GenePharma (Shanghai, China). The mimic/inhibitor miR-129-5p and the negative control were also obtained from GenePharma. Short hairpin (sh) RNA targeting circ-CCDC66 (sh-circ-CCDC66) and sh-NC were designed and purchased from Genecopoeia (Guangzhou, China). The lentiviral vector for circ-CCDC66 was purchased from GeneCreate Biological Engineering (Wuhan, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions, and the transfection efficacy was tested at 48 h. The sequence of siRNAs used was: si-circ-CCDC66 #1, sense: 5ʹ-AUUUUCUUUGCAGUUCUUGUU-3ʹ, antisense: 5ʹ-CAAGAACUGCAAAGAAAAUGG-3ʹ; si-circ-CCDC66 #2, sense: 5ʹ-AAUAUAUAAUUUUUUCCUCUA-3ʹ, antisense: 5ʹ-GAGGAAAAAAUUAUAUAUUCA-3ʹ; LARP1 siRNA, sense: 5ʹ-AUAGUUAAAACUUCAGAACAA-3ʹ, antisense: 5ʹ- GUUCUGAAGUUUUAACUAUUA-3ʹ. Transfection ef-ficiency of more than 70% was considered as an effective transfection.
2.7. Proliferation assay
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was performed as described previously [Citation46]. Briefly, approximately 2 × 103 transfected cells were seeded per well in a 96-well plate and cultivated for 0, 24, 48, and 72 h, respectively. The cells were exposed to CCK-8 solution for 2 h, followed by detection at 450 nm using a microplate reader (BioTek Instruments, Winooski, VT, USA).
EdU assay
EdU assay was performed as described previously [Citation47]. Briefly, 3 × 103 transfected cells were seeded per well in a 96-well plate and exposed to 50 mM EdU (RiboBio, Guangzhou, China) for 24 h. Cells were then fixed using 4% methanol and permeabilized with Triton X-100. The cells were then cultivated in EdU mixed buffer and counterstained with DAPI. EdU-positive and DAPI-positive cells in five random fields per well were captured under a fluorescence microscope.
2.8. Transwell assay
Transwell assay was performed as described previously [Citation47]. Transwell inserts (8 μm, Corning, Corning, NY, USA) were placed in a 24-well plate. The upper insert was pre-coated with 100 μg Matrigel, and then 4 × 104 cells suspended in FBS-free medium were seeded in the upper chamber, and 500 μL of medium containing 10% FBS was added to the bottom chamber. Cells were allowed to penetrate for 24 h, and those in the bottom chamber were fixed with methanol for 15 min and stained with crystal violet for 20 min for visualization. Invasive cells were quantified by capturing five random fields per well (×200). Migratory cells were similarly measured in transwell inserts without pre-coating with Matrigel. The number of migratory or invaded cells was determined using ImageJ v1.5.
2.9. Subcellular fractionation
Subcellular fractionation was performed as described previously [Citation48]. Briefly, approximately 4 × 104 cells were transferred to a 1.5 ml EP tube and lysed in RLA on ice for 20 min, followed by centrifugation at 3,000 rpm for 15 min. The supernatant was collected as the cytoplasmic fraction. Next, the precipitant was washed in RLA three times and induced in RIPA on ice for 20 min, with 30 s vortex oscillation at 5 min intervals. The mixture was then centrifuged at 12,500 rpm for 15 min, and the supernatant was collected as the nuclear fraction.
2.10. Dual-luciferase reporter assay
Dual-luciferase reporter assay was performed as described previously [Citation49]. The wild-type (WT) luciferase reporter vectors circ-CCDC66-WT and LARP1 3ʹ UTR-WT were constructed by inserting the sequence of circ-CCDC66 or LARP1 3 UTR containing miR-129-5p binding sites into the pmir-GLO vector. The mutant (MUT) circ-CCDC66-MUT and LARP1 3 UTR-MUT were constructed by mutating the complementary sites (CGTTTTT) of miR-136-5p to (GTTTTT). MUT or WT circ-CCDC66 (or LARP1) luciferase vector was co-transfected in K1 and TPC-1 cells with either negative control or miR-129-5p mimics for 48 h. Cells were processed using the dual-luciferase reporter assay kit (Promega, Madison, WI, USA) to measure relative luciferase activity.
2.11. Western blot
Western blot was performed as described previously [Citation50]. Briefly, PTC cells were lysed in PMSF on ice to extract protein and the protein concentrations were measured using BCA protein kit (Beyotime, Shanghai, China). Protein samples (30 μg) were denatured at 100°C and then separated by 10% SDS-PAGE, and loaded onto PVDF membranes, which were cut into pieces according to the molecular size of the protein of interest. Membranes were immunoblotted with primary and secondary antibodies and exposed to analyze the protein bands. The primary antibodies used were as follows: LARP1 (ab86359, 1:3000, Abcam, Cambridge, UK) and GAPDH (ab8245, 1:1000, Abcam).
2.12. Immunohistochemistry (IHC)
IHC was performed as described previously [Citation51]. Briefly, the PTC tissues were heated in citrate buffer and cooled to room temperature for antigen retrieval. The specimens were then incubated with LARP1 antibody (ab86359, 1:200, Abcam) overnight at 4°C, followed by incubation with a secondary antibody at room temperature for 2 h. The specimens were further stained with streptavidin-biotin-peroxidase reagents and fixed on gelatin-coated glass slides. Then, the average number of LARP1-positive cells was calculated.
2.13. Tumor xenograft assay
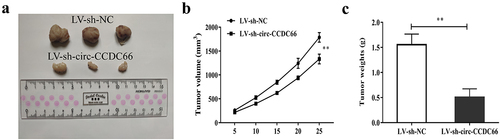
BALB/c male nude mice (4 weeks old) were purchased from Vital River Laboratory Animal Technology (Beijing, China). Tumor xenograft assay was performed as described previously [Citation52]. Briefly, 5 × 106 TPC-1 cells were stably transfected with lentivirus-carrying sh-circ-CCDC66 or sh-NC and subcutaneously injected into the right flank of nude mice. The volume (length × width2 × 0.5) of the tumor was calculated on days 0, 5, 10, 15, 20, and 25. The weight of the tumor was counted the 25th day. All procedures involving animals were approved by the Animal Care and Use Committee of the Fourth Affiliated Hospital of Anhui Medical University.
2.14. Statistical analysis
All experiments were performed three times. The GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA) and SPSS 22.0 (IBM, Armonk, NY, USA) were used to process data that were expressed as mean ± standard deviation. Differences between groups were compared using Student’s t-test and one-way ANOVA. Pearson correlation tests were conducted to assess the correlation between the genes. Kaplan–Meier curves were used for survival analysis. Statistical significance was set at p < 0.05.
3. Results
This study aimed to demonstrate the expression level, biological role, and potential mechanism of circ-CCDC66 in PTC. We demonstrated that circ-CCDC66 may play an essential role in the progression of PTC. Our results indicated that circ-CCDC66 expression levels were markedly increased in PTC tissues and cell lines, and circ-CCDC66 could sponge miR-129-5p to promote LARP1 expression, which can potentially accelerate malignant progression of PTC.
3.1. Circ-CCDC66 is upregulated and correlated with poor prognosis in PTC
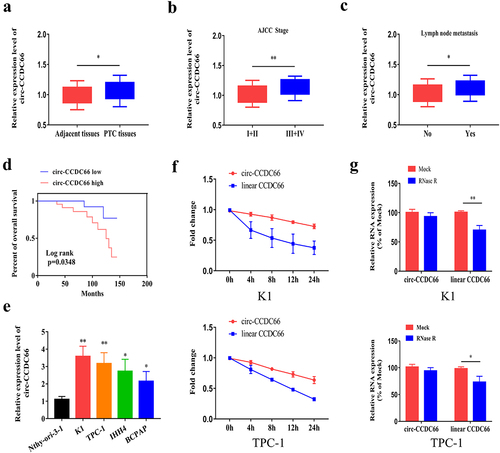
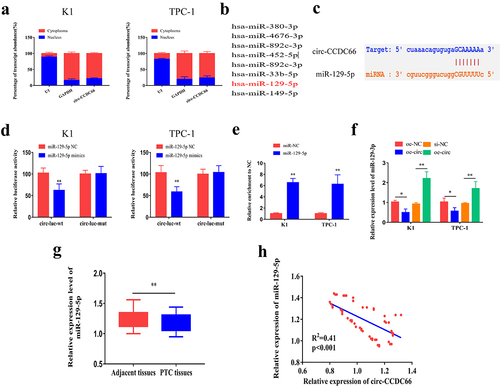
To explore the role of circ-CCDC66 in PTC, we detected relative levels of circ-CCDC66 by qRT-PCR in PTC samples and observed that circ-CCDC66 was highly expressed in PTC samples compared to the controls (). We further analyzed the correlation between circ-CCDC66 and the clinical data of patients with PTC. High levels of circ-CCDC66 were closely correlated with AJCC staging and lymphatic metastasis of PTC (–C). Survival curves were depicted using the Kaplan–Meier method based on the follow-up data of patients with PTC. As shown in , high levels of CCDC66 were predictive of poor prognosis in patients with PTC. We also analyzed the relationship between the expression levels of circ-CCDC66 and clinical pathological features. As shown in , the expression level of circ-CCDC66 was significantly associated with tumor size, TNM stage, and lymph node metastasis in patients with PTC. Additionally, circ-CCDC66 was upregulated in PTC cell lines compared to the normal thyroid cell line (). Among them, the expression of circ-CCDC66 was most significantly increased in K1 and TPC-1 cells, so we selected these two cell lines for in vitro experiments. Based on the features of stably expressed circRNAs, we examined the stability of circ-CCDC66 by exposing PTC cells to actinomycin D. Compared with the half-life of linear CCDC66 mRNA (<8 h), the half-life of circ-CCDC66 was longer than 24 h, confirming the stability of circ-CCDC66 (). Consistent with these findings, RNase R treatment did not influence the expression level of circ-CCDC66, whereas linear CCDC66 mRNA was markedly degraded ().
Table 2. Relationship between circ-CCDC66 expression and the clinical pathological characteristics of PTC patients (n = 60)
Figure 1. Circ-CCDC66 is upregulated in PTC and correlated to poor prognosis.
3.2. Knockdown of circ-CCDC66 suppresses proliferative, migratory and invasive capacities of PTC cells
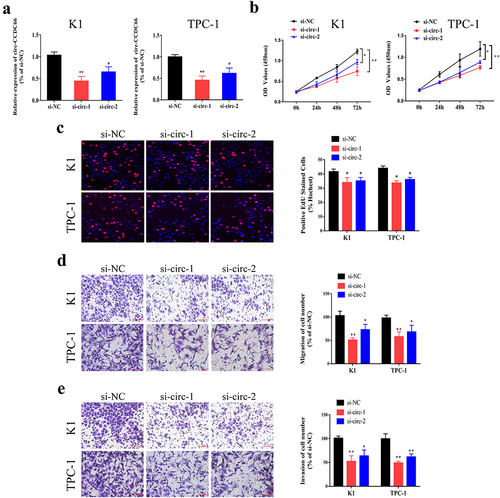
To explore the role of circ-CCDC66 in PTC, the expression of circ-CCDC66 was silenced by siRNA transfection, and the transfection efficacy of circ-CCDC66 siRNAs in K1 and TPC-1 cells was examined by qRT-PCR (). CCK-8 and EdU assays showed that knockdown of circ-CCDC66 in PTC cells markedly attenuated proliferative capacity (). Furthermore, the Transwell assay revealed that inhibition of circ-CCDC66 suppressed the migratory and invasive capacities of K1 and TPC-1 cells (). Taken together, these results suggest that circ-CCDC66 may play an oncogenic role in the development of PTC.
Figure 2. Knockdown of circ-CCDC66 suppressed proliferative, migratory and invasive capacities of PTC.
3.3. Circ-CCDC66 can bind to miR-129-5p
Furthermore, we investigated the mechanism through which circ-CCDC66 promotes malignant progression of PTC. First, we explored the subcellular distribution of circ-CCDC66. We observed that circ-CCDC66 was mainly distributed in the cytoplasm of PTC cells, suggesting a role of circ-CCDC66 in mediating post-transcriptional levels of genes (). By analyzing the Starbase database (http://starbase.sysu.edu.cn/), the following candidate miRNAs were identified to bind circ-CCDC66 in the promoter region: miR-129-5p, miRNA-380-3p, miRNA-4676-3p, miRNA-892c-3p, miRNA-452-5p, miRNA-33b-5p, and miRNA-149-5p (). Among the screened miRNAs, miR-129-5p was the most likely to bind circ-CCDC66. We constructed WT and MUT circ-CCDC66 vectors based on the binding sites of circ-CCDC66 and miR-129-5p (). Overexpression of miR-129-5p in K1 and TPC-1 cells markedly decreased luciferase activity in the WT circ-CCDC66 vector, while the luciferase activity in the MUT vector was not affected, confirming that circ-CCDC66 bound miR-129-5p (). Furthermore, the interaction between the two was also confirmed by RNA immunoprecipitation (RIP) (). We also confirmed that circ-CCDC66 could bind to miR-129-5p in 293 T cells by dual-luciferase reporter gene and RIP assays (Figure S1A–B). In K1 and TPC-1 cells, circ-CCDC66 negatively regulated expression of miR-129-5p (). Additionally, qRT-PCR data revealed that miR-129-5p expression in PTC specimens was low (). The expression level of miR-129-5p was also significantly associated with tumor size, TNM stage, and lymph node metastasis in patients with PTC (). miR-129-5p expression was found to be negatively correlated with circ-CCDC66 levels in PTC specimens (R2 = 0.41, p < 0.001) (). Through in vitro experiments, we found that overexpression of circ-CCDC66 might promote the EMT signaling, while overexpression of miR-129-5p could inhibit the EMT pathway (Figure S2). We speculated that circ-CCDC66 might affect the malignant progression of PTC by regulating the EMT pathway.
Table 3. Relationship between miR-129-3p expression and the clinical pathological characteristics of PTC patients (n = 60)
Figure 3. Circ-CCDC66 could bind miR-129-5p.
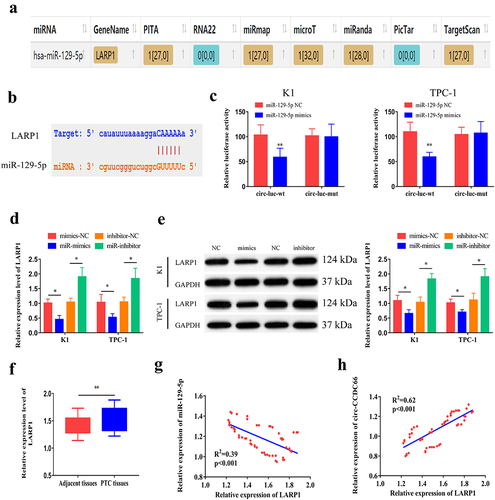
3.4. miR-129-5p can bind to LARP1
By analyzing microT, PITA, miRmap, microT, miRanda, and Targetscan databases, we searched for potential genes that could bind to miR-129-5p. After cross-matching the predicted data, LARP1 was identified as a potential target (). We then constructed WT and MUT LARP1 vectors () to examine the binding relationship between miR-129-5p and LARP1 using a dual-luciferase reporter assay (). In K1 and TPC-1 cells, overexpression of miR-129-5p downregulated the mRNA and protein levels of LARP1, while knockdown of miR-129-5p resulted in upregulation of LARP1 at the mRNA and protein levels (). Compared with paracancerous specimens, LARP1 was highly expressed in PTC tissues (). In addition, the expression level of LARP1 was significantly associated with tumor size, TNM stage, and lymph node metastasis in patients with PTC (). Moreover, we also performed IHC to examine LARP1 protein expression in normal and PTC tissues. We observed that LARP1 protein expression levels were higher in PTC tissues than in normal tissues (Figure S3A). Kaplan–Meier survival analysis from our data and The Cancer Genome Atlas (TCGA) dataset (http:// www.oncolnc.org/) revealed that LARP1 expression levels were not significantly correlated with the prognosis of PTC patients (Figure S3B–C). However, LARP1 expression was negatively correlated with miR-129-5p levels (R2 = 0.39, p < 0.001) and positively correlated with circ-CCDC66 levels in PTC specimens (R2 = 0.62, p < 0.001) (). Taken together, these results suggest that, circ-CCDC66 competitively binds to miR-129-5p to upregulate LARP1 expression.
Table 4. Relationship between LARP1 expression and the clinical pathological characteristics of PTC patients (n = 60)
Figure 4. miR-129-5p could bind LARP1.
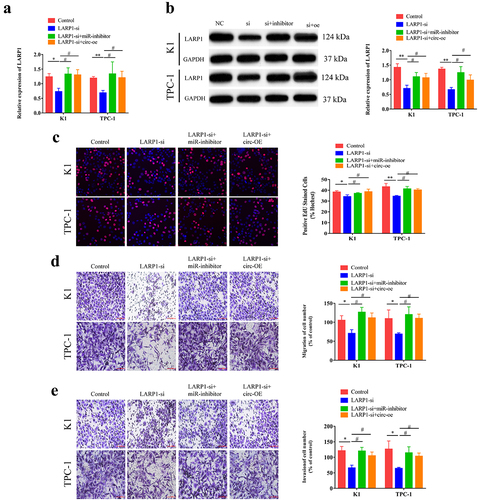
3.5. Circ-CCDC66 is involved in the development of PTC through the miR-129-5p/LARP1 axis
To ascertain the role of the circ-CCDC66/miR-129-5p/LARP1 axis in the development of PTC, we co-transfected LARP1-siRNA, or miR-129-5p inhibitor and circ-CCDC66-OE in K1 and TPC-1 cells. Higher levels of LARP1 were observed in cells co-transfected with LARP1-siRNA and circ-CCDC66-OE, or LARP1-siRNA and miR-129-5p inhibitor than in cells transfected with LARP1-siRNA alone (). Subsequently, we examined the influence of the circ-CCDC66/miR-129-5p/LARP1 axis on PTC cell behavior. LARP1 knockdown attenuated the proliferative, migratory, and invasive capacities of PTC cells. These effects were partially relieved by co-knockdown of LARP1 and miR-129-5p, or co-transfection of LARP1-siRNA and circ-CCDC66-OE (). These results suggested that circ-CCDC66 competitively binds miR-129-5p to upregulate LARP1, thus promoting the proliferative, migratory, and invasive capacities of PTC cells.
Figure 5. Circ-CCDC66 was involved in the development of PTC through the miR-129-5p/LARP1 axis.
3.6. Inhibition of circ-CCDC66 can inhibit the growth of PTC tumor
We also explored the effect of circ-CCDC66 inhibition on PTC tumor growth in a mouse xenograft model. As shown in , after 25 days of cancer cell inoculation, the tumor growth volume and weight of nude mice injected with PTC cells transfected with LV-sh-circ-CCDC66 were significantly smaller than those of LV-sh-NC.
4. Discussion
CircRNAs can act as ceRNAs that bind to miRNAs and participate in the progression and development of various cancers [Citation53,Citation54]. Cancer is a critical disease that threatens human lives worldwide and has been highlighted in life science research because of its complicated pathogenesis and limited therapeutic strategies. Many studies have shown that circRNAs are involved in cancer development and are promising novel biomarkers and therapeutic targets. The biological functions of circRNAs in the development of PTC have been reported previously. It has been reported that circ_PSD3 can stimulate the progression of PTC by modulating the miRNA-637/HEMGN axis and activating the PI3K/Akt signaling pathway [Citation55]. CircEIF3I can competitively bind to miRNA-149 to upregulate KIF2A, thus accelerating the development of PTC [Citation56]. The hsa_circ_0058124/NOTCH3/GATAD2A regulatory axis is critical to the carcinogenesis of PTC and its invasiveness and is a promising target for the management of PTC [Citation57]. Our study revealed that circ-CCDC66 is upregulated in PTC specimens and correlates with poor clinical characteristics in PTC cases. In vitro experiments also demonstrated the oncogenic role of circ-CCDC66 in modulating the proliferative, migratory, and invasive capacities of PTC cells.
We investigated the mechanism through which circ-CCDC66 regulates PTC cell functions. Our results suggest that circ-CCDC66 is able to bind to miR-129-5p and inhibit its expression. miR-129-5p expression was also found to be markedly downregulated in PTC specimens. miR-129-5p is a functional miRNA involved in several types of tumors, including breast cancer [Citation58], glioblastoma [Citation59], gastric cancer [Citation60,Citation61], lung cancer [Citation62], bladder cancer [Citation63] and prostate cancer [Citation64]. Zhang et al. [Citation65] hypothesized that lncRNA NEAT1 participates in the development of PTC through the miR-129-5p/KLK7 axis.
By analyzing online bioinformatics databases, we identified LARP1 as a potential target gene of miR-129-5p. LARP1 is an RNA-binding protein that was first discovered in Drosophila melanogaster. LARP1 has vital biological functions, especially in embryogenesis and cell cycle progression [Citation66,Citation67]. Members of the LARP family include LARP1, LARP1b, LARP4, LARP4b, LARP6, and LARP7, which are important for mRNA transcription or translation [Citation66,Citation67]. LARP1 is located on human chromosome 5 and encodes a protein containing 1,096 amino acids. LARP1 lacks an enzymatic domain, but the presence of LA structure combined with an RNA recognition motif, as well as a highly conserved DM15 domain is attributed to the function of LARP1 in protein translation and mRNA translation, respectively, via recognition of certain nuclear fragments [Citation68,Citation69]. So far, the function of the DM15 domain is inconsistent, which may be associated with the nuclear acid regulation and gene translation that are responsible for carcinogenesis. LARP1 is of significance in osteosarcoma [Citation70], prostate cancer [Citation71], non-small cell lung cancer [Citation72], CRC [Citation73] and ovarian cancer [Citation74]. However, the role of LARP1 in PTC requires further investigation.
Furthermore, we conducted a dual-luciferase reporter, RIP and Western blot assays, and found that miR-129-5p could target LARP1, while circ-CCDC66 promotes LARP1 expression by competitively binding to miR-129-5p. In addition, knockdown of LARP1 suppressed the proliferative, migratory, and invasive capacities of PTC cells as well as PTC tumor growth in a mouse xenograft model. Moreover, the oncogenic function of LARP1 could be partially reversed by knockdown of miR-129-5p or overexpression of circ-CCDC66.
Nevertheless, the present study has several limitations. First, we did not screen the differentially expressed circRNAs in PTC tissues using high-throughput sequencing technology, and the study lacked innovation. Second, circ-CCDC66 can bind to many miRNAs. We only investigated miR-129-5p, which has the strongest binding ability. Whether circ-CCDC66 can coordinately regulate the expression of LARP1 by binding to other miRNAs remains to be investigated. In addition, cell proliferation, migration, and invasion involve many related pathways, and whether circ-CCDC66 can regulate the biological effects of tumor cells by regulating the expression of these pathways remains to be explored further.
5. Conclusion
In conclusion, this study demonstrated that circ-CCDC66 was upregulated in PTC tissues and correlated with the poor prognosis in PTC patients. Circ-CCDC66 was also found to promote PTC cell proliferation, migration, and invasion by modulating the miR-129-5p/LARP1 regulatory axis. The circ-CCDC66/miR-129-5p/LARP1 axis may be a promising target for prognosis and therapy of PTC.
Author contributions
Guarantor of integrity of the entire study: Hongwu Li; Study concepts: Peipei Li and Junhui Chen; Study design: Peipei Li and Junhui Chen; Definition of intellectual content: Hongwu Li and Peipei Li; Literature research: Peipei Li, Junhui Chen and Jun Zou; Clinical studies: Peipei Li, Junhui Chen and Jun Zou; Experimental studies: Peipei Li and Junhui Chen; Data acquisition: Peipei Li, Jun Zou and Wei Zhu; Data analysis: Junhui Chen and Yan Zang; Statistical analysis: Junhui Chen and Yan Zang.
Supplemental Material
Download Zip (8 MB)Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
All data included in this study are available upon request by contacting the corresponding author.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019 Jan;69(1):7–34.
- Fillon M. Both leading molecular thyroid tests may reduce the need for diagnostic surgery. CA Cancer J Clin. 2021 May;71(3):193–194.
- Miranda-Filho A, Lortet-Tieulent J, Bray F, et al. Thyroid cancer incidence trends by histology in 25 countries: a population-based study. Lancet Diabetes Endocrinol. 2021 Apr;9(4):225–234.
- Rossi ED, Pantanowitz L, Hornick JL. A worldwide journey of thyroid cancer incidence centred on tumour histology. Lancet Diabetes Endocrinol. 2021 Apr;9(4):193–194.
- Morton LM, Karyadi DM, Stewart C, et al. Radiation-related genomic profile of papillary thyroid carcinoma after the Chernobyl accident. Science. 2021 May 14;372(6543). 10.1126/science.abg2538.
- Krishnan A, Berthelet J, Renaud E, et al. Proteogenomics analysis unveils a TFG-RET gene fusion and druggable targets in papillary thyroid carcinomas. Nat Commun. 2020 Apr 28;11(1):2056.
- Samimi H, Haghpanah V. Gut microbiome and radioiodine-refractory papillary thyroid carcinoma pathophysiology. Trends Endocrinol Metab. 2020 Sep;31(9):627–630.
- Lee SH, Roh JL, Gong G, et al. Risk factors for recurrence after treatment of N1b papillary thyroid carcinoma. Ann Surg. 2019 May;269(5):966–971.
- Angell TE, Lechner MG, Jang JK, et al. MHC class I loss is a frequent mechanism of immune escape in papillary thyroid cancer that is reversed by interferon and selumetinib treatment in vitro. Clin Cancer Res. 2014 Dec 1; 20(23):6034–6044.
- Adam MA, Pura J, Gu L, et al. Extent of surgery for papillary thyroid cancer is not associated with survival: an analysis of 61,775 patients. Ann Surg. discussion 5-7. 2014 Oct;260(4):601–605.
- Di Maro G, Salerno P, Unger K, et al. Anterior gradient protein 2 promotes survival, migration and invasion of papillary thyroid carcinoma cells. Mol Cancer. 2014 Jun 30;13(1):160.
- Okholm TLH, Sathe S, Park SS, et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020 Dec 7;12(1):112.
- Schreiner S, Didio A, Hung LH, et al. Design and application of circular RNAs with protein-sponge function. Nucleic Acids Res. 2020 Dec 2; 48(21):12326–12335.
- Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2020 Nov 1; 12(12):911–946.
- Wang Y, Zhang Y, Wang P, et al. Circular RNAs in renal cell carcinoma: implications for tumorigenesis, diagnosis, and therapy. Mol Cancer. 2020 Oct 14; 19(1):149.
- Gokool A, Loy CT, Halliday GM, et al. Circular RNAs: the brain transcriptome comes full circle. Trends Neurosci. 2020 Oct;43(10):752–766.
- Xu T, Wang M, Jiang L, et al. CircRNAs in anticancer drug resistance: recent advances and future potential. Mol Cancer. 2020 Aug 17;19(1):127.
- Ma C, Wang X, Yang F, et al. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020 Nov 10;19(1):157.
- Peng L, Sang H, Wei S, et al. Circcul2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020 Nov 5;19(1):156.
- Saaoud F, Drummer IVC, Shao Y, et al. Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers. Pharmacol Ther. 2020 Oct 24;220:107715.
- Muhammad N, Bhattacharya S, Steele R, et al. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016 Sep 6; 7(36):58595–58605.
- Wu YH, Huang YF, Chang TH, et al. miR-335 restrains the aggressive phenotypes of ovarian cancer cells by inhibiting COL11A1. Cancers (Basel). 2021 Dec 13;13(24):6257.
- Boukrout N, Souidi M, Lahdaoui F, et al. Antagonistic Roles of the Tumor Suppressor miR-210-3p and Oncomucin MUC4 Forming a Negative Feedback Loop in Pancreatic Adenocarcinoma. Cancers (Basel). 2021 Dec 9;13(24):6197.
- Zhang M, Jiang D, Xie X, et al. miR-129-3p inhibits NHEJ pathway by targeting SAE1 and represses gastric cancer progression. Int J Clin Exp Pathol. 2019;12(5):1539–1547.
- Fu R, Yang P, Sajid A, et al. Avenanthramide A induces cellular senescence via miR-129-3p/Pirh2/p53 signaling pathway to suppress colon cancer growth. J Agric Food Chem. 2019 May 1; 67(17):4808–4816.
- Cui S, Zhang K, Li C, et al. Methylation-associated silencing of microRNA-129-3p promotes epithelial-mesenchymal transition, invasion and metastasis of hepatocelluar cancer by targeting Aurora-A. Oncotarget. 2016 Nov 22;7(47):78009–78028.
- Bijnsdorp IV, Hodzic J, Lagerweij T, et al. miR-129-3p controls centrosome number in metastatic prostate cancer cells by repressing CP110. Oncotarget. 2016 Mar 29;7(13):16676–16687.
- Zhang Y, Wang Y, Wei Y, et al. MiR-129-3p promotes docetaxel resistance of breast cancer cells via CP110 inhibition. Sci Rep. 2015 Oct 21;5(1):15424.
- Ma S, Kong S, Wang F, et al. CircRNAs: biogenesis, functions, and role in drug-resistant tumours. Mol Cancer. 2020 Aug 5; 19(1):119.
- Huang JL, Su M, Wu DP. Functional roles of circular RNAs in Alzheimer’s disease. Ageing Res Rev. 2020 Jul;60:101058.
- Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020 Apr;6(4):319–336.
- Wu P, Mo Y, Peng M, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020 Feb 4;19(1):22.
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020 Feb 15; 19(1):30.
- Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020 Mar;30(3):226–240.
- Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017 May 1;77(9):2339–2350.
- Yang J, Yang L, Li S, et al. HGF/c-Met promote renal carcinoma cancer stem cells enrichment through upregulation of Cir-CCDC66. Technol Cancer Res Treat. 2020 Jan-Dec;19:1533033819901114.
- Qi L, Wang W, Zhao G, et al. Circular RNA circCCDC66 promotes glioma proliferation by acting as a ceRNA for miR-320a to regulate FOXM1 expression. Aging (Albany NY). 2021 Jul 12;13(13):17673–17689.
- Zhang Y, Li X, Zhang J, et al. Circ-CCDC66 upregulates REXO1 expression to aggravate cervical cancer progression via restraining miR-452-5p. Cancer Cell Int. 2021 Jan 6; 21(1):20.
- Federico C, Sun J, Muz B, et al. Localized delivery of cisplatin to cervical cancer improves its therapeutic efficacy and minimizes its side effect profile. Int J Radiat Oncol Biol Phys. 2021 Apr 1;109(5):1483–1494.
- Suhail M, Tarique M, Muhammad N, et al. A critical transcription factor NF-kappaB as a cancer therapeutic target and its inhibitors as cancer treatment options. Curr Med Chem. 2021;28(21):4117–4132.
- Floberg JM, Zhang J, Muhammad N, et al. Standardized uptake value for (18)F-Fluorodeoxyglucose is a marker of inflammatory state and immune infiltrate in cervical cancer. Clin Cancer Res. 2021 Aug 1;27(15):4245–4255.
- Rashmi R, Jayachandran K, Zhang J, et al. Glutaminase inhibitors induce thiol-mediated oxidative stress and radiosensitization in treatment-resistant cervical cancers. Mol Cancer Ther. 2020 Dec;19(12):2465–2475.
- Singh SV, Chaube B, Mayengbam SS, et al. Metformin induced lactic acidosis impaired response of cancer cells towards paclitaxel and doxorubicin: role of monocarboxylate transporter. Biochim Biophys Acta Mol Basis Dis. 2021 Mar 1;1867(3):166011.
- Cen Y, Zhu T, Zhang Y, et al. hsa_circ_0005358 suppresses cervical cancer metastasis by interacting with PTBP1 protein to destabilize CDCP1 mRNA. Mol Ther Nucleic Acids. 2022 Mar 8;27:227–240.
- Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019 Jul 19;10(1):3200.
- Chioccarelli T, Falco G, Cappetta D, et al. FUS driven circCNOT6L biogenesis in mouse and human spermatozoa supports zygote development. Cell Mol Life Sci. 2021 Dec 22;79(1):50.
- Niu R, Li D, Chen J, et al. Circ_0014235 confers Gefitinib resistance and malignant behaviors in non-small cell lung cancer resistant to Gefitinib by governing the miR-146b-5p/YAP/PD-L1 pathway. Cell Cycle. 2021 Dec;17:1–15.
- Callaghan MM, Koch B, Hackett KT, et al. Expression, localization, and protein interactions of the partitioning proteins in the gonococcal type IV secretion system. Front Microbiol. 2021;12:784483.
- Chen Z, Zheng Z, Xie Y, et al. Circular RNA circPPP6R3 upregulates CD44 to promote the progression of clear cell renal cell carcinoma via sponging miR-1238-3p. Cell Death Dis. 2021 Dec 21;13(1):22.
- Min X, Cai MY, Shao T, et al. A circular intronic RNA ciPVT1 delays endothelial cell senescence by regulating the miR-24-3p/CDK4/pRb axis. Aging Cell. 2021 Dec 13;21(1):e13529.
- Yuan Y, Yan G, He M, et al. ALKBH5 suppresses tumor progression via an m(6)A-dependent epigenetic silencing of pre-miR-181b-1/YAP signaling axis in osteosarcoma. Cell Death Dis. 2021 Jan 11;12(1):60.
- Ji Y, Yang S, Yan X, et al. CircCRIM1 promotes hepatocellular carcinoma proliferation and angiogenesis by sponging miR-378a-3p and regulating SKP2 expression. Front Cell Dev Biol. 2021;9:796686.
- Shang BQ, Li ML, Quan HY, et al. Functional roles of circular RNAs during epithelial-to-mesenchymal transition. Mol Cancer. 2019 Sep 16;18(1):138.
- Shan C, Zhang Y, Hao X, et al. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. 2019 Sep 13; 18(1):136.
- Li Z, Huang X, Liu A, et al. Circ_PSD3 promotes the progression of papillary thyroid carcinoma via the miR-637/HEMGN axis. Life Sci. 2020 Oct 22;264:118622.
- Wang YF, Li MY, Tang YF, et al. Circular RNA circEIF3I promotes papillary thyroid carcinoma progression through competitively binding to miR-149 and upregulating KIF2A expression. Am J Cancer Res. 2020;10(4):1130–1139.
- Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 2019 Jul 19;38(1):318.
- Wu D, Zhu J, Fu Y, et al. LncRNA HOTAIR promotes breast cancer progression through regulating the miR-129-5p/FZD7 axis. Cancer Biomark. 2020 Oct 13.
- Wu L, Zhu X, Song Z, et al. FGD5-AS1 facilitates glioblastoma progression by activation of Wnt/beta-catenin signaling via regulating miR-129-5p/HNRNPK axis. Life Sci. 2020 Sep 1; 256:117998
- Yu J, Zhang X, Ma Y, et al. MiR-129-5p restrains apatinib resistance in human gastric cancer cells via downregulating HOXC10. Cancer Biother Radiopharm. 2020 Jun 16;36(1):95–105.
- He J, Ge Q, Lin Z, et al. MiR-129-5p induces cell cycle arrest through modulating HOXC10/Cyclin D1 to inhibit gastric cancer progression. FASEB J. 2020 Jun;34(6):8544–8557.
- Xu C, Du Z, Ren S, et al. MiR-129-5p sensitization of lung cancer cells to etoposide-induced apoptosis by reducing YWHAB. J Cancer. 2020;11(4):858–866.
- Liao C, Long Z, Zhang X, et al. LncARSR sponges miR-129-5p to promote proliferation and metastasis of bladder cancer cells through increasing SOX4 expression. Int J Biol Sci. 2020;16(1):1–11.
- Wu C, Miao C, Tang Q, et al. MiR-129-5p promotes docetaxel resistance in prostate cancer by down-regulating CAMK2N1 expression. J Cell Mol Med. 2020 Feb;24(3):2098–2108.
- Zhang H, Cai Y, Zheng L, et al. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J Cell Physiol. 2018 Oct;233(10):6638–6648.
- Fonseca BD, Lahr RM, Damgaard CK, et al. LARP1 on TOP of ribosome production. Wiley Interdiscip Rev RNA. 2018 May 2.
- Deragon JM, Bousquet-Antonelli C. The role of LARP1 in translation and beyond. Wiley Interdiscip Rev RNA. 2015 Jul-Aug;6(4):399–417.
- Hong S, Freeberg MA, Han T, et al. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife. 2017 Jun 26;6: 10.7554/eLife.25237
- Fonseca BD, Zakaria C, Jia JJ, et al. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem. 2015 Jun 26;290(26):15996–16020.
- Zhang Y, Cai W, Zou Y, et al. Knockdown of KCNQ1OT1 inhibits proliferation, invasion, and drug resistance by regulating miR-129-5p-Mediated LARP1 in osteosarcoma. Biomed Res Int. 2020;2020:7698767.
- Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018 Sep 18;503(4):2429–2435.
- Xu Z, Xu J, Lu H, et al. LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma. Oncol Rep. 2017 Dec;38(6):3659–3667.
- Ye L, Lin S-T, Mi Y-S, et al. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumour Biol. 2016 Nov;37(11):14585–14594.
- Hopkins TG, Mura M, Al-Ashtal HA, et al. The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res. 2016 Feb 18;44(3):1227–1246.