ABSTRACT
Circular RNAs (circRNAs) are associated with the progression of gastric cancer (GC). This study investigates the regulation of the circular RNA, hsa_circ_0119412 in GC and its effects on GC cells. The expression of hsa_circ_0119412, microRNA (miR)-1298-5p, and zinc finger BED-type containing 3 (ZBED3) were measured by quantitative reverse transcription-PCR (qRT-PCR) and Western blotting. The cell counting kit-8 (CCK-8) assay, flow cytometry, transwell, and animal assays were performed to identify the roles of hsa_circ_0119412, miR-1298-5p, and ZBED3 in the viability, apoptosis, invasion, and growth of GC cells. The relationship between hsa_circ_0119412, miR-1298-5p, and ZBED3 was confirmed by luciferase, RNA immunoprecipitation (RIP), and RNA pull-down assays. Our data revealed that hsa_circ_0119412 and ZBED3 expression was upregulated in GC, while miR-1298-5p expression was downregulated. Both the knockdown of hsa_circ_0119412/ZBED3 and miR-1298-5p overexpression inhibited GC cell growth and invasion, and enhanced cell apoptosis, while miR-1298-5p interference or ZBED3 overexpression showed the opposite trend. Mechanistically, hsa_circ_0119412 sponges miR-1298-5p, which regulates ZBED3 expression. Silencing hsa_circ_0119412 inhibits the progression of GC, at least in part, by targeting the miR-1298-5p/ZBED3 axis.
graphic abstract

Introduction
Gastric cancer (GC) is the fourth most common malignant cancer and the second leading cause of cancer-related deaths [Citation1]. Despite advances in surgical techniques, radiotherapy, chemotherapy, and neoadjuvant therapy, the early diagnosis rate is low, and most patients are first diagnosed with advanced GC [Citation2]. Therefore, it is imperative to improve the primary detection of GC and identify effective molecular targets for early cancer screening.
Circular RNAs (circRNAs) are covalently closed loop structures produced by post-mRNA splicing that have 5′ caps and 3′ tails. CircRNAs are functional RNAs that regulate a variety of cellular activities and pathological processes [Citation3,Citation4]. Recently, the critical role of circRNAs in coding mechanisms found in human cancers has attracted extensive attention [Citation5]. For instance, the expression of hsa_circ_0000285 is overexpressed in cervical cancer, and its knockout inhibits the survival and migration of cancer cells [Citation6]. In addition, high hsa_circ_0003221 (circPTK2) expression in colorectal cancer (CRC) is associated with accelerated tumor growth and metastasis [Citation7], and hsa_circ_100395 was negatively correlated with increased disease-free survival in liver cancer patients [Citation8]. Hsa_circ_0119412, also known as hsa_circRNA_102958, has recently been reported to play a role in tumors; it is linked to poor prognosis in colorectal and ovarian cancers, and promotes the malignant behavior of several cancer cells in vitro [Citation9,Citation10]. In addition, circ_0119412 is overexpressed in GC tissues and is positively correlated with clinicopathological differences [Citation11]. However, the effect of hsa_circ_0119412 on the malignant behavior of GC has rarely been reported. Therefore, this study aims to investigate whether circ_0119412 could be a new molecular target for GC screening.
The recently discovered circRNA-miRNA code, which regulates gene expression through the interaction of two RNA molecules, has proven to be a promising area of research for the early detection and prognosis of cancer [Citation12]. For instance, hsa_circ_100859 behaves as an oncogene in colon cancer and acts as a miR-217 sponge targeting HIF-1α [Citation13]. Circular RNA-BTG3 associated nuclear protein (circ-BANP) induces miR-503 suppression, which leads to increased expression of La-related protein 1 (LARP1) in lung cancer, and ultimately promotes the development of lung cancer [Citation14]. miR-1298-5p is negatively regulated by the hsa_circ_0003028 sponge to inhibit tumor progression in non-small cell lung cancer [Citation15]. Additionally, zinc finger BED-type containing 3 (ZBED3) plays a cancer-promoting role in lung [Citation16] and pancreatic cancer [Citation17]. Nevertheless, the hsa_circ_0119412/miR-1298-5p/ZBED3 regulatory mechanism in GC remains to be elucidated.
This study aims to elucidate the effect of hsa_circ_0119412 on the function of GC cells, and determine the regulatory mechanism of hsa_circ_0119412/miR-1298-5p/ZBED3 in GC. We hypothesize that hsa_circ_0119412 promotes the malignant behavior of GC cells by targeting miR-1298-5p and upregulating ZBED3. In addition, the purpose of this study was to provide a valuable theoretical basis for the clinical study of GC.
Methods
Tissue samples
GC tissues and paracancerous normal tissues were collected from 36 patients in our hospital, and all cancer tissue specimens were confirmed by histopathology. The ethics committee of our hospital approved this study, and all participants provided informed consent. Once harvested, the tissues were immediately stored in liquid nitrogen.
Cell culture
The human gastric epithelial cell line, GES-1 and GC cell lines: MKN74, GES-1, HGC-27, and GTL-16, were purchased from the Institute of Biochemistry and Cell Biology (Shanghai, China). The cells were cultured in RPMI-1640 containing 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (HyClone, USA). Cells were incubated in an atmosphere containing 5% CO2 at 37°C.
qRT-PCR assay
Total RNA was extracted using the Norgen Biotek total RNA purification kit (PA, USA) and reverse transcribed to cDNA using the PrimeScript RT reagent kit (TaKaRa, Japan) according to the manufacturer’s instructions. Total miRNA was extracted using the mirVanaTM miRNA isolation kit (Ambion, USA) and synthesized into cDNA using a TaqMan miRNA reverse transcription kit (ABI, USA) according to the manufacturer’s instructions. qPCR was performed using the SYBR premix ex Taq II kit (Takara) on an IQ5 thermal cycler (Bio-Rad, USA). The relative expression of each gene was determined by 2−ΔΔCT [Citation18]. Uracil 6 (U6) was used as an endogenous control for miR-1298-5p, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control for hsa_circ_0119412 and ZBED3 expression. The primer sequences used in this study are listed in .
Table 1. The Sequences of the Primers in This Study
Subcellular fractionation assay
Cytoplasmic and nucleic RNA was isolated from MKN74 and GTL-16 cells using the PARIS nuclear/cytoplasmic separation kit (Life Technologies, USA). Briefly, the nucleus and cytoplasm were separated by centrifugation after cell lysis in the cell fractionation buffer and the cytoplasmic fraction (supernatant) was extracted. The remaining precipitate was resuspended in a cell disruption buffer and then centrifuged to separate the lysate components. The levels of hsa_circ_0119412 in the cytoplasm and nucleus were detected by qRT-PCR, and U6 and GAPDH were used as internal reference of nucleus and cytoplasm, respectively [Citation19].
RNase R treatment
RNA isolated from the nucleus and cytoplasm of MKN74 and GTL-16 cells was incubated at 37°C for 30 min with 4 U/μg RNase R (Epicenter Biotechnologies, USA). The RNEasy Minelute Cleanup Kit (Qiagen, USA) was used to purify the incubated RNA, and the stability of hsa_circ_0119412 and its linear transcript, period circadian regulator 2 (PER2) were detected by qRT-PCR [Citation20].
Cell transfection
A miR-1298-5p inhibitor, miR-1298-5p mimic, and their negative control (inhibitor-NC and mimic-NC) were obtained from Switchgear Genomics (USA). Small interfering RNAs (siRNAs) for hsa_circ_0119412 (si-circ-1 or si-circ-2), ZBED3 (si-ZBED3) and control (si-NC), as well as the plasmid for ZBED3 overexpression (OE-ZBED3), and the control (OE-NC) were obtained from Ribobio (China). MKN74 and GTL-16 cells were stably passaged to the third generation, then transfected with 50 nM siRNAs, 2 μg/mL OERNA, 75 nM miRNA inhibitor, or 100 nM miRNA mimic using lipofectamine RNAiMAX (Invitrogen, USA).
Cell Counting Kit-8 (CCK-8) assay
MKN74 and GTL-16 cells were seeded into 96-well plates at a density of 1 × 103 cells per well, and cell viability was measured using the CCK-8 kit purchased from Dojindo (Japan). Briefly, the cells were incubated with 10 μL CCK-8 at 37°C for 4 h at 24, 48, 72, or 96 h, and then the absorbance was read at 450 nm using a microplate reader (Bio-Rad) [Citation21].
Invasion assay
This assay was conducted in a Matrigel-coated transwell chamber (BD Biocoat, USA). Cells (2 × 104) were resuspended in 200 μL serum-free medium and added to the upper chamber, whereas the 500 μL complete medium was added in the lower chamber. After 24 h, the noninvasive cells were removed, and the invasive cells were stained with crystal violet. Subsequently, the cells were imaged under a microscope and the number of invasions was quantified [Citation22].
Flow cytometry assay
Apoptosis was detected by an Annexin V/PI apoptosis detection kit (Yeasen Biotech, China) according to manufacturer’s instructions. Transfected MKN74 and GTL-16 cells (5 × 105) were digested and resuspended in binding buffer and incubated with 5 μL Annexin V-APC and PI in the dark at 25°C for 15 min. After labeling, the apoptosis rate was analyzed using flow cytometry (BD Biosciences, USA) [Citation23].
Xenograft tumor experiment
The short hairpin structure of hsa_circ_0119412 (sh-circ) and a control (sh-NC) (Ribobio) were transfected into MKN74 cells at a concentration of 50 nM. MKN74 cells (2 × 106) that contained either sh-circ or sh-NC were subcutaneously inoculated into five-week-old female BALB/c mice (Hunan SJA Laboratory Animal, China). A vernier caliper was used once a week to monitor the length and width of the tumor and to calculate the tumor volume. Five weeks later, the mice were euthanized with excessive carbon dioxide. The tumor was then removed, imaged, and weighed [Citation24].
Luciferase assay
The wild-type hsa_circ_0119412 or ZBED3 fragment containing the miR-1298-5p binding site were cloned into the pLG3 vector (circ-WT or ZBED3-WT). The Quik-Change site-directed mutagenesis kit (Stratagene, USA) was used to mutate the miR-1298-5p binding sites in hsa_circ_0119412 or ZBED3 to generate mutant reporter vectors (circ-MUT or ZBED3-MUT). Following this, 200 ng report vector and 50 nM miR-1298-5p mimic were transfected into MKN74 and GTL-16 cells using Lipofectamine 2000 (Invitrogen). A dual-luciferase reporter kit (Promega) was used to measure luciferase activity according to the manufacturer’s instructions after 48 h of transfection [Citation25].
RNA immunoprecipitation (RIP) assay
This assay was performed using the Magna RNA-binding protein immunoprecipitation (RIP) kit (Millipore, USA). MKN74 and GTL-16 cells were lysed and incubated overnight with anti-argonaute RISC catalytic component 2 (AGO2) antibody (Millipore) or anti-IgG-coated magnetic beads at 4°C. Subsequently, the magnetic beads were removed and eluted. The co-immunoprecipitated RNA was extracted using RNAiso plus (Takara, Japan), and the levels of miR-1298-5p and hsa_circ_0119412 were measured by qRT-PCR [Citation26].
RNA pull-down assay
An RNA pull-down assay was performed to detect the interaction between miR-1298-5p and ZBED3. Biotin-labeled miR-1298-5p (Bio-miR-1298-5p) and a negative control probe (Bio-NC; Sangon, China) were transfected into MKN74 and GTL-16 cells. Following 48 h transfection, cells were collected, lysed, and then incubated with Dynabeads M-280 Streptavidin (Invitrogen) according to the manufacturer’s protocol. In short, streptomycin affinity magnetic beads were cleaned and treated in RNAse-free solutions and then incubated with cell lysates at room temperature by gentle rotation. Subsequently, the expression of ZBED3 in the elution RNA complex was detected using qRT-PCR [Citation27].
Western blot assay
MKN74 and GTL-16 cells were lysed with radioimmunoprecipitation assay lysis buffer (Beyotime, China), and the protein concentration was determined using the BCA protein detection kit (Beyotime, China). The 20 µg protein was electrophoresed and transferred to a polyvinylidene fluoride membrane, and then blocked for 1 h with 5% skimmed milk in 0.1% tris-buffered saline with tween (TBST) buffer. Subsequently, the membrane was incubated with anti-ZBED3 (ab106383; Abcam, UK) or anti-GAPDH (ab8245; Abcam, UK) overnight at 4°C, followed by treatment with a secondary antibody (ab205718; Abcam, UK). The signal intensities of the bands were observed using an ECL kit (Millipore, USA) [Citation28].
Statistical analysis
Data expressed as the mean ± standard deviation were analyzed with SPSS 22.0 (IBM, USA) and consisted of at least three independent experiments. Statistical significance was set at p < 0.05. The differences between groups were compared using the Student’s t-test or one-way analysis of variance (ANOVA). Pearson analysis was used to analyze the correlation between the expression levels.
Results
Hsa_circ_0119412 was identified to be highly expressed in GC
To clarify the differential expression of hsa_circ_0119412 in GC, we performed qRT-PCR analysis in tissues and cell lines. Hsa_circ_0119412 expression was up-regulated approximately 2.5 times in GC tissues when compared with matched adjacent normal tissues (). Moreover, the expression of hsa_circ_0119412 was higher in GC cells compared to GES-1 cells, and was also increased in MKN74 and GTL-16 cells (). In addition, hsa_circ_0119412 was mainly distributed in the cytoplasm of GC cells (). Additionally, the exonuclease RNase R was found to degrade linear PER2 but not has_circ_0119412 (). Therefore, hashsa_circ_0119412 is a stably upregulated circRNA in GC and functions mainly in the cytoplasm.
Figure 1. Hsa_circ_0119412 was identified to be highly expressed in GC (a) Relative expression of hsa_circ_0119412 in GC tissues measured by qRT-PCR assay compared to adjacent normal tissues. **P < 0.001. (b) Relative expression of hsa_circ_0119412 in GC cells measured by qRT-PCR assay compared to GES-1 cells. **P < 0.001 compared to GES-1 cells. (c) Nuclear-cytoplasmic fractionation assay showed that hsa_circ_0119412 was mainly localized in the cytoplasm of MKN74 and GTL-16 cells. (d) RNase R treatment was used to evaluate the exonuclease resistance of hsa_circ_0119412 in MKN74 and GTL-16 cells. **P < 0.001 compared to control.
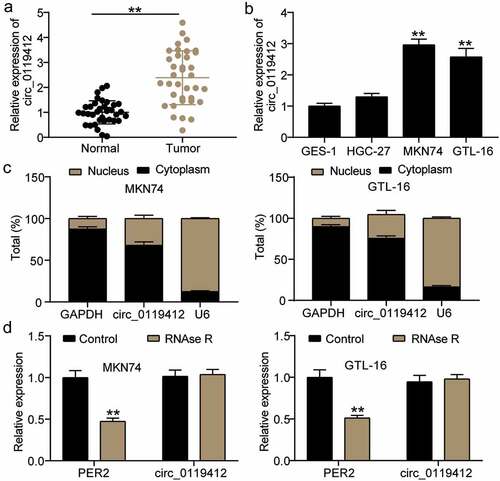
Silencing hsa_circ_0119412 inhibits the survival and invasion of GC cells and promotes apoptosis
siRNAs, si-circ-1 and si-circ-2 were transfected into MKN74 and GTL-16 cells and used to elucidate the biological function of hsa_circ_0119412 in GC cells. hashsa_circ_0119412 was reduced by approximately 70% and 60% in cells treated with si-circ-1 and si-circ-2, respectively (). The growth curve for the CCK-8 assay demonstrated that hsa_circ_0119412 knockdown inhibited the viability of GC cells (). In addition, the effect of hsa_circ_0119412 on GC cell invasion was evaluated using a transwell assay. The results showed that cell invasion levels were repressed in the si-circ-1 and si-circ-2 groups (). Flow cytometric analysis revealed a higher apoptosis rate in si-circ-1 and si-circ-2 groups than in the si-NC group (). Moreover, we investigated the effect of hsa_circ_0119412 on tumor growth by subcutaneously injecting MKN74 cells with hsa_circ_0119412 knockdown into nude mice. The growth curve of the tumor volume in the hsa_circ_0119412 knockdown mice was slower than that of the sh-NC group, and the tumor weight was also significantly lower (). In addition, we detected hsa_circ_0119412 expression in tumor tissues, and observed that the level of hsa_circ_0119412 in the sh-circ group was 50% of that in the sh-NC group (). This suggests that hsa_circ_0119412 plays a crucial role in the survival, invasion, and apoptosis of GC cells, and ultimately promotes tumorigenesis.
Figure 2. Silencing hsa_circ_0119412 inhibits the survival and invasion of GC cells and promotes apoptosis (a) Relative expression of hsa_circ_0119412 was determined in MKN74 and GTL-16 cells transfected with si-circ (si-circ-1, si-circ-2) or si-NC by qRT-PCR. (b) The cell viability was measured in MKN74 and GTL-16 cells transfected with si-circ (si-circ-1, si-circ-2) or si-NC by CCK-8 assay. (c) The cell invasion ability was determined by transwell assay after knockdown of hsa_circ_0119412 (si-circ-1, si-circ-2) in MKN74 and GTL-16 cells. (d) The cell apoptosis rate was detected in MKN74 and GTL-16 cells after transfection with hsa_circ_0119412 siRNA plasmids (si-circ-1, si-circ-2) by flow cytometry assay. **P < 0.001 compared to si-NC. (e) Representative images of xenograft tumors of sh-circ and sh-NC group, tumor growth curves and tumor weight analysis were shown. (f) Relative expression of hsa_circ_0119412 was determined in xenograft tumors of sh-circ and sh-NC group. **P < 0.001 compared to sh-NC.
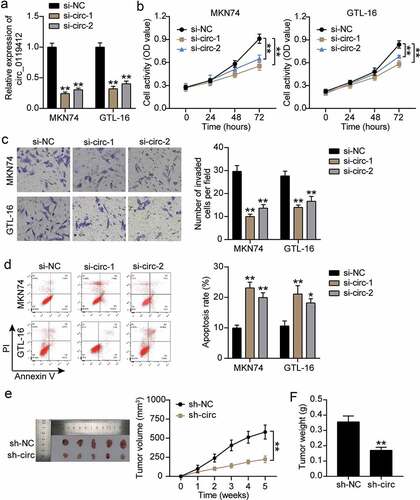
Hsa_circ_0119412 acts as an efficient sponge for miR-1298-5p in GC
To explore the molecular mechanism associated with hsa_circ_0119412-mediated function, we first predicted hsa_circ_0119412 potential target genes using the GSE93415 GEO dataset. Using the parameters, adj. p < 0.05, and logFC<-1, 19 downregulated miRNAs were screened. In addition, the circInteractome tool was used to predict the miRNAs targeted by circ_0119412. By overlapping the results of GSE93415 and circInteractome, miR-1298-5p was identified as a common miRNA (). Moreover, as illustrated in , the prediction of circInteractome indicated that miR-1298-5p had binding sites for hsa_circ_0119412. Subsequently, the luciferase reporter genes, circ-WT and circ-MUT were constructed, and the binding of hsa_circ_0119412 to miR-1298-5p were measured by luciferase analysis. The data revealed a significant reduction in circ-WT reporter luciferase activity with the miR-1298-5p mimic, and no change in circ-MUT luciferase activity (). Furthermore, we used an anti-Ago2 RIP to evaluate the Ago2-binding RNA transcripts in MKN74 and GTL-16 cells. As expected, miR-1298-5p and hsa_circ_0119412 were significantly enhanced by the anti-Ago2 antibody (). Subsequently, miR-1298-5p expression in tissues was evaluated, and qRT-PCR data showed that miR-1298-5p was reduced by approximately 50% in GC tissues compared to the adjacent normal tissues (). Detection at the cellular level also showed that miR-1298-5p levels were lower in GC cells than in normal cells (). Additionally, miR-1298-5p expression was inversely correlated with hsa_circ_0119412 in GC tissues (). Therefore, our results indicate that hsa_circ_0119412 serves as a sponge for miR-1298-5p.
Figure 3. Hsa_circ_0119412 acts as an efficient sponge for miR-1298-5p in GC (a) miR-1298 was overlapped from GSE93415 and circInteractome. (b) The miR-1298-5p binding site on hsa_circ_0119412 was predicted by circInteractome. (c) The relative luciferase activities were detected in MKN74 and GTL-16 cells after transfection with circ-WT or circ-MUT and miR-1298-5p mimic or miR-NC, respectively. **P < 0.001 compared to mimic-NC. (d) Anti-Ago2 RIP was executed in MKN74 and GTL-16 cells by qRT-PCR assay to detect hsa_circ_0119412 and miR-1298-5p. **P < 0.001 compared to Anti-IgG. (e) Relative expression of miR-1298-5p in GC tissues measured by qRT-PCR assay compared to adjacent normal tissues. **P < 0.001. (f) Relative expression of miR-1298-5p in GC cells measured by qRT-PCR assay compared to GES-1 cells. **P < 0.001 compared to GES-1 cells. (g) The correlation between hsa_circ_0119412 and miR-1298-5p expression in GC samples analyzed by Pearson analysis.
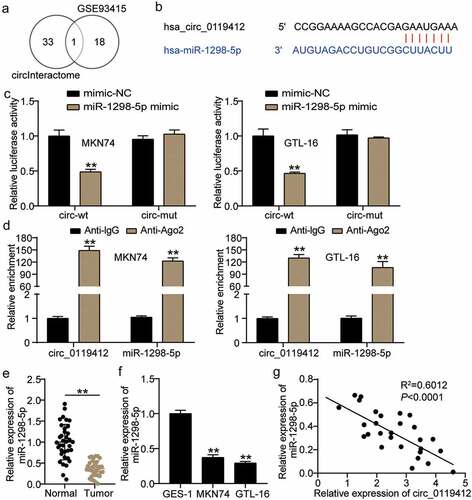
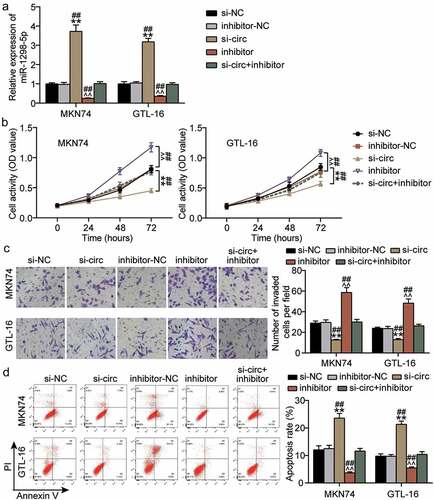
miR-1298-5p partially eliminates the functional effect of hsa_circ_0119412 on GC cells
Knockdown of hsa_circ_0119412 upregulated the level of miR-1298-5p, and as expected, knockdown of miR-1298-5p resulted in downregulation of the miRNA (). To clarify whether hsa_circ_0119412 regulates the biological function of GC cells through miR-1298-5p, we conducted a series of rescue experiments. The results of CCK-8 and transwell assays showed that reduced miR-1298-5p expression significantly augmented the viability and invasion of GC cells, and weakened the inhibitory effect of hsa_circ_0119412 knockdown on the viability and invasion of GC cells (Figure4(bc)). Flow cytometry showed that the miR-1298-5p inhibitor reduced apoptosis by approximately 60% and reversed the apoptosis-promoting effect induced by hsa_circ_0119412 silencing (). Overall, the interference of miR-1298-5p partially abolished the functional effect of hsa_circ_0119412 knockdown on GC cells.
Figure 4. miR-1298-5p partially eliminated the functional effect of hsa_circ_0119412 on GC cells (a) Relative expression of miR-1298-5p was determined in MKN74 and GTL-16 cells transfected with si-circ or miR-1298-5p inhibitor by qRT-PCR. (b) The cell viability was measured in MKN74 and GTL-16 cells transfected with si-circ or miR-1298-5p inhibitor by CCK-8 assay. (c) The cell invasion ability was determined by transwell assay after knockdown of hsa_circ_0119412 or miR-1298-5p in MKN74 and GTL-16 cells. (d) The cell apoptosis rate was detected in MKN74 and GTL-16 cells after transfection with hsa_circ_0119412 siRNA or miR-1298-5p inhibitor plasmids by flow cytometry assay. **P < 0.001 compared to si-NC; ^^P < 0.001 compared to inhibitor-NC; ##P < 0.001 compared to si-circ+inhibitor.
Overexpression of miR-1298-5p inhibits the malignant phenotype of GC cells
Next, we investigated the effect of miR-1298-5p overexpression on GC cells. qRT-PCR showed an approximately four-fold upregulation of miR-1298-5p levels in the mimic group compared to that in the control group (Supplementary Figure 1A). In addition, functional experiments showed that the miR-1298-5p mimic inhibited GC cell viability and invasion but promoted apoptosis in contrast to the control group (Supplementary Figure 1B, C, and D). Overall, the results showed that miR-1298-5p overexpression inhibited the malignant phenotype of GC cells.
ZBED3 is a direct target of miR-1298-5p
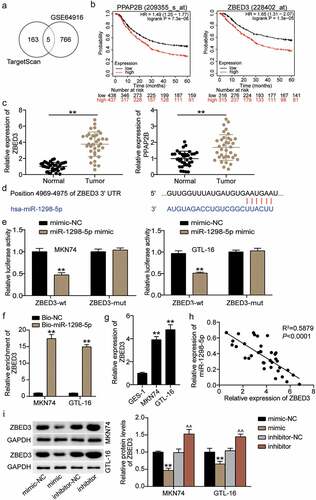
TargetScan was performed to predict miR-1298-5p target genes, while the mRNA microarray GSE64916 from GEO DataSets was used to screen for upregulated genes with p < 0.05, and logFC>1. Overlapping the results of TargetScan and GSE64916 identified five downregulated genes as the common mRNAs: solute carrier family 30 member 5 (SLC30A5), phosphatidic acid phosphatase type 2 B (PPAP2B), phosphodiesterase 4D (PDE4D), lysine methyltransferase 5 B (KMT5B/SUV420H1), and ZBED3 (). Among the five genes, only the high expression of PPAP2B and ZBED3 were linked to a poor five-year survival rate of GC according to the Kaplan–Meier plot analysis (). Subsequently, the expression of both PPAP2B and ZBED3 was upregulated in cancer samples compared to normal tissues, and ZBED3 was upregulated more significantly in GC (). Therefore, ZBED3 was selected for follow-up studies. Using TargetScan for bioinformatics analysis, the data showed that ZBED3 contained a conserved target site for miR-1298-5p (). Luciferase analysis results showed that the miR-1298-5p mimic significantly reduced the activity of the ZBED3 luciferase reporter in comparison with the control group (). Moreover, RNA pull-downs revealed that the enrichment level of ZBED3 in the Bio-miR-1298-5p group was increased by more than 15 times compared with the control (). ZBED3 expression was significantly upregulated in GC cells (). Next, an inverse relationship between ZBED3 and miR-1298-5p levels was confirmed using Pearson correlation analysis (). Furthermore, Western blotting revealed that ZBED3 protein expression was downregulated in the mimic group and upregulated in the inhibitor group (). Together, these data revealed that miR-1298-5p targets and negatively regulates ZBED3 expression.
Figure 5. ZBED3 is a direct target of miR-1298-5p (a) five downregulated genes (SLC30A5, PPAP2B, PDE4D, SUV420H1 and ZBED3) was overlapped from TargetScan and GSE64916. (b) PPAP2B and ZBED3 with high expression indicated the poor 5-year survival rate of GC. (c) Relative expression of PPAP2B and ZBED3 in GC tissues measured by qRT-PCR assay compared to adjacent normal tissues. **P < 0.001.(d) The miR-1298-5p binding site on ZBED3 was predicted by TargetScan. (e) The relative luciferase activities were detected in MKN74 and GTL-16 cells after transfection with ZBED3-WT or ZBED3-MUT and miR-1298-5p mimic or miR-NC, respectively. **P < 0.001 compared to mimic-NC. (f) The relative ZBED3 enrichment level were detected in MKN74 and GTL-16 cells after transfection with Bio-miR-1298-5p and Bio-NC by RNA pull-down assay. **P < 0.001 compared to Bio-NC. (g) Relative expression of ZBED3 in GC cells measured by qRT-PCR assay compared to GES-1 cells. **P < 0.001 compared to GES-1 cells. (h) The correlation between ZBED3 and miR-1298-5p expression in GC samples analyzed by Pearson analysis. (i) Relative protein expression of ZBED3 in MKN74 and GTL-16 cells was measured by Western blot after transfection with miR-1298-5p mimic or inhibitor. **P < 0.001 compared to mimic-NC; ^^P < 0.001 compared to inhibitor-NC.
miR-1298-5p plays a role in the proliferation, invasion, and apoptosis of GC cells by negatively regulating ZBED3 level
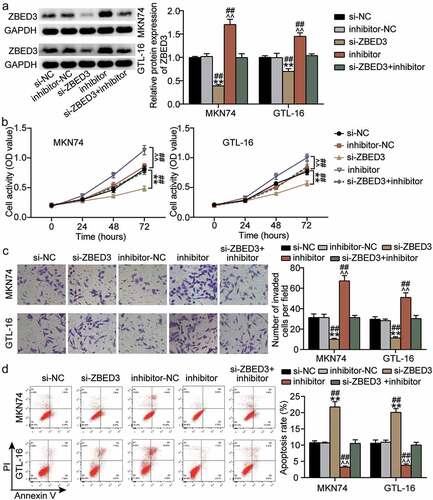
The regulatory effects of miR-1298-5p on ZBED3 were investigated. Western blotting showed that ZBED3 expression was enhanced in MKN74 and GTL-16 cells after treatment with the miR-1298-5p inhibitor, but decreased after ZBED3 knockdown (). Additionally, the effect of miR-1298-5p/ZBED3 on the growth of GC cells was examined. Both the viability and invasiveness of GC cells were reduced by ZBED3 knockdown, while the miR-1298-5p inhibitor restored this decrease (Figure 6BC). In addition, flow cytometry data revealed that ZBED3 deficiency increased the apoptosis of MKN74 and GTL-16 cells and reversed the promotion of apoptosis induced by miR-1298-5p downregulation in GC cells (). Therefore, it can be concluded that the functional changes in GC cells mediated by miR-1298-5p require ZBED3.
Figure 6. miR-1298-5p plays a role in the proliferation, invasion and apoptosis of GC cells by negatively regulating ZBED3 level (a)Relative protein expression of ZBED3 was determined in MKN74 and GTL-16 cells transfected with si-ZBED3 or miR-1298-5p inhibitor by Western blot. (b) The cell viability was measured in MKN74 and GTL-16 cells transfected with si-ZBED3 or miR-1298-5p inhibitor by CCK-8 assay. (c) The cell invasion ability was determined by transwell assay after knockdown of ZBED3 or miR-1298-5p in MKN74 and GTL-16 cells. (d) The cell apoptosis rate was detected in MKN74 and GTL-16 cells after transfection with ZBED3 siRNA or miR-1298-5p inhibitor plasmids by flow cytometry assay. **P < 0.001 compared to si-NC; ^^P < 0.001 compared to inhibitor-NC; ##P < 0.001 compared to si-ZBED3+ inhibitor.
Overexpression of ZBED3 reduced GC malignant phenotype and reversed the effect of hsa_circ_0119412 knockdown
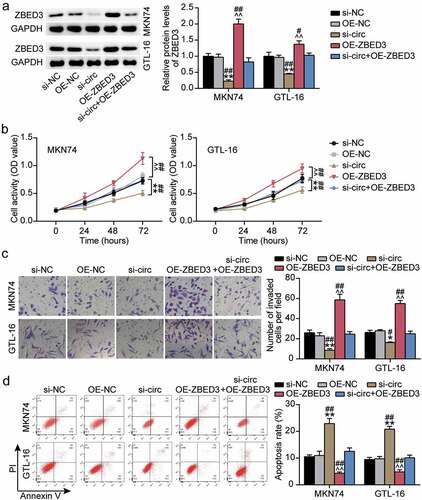
Next, we investigated the effects of hsa_circ_0119412 knockdown and ZBED3 overexpression on the biological function of GC cells. hsa_circ_0119412 knockdown resulted in the downregulation of ZBED3 protein levels, whereas ZBED3 overexpression resulted in an upregulation (). In addition, ZBED3 overexpression reversed the effect of hsa_circ_0119412 knockdown on ZBED3 protein levels (). Moreover, the cell viability and invasion ability of GC cells in the OE-ZBED3 group were upregulated compared to the OE-NC and si-circ+OE-ZBED3 groups. In addition, ZBED3 overexpression reversed the negative effect of hsa_circ_0119412 knockdown on cell viability and invasiveness of GC cells (). Apoptosis was downregulated in the ZBED3 overexpression group, and the increase in apoptosis caused by hsa_circ_0119412 knockdown was reversed by ZBED3 overexpression (). Overall, these results suggest that ZBED3 overexpression promotes GC malignant behavior and eliminates the effect of hsa_circ_0119412 knockdown on GC cells.
Figure 7. Overexpression of ZBED3 worsened GC malignant phenotype and reversed the effect of hsa_circ_0119412 knockdown (a) Relative protein expression of ZBED3 was determined in MKN74 and GTL-16 cells transfected with si-circ or OE-ZBED3 by Western blot. (b) The cell viability was measured in MKN74 and GTL-16 cells transfected with si-circ or OE-ZBED3 by CCK-8 assay. (c) The cell invasion ability was determined by transwell assay in MKN74 and GTL-16 cells transfected with si-circ or OE-ZBED3. (d) The cell apoptosis rate was detected in MKN74 and GTL-16 cells after transfection with si-circ or OE-ZBED3 by flow cytometry assay. *P < 0.05, **P < 0.001 compared to si-NC; ^^P < 0.001 compared to OE-NC; #P < 0.05, ##P < 0.001 compared to si-circ+OE-ZBED3.
Discussion
This study elucidates the influence of the hsa_circ_0119412/miR-1298-5p/ZBED3 regulatory network on the function of GC cells. We demonstrated for the first time that highly expressed hsa_circ_0119412 was observed in GC, and its knockdown reduced the survival and invasion activity of GC cells, and induced apoptosis. At the same time, we confirmed the relationship between the components of the hsa_circ_0119412/miR-1298-5p/ZBED3 network. Further, we demonstrated that hsa_circ_0119412 regulates ZBED3 by sponging miR-1298-5p, thereby affecting the function of GC cells.
Aberrant levels of circRNAs have been detected in various human diseases, including GC and are showing great potential as cancer biomarkers [Citation29]. Zhang et al. [Citation30], showed that hsa_circ_100269 is downregulated in GC, and the overexpression of hsa_circ_100269 inhibits cell proliferation. Shao et al. [Citation31], recently reported that high hsa_circ_0065149 expression is associated with low overall survival in GC. In this study, hsa_circ_0119412 was overexpressed in both GC tissues and cell lines, which is similar to the reports by Wei et al. [Citation11]. In addition, the covalent loop structure of hsa_circ_0119412 was stably located in the cytoplasm of GC cells, which is consistent with the finding that circRNAs are mainly located in the cytoplasm [Citation5]. Additionally, hsa_circ_0119412 knockdown reduced GC cell viability, invasion capacity, induced apoptosis, and inhibited cell growth in vivo. Therefore, hsa_circ_0119412 has the potential to be a molecular marker for GC screening.
CircRNAs negatively regulate miRNA activity through binding to miRNA response elements [Citation12]. Microarray analysis showed that hsa_circ_0119412 was likely to bind to miR-1298-5p. Furthermore, luciferase and RIP analyses confirmed that hsa_circ_0119412 served as a sponge for miR-1298-5p. In addition, the interference of miR-1298-5p in GC was shown to augment the survival and invasion of GC cells and inhibit apoptosis. In support of this finding, miR-1298-5p can act as a tumor suppressor in various cancers, inhibiting the viability, proliferation, and metastasis of breast cancer and glioma cells [Citation32,Citation33]. Therefore, we hypothesized that the pro-cancerous effect of hsa_circ_0119412 on GC cells might be mediated by the sponging of miR-1298-5p.
Zinc finger proteins are associated with various biological functions, including cell differentiation, development, chromatin remodeling, and regulation of cellular function [Citation34]. ZBED3 has a critical role in mammalian embryogenesis and carcinogenesis and is a member of the zinc finger family [Citation35]. It is worth noting that ZBED3 participates in carcinogenesis by regulating biological processes, including apoptosis, proliferation, and invasion of cancer cells [Citation16,Citation17]. Similar to previous studies, this study showed that ZBED3 expression was upregulated in GC, and knocking down ZBED3 inhibited the vitality and invasion of GC cells and promoted apoptosis. Additionally, mechanistic analysis showed that ZBED3 was the target of miR-1298-5p, and its expression was induced by hsa_circ_0119412.
However, there are some limitations to our study. First, the clinical relevance of hsa_circ_0119412 in GC tissue pathology and the prognosis of patients requires further investigation. In addition, the regulation of hsa_circ_0119412 on the miR-1298-5p/ZBED3 axis in vivo is still unclear.
Conclusions
Knocking down hsa_circ_0119412 decreased the viability and invasion of GC cells and promoted apoptosis by sponging miR-1298-5p and regulating ZBED3. This study reveals a novel regulatory mechanism of GC cells based on hsa_circ_0119412, which may be a promising molecular target for the screening and treatment of GC.
Authors’ contributions
Ting Huang: Conceptualization; Formal Analysis; Writing – original draft
Yacheng Wang: Investigation; Methodology; Writing – review & editing
Miao Li: Project administration; Data curation
Wenjie Wang: Software; Validation
Zhaozhen Qi: Visualization; Supervision
Jun Li: Funding acquisition; Resources
Availability of materials and data
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent to publish statement
Consent for publication was obtained from the participants.
Ethical approval
The present study was approved by the Ethics Committee of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China) (Approved No. of ethic committee:TJ-IRB202009112). The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki.
Informed consent from participants
All patients signed written informed consent.
Supplemental Material
Download TIFF Image (961.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- RL S, KD M, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
- Das M. Neoadjuvant chemotherapy: survival benefit in gastric cancer. Lancet Oncol. 2017;18(6):e307.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
- Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. 2020;22(12):2162–2169.
- Chen RX, Liu HL, Yang LL, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci. 2019;23(20):8771–8778.
- Yan Z, Yang Q, Xue M, et al. YY1-induced lncRNA ZFPM2-AS1 facilitates cell proliferation and invasion in small cell lung cancer via upregulating of TRAF4. Cancer Cell Int. 2020;20(1):108.
- Foulkes EC. On the mechanism of transfer of heavy metals across cell membranes. Toxicology. 1988;52(3):263–272.
- Li R, Wu B, Xia J, et al. Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res. 2019;11:6887–6893.
- Wang G, Zhang H, Li P, Erratum: Upregulation of hsa_circRNA_102958. Indicates Poor Prognosis and Promotes Ovarian Cancer Progression Through miR-1205/SH2D3A Axis [Corrigendum]. Cancer Manag Res. 2020;12:6863.
- Wei J, Wei W, Xu H, et al. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2020;27(2):139–145.
- Verduci L, Strano S, Yarden Y, et al. The circRNA–micro RNAcode: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680.
- Zhou P, Xie W, Huang HL, et al. CircRNA_100859 functions as an oncogene in colon cancer by sponging the miR-217-HIF-1α pathway. Aging (Albany NY). 2020;12(13):13338–13353.
- Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–2435.
- Guan H, Sun C, Gu Y, et al. RNA circ_0003028 contributes to tumorigenesis by regulating GOT2 via miR-1298-5p in non-small cell lung cancer. Bioengineered. 2021;12(1):2326–2340.
- Shi X, Zhao Y, Fan C. Zbed3 promotes proliferation and invasion of lung cancer partly through regulating the function of Axin-Gsk3β complex. J Cell Mol Med. 2019;23(2):1014–1021.
- Somerville TDD, Xu Y, Wu XS, et al. ZBED2 is an antagonist of interferon regulatory factor 1 and modifies cell identity in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America 2020; 117:11471–11482.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Chen L, Shi J, Wu Y, et al. CircRNA CDR1as promotes hepatoblastoma proliferation and stemness by acting as a miR-7-5p sponge to upregulate KLF4 expression. Aging (Albany NY). 2020;12(19):19233–19253.
- Jia Q, Ye L, Xu S, et al. Circular RNA 0007255 regulates the progression of breast cancer through miR-335-5p/SIX2 axis. Thorac Cancer. 2020;11(3):619–630.
- Liu K, Zhao D, Wang D. LINC00528 regulates myocardial infarction by targeting the miR-143-3p/COX-2 axis. Bioengineered. 2020;11(1):11–18.
- Wang X, Li T. Ropivacaine inhibits the proliferation and migration of colorectal cancer cells through ITGB1. Bioengineered. 2021;12(1):44–53.
- Tian F, Wang J, Zhang Z, et al. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res. 2020;53(1):9.
- Wang CJ, Zhu CC, Xu J, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18(1):115.
- Yin D, Lu X. Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered. 2021;12(1):225–239.
- Pan G, Mao A, Liu J, et al. RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53(2):e12720.
- Tian F, Tang P, Sun Z, et al. miR-210 in Exosomes Derived from Macrophages under High Glucose Promotes Mouse Diabetic Obesity Pathogenesis by Suppressing NDUFA4 Expression. J Diabetes Res. 2020;2020:6894684.
- Hamano R, Miyata H, Yamasaki M, et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17(9):3029–3038.
- Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661–1680.
- Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9(6):1585–1594.
- Shao Y, Tao X, Lu R, et al. Hsa_circ_0065149 is an Indicator for Early Gastric Cancer Screening and Prognosis Prediction. Pathol Oncol Res. 2020;26(3):1475–1482.
- Zhang J, Hu D. miR-1298-5p Influences the Malignancy Phenotypes of Breast Cancer Cells by Inhibiting CXCL11. Cancer Manag Res. 2021;13:133–145.
- Liu X, Ju J, Liu Q, et al. The Chinese Medicine, Shezhi Huangling Decoction, Inhibits the Growth and Metastasis of Glioma Cells via the Regulation of miR-1298-5p/TGIF1 Axis. Cancer Manag Res. 2020;12:5677–5687.
- Razin SV, Borunova VV, Maksimenko OG, et al. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochem Biokhimiia. 2012;77(3):217–226.
- Jin Y, Li R, Zhang Z, et al. ZBED1/DREF: a transcription factor that regulates cell proliferation. Oncol Lett. 2020;20(5):137.
