ABSTRACT
The excessive proliferation and the deposition of extracellular matrix (ECM) of airway smooth muscle (ASM) cells facilitates airway remodeling in asthma. This study explores how microRNA-15b-5p (miR-15b-5p) functions in modulating the proliferation, migration, inflammatory response, and ECM deposition of ASM cells. MiR-15b-5p and yes-associated protein 1 (YAP1) mRNA expression levels in tumor necrosis factor alpha (TNF-α)-induced ASM cells were, respectively, examined by real-time quantitative polymerase-chain reaction. Besides, the proliferative ability and migrative potential of ASM cells were examined by cell counting kit-8 assay, 5-bromo-2 ‘-deoxyuridine assay, and transwell assays, respectively. Interleukin-6 and interleukin-8 levels in ASM cells were detected by enzyme-linked immunosorbent assay. YAP1, collagen I, and collagen III expressions in ASM cells were detected by Western blot. With dual-luciferase reporter gene assay, the relations between miR-15b-5p and YAP1 3ʹUTR in ASM cells was examined. MiR-15b-5p expression level was reduced in ASM cells treated with TNF-α. MiR-15b-5p repressed TNF-α-initiated growth and migration of ASM cells and also suppressed IL-6 and IL-8 secretion, and inhibited collagen I and collagen III expressions in ASM cells. Furthermore, it was validated that YAP1 was a downstream target of miR-15b-5p in ASM cells. Notably, YAP1 overexpression attenuated the inhibitory effects of miR-15b-5p up-regulation on the proliferation, migration, and inflammatory response, as well as ECM deposition of TNF-α-induced ASM cells. In conclusion, miR-15b-5p/YAP1 axis modulates the growth, migration, inflammatory response, and ECM deposition of ASM cells, thus participating in the pathogenesis of asthma.
Graphical Abstract
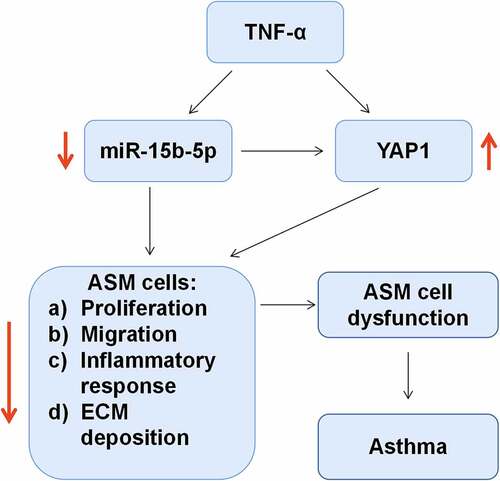
Introduction
Asthma is a common respiratory disease characterized by chronic airway inflammation, airway hyperresponsiveness (AHR), and airway remodeling, affecting 400 million people worldwide [Citation1,Citation2]. Multiple inflammatory cytokines are pertinent to airway inflammation and AHR in asthma [Citation3]. Among them, tumor necrosis factor α (TNF-α) significantly highly expressed in the lung tissue of asthmatic mice and bronchoalveolar lavage fluid of asthmatic children, has been reported to induce the proliferation, migration, and extracellular matrix (ECM) deposition of airway smooth muscle (ASM) cells [Citation4,Citation5]. ASM cells are the main structural components of the airways, and their excessive proliferation causes airway remodeling and can exacerbate the airway inflammatory response by secreting pro-inflammatory cytokines/chemokines, such as interleukin-6 (IL-6) and interleukin-8 (IL-8) [Citation6]. However, the molecular mechanisms of ASM cell hyperproliferation are not fully clarified.
MicroRNAs (miRNAs/miRs) feature prominently in regulating biological processes such as cell growth, migration, autophagy, and apoptosis [Citation7,Citation8]. Multiple miRNAs, such as miR-221, miR-133a, miR-375, miR-142-3p, miR-146a-5p, miR-26a, miR-31 are important regulators in asthma [Citation4,Citation9–13]. For example, miR-26a, miR-146a, and miR-31 are highly expressed in the lung tissue of asthmatic mice and bronchoalveolar lavage fluid of asthmatic children, and this disorder is involved in the pathogenesis of asthma via regulating inflammatory cytokines contents [Citation4]. MiR-192-5p is lowly expressed in asthmatic mouse lung tissue and alleviates airway remodeling in asthma via targeting matrix metalloproteinases 16 and autophagy-related 7 [Citation14]. Interestingly, it is reported that miR-15b-5p expression was declined in the plasma of asthmatics [Citation15]. However, how miR-15b-5p functions in ASM cells are worth exploring.
Yes-associated protein 1 (YAP1) is a pivotal protein in Hippo pathway and is involved in intracellular signal transduction and transcriptional co-activation of downstream genes [Citation16]. YAP1 is vital in regulating biological processes, including cell proliferation, apoptosis, and differentiation [Citation17]. Importantly, YAP1 is also an important modulator of asthma pathogenesis [Citation11,Citation18]. Fibulin-5 can promote ASM cell growth and migration via modulating Hippo-YAP/TAZ signaling pathway [Citation18]. Additionally, YAP1 enhances the pro-angiogenesis of ASM cells by activating STAT3 and VEGF signaling pathways [Citation11]. However, the interaction of YAP1 with miR-15b-5p in ASM cells has not been elucidated.
In this work, we hypothesized that miR-15b-5p played an important role in TNF-α-induced dysfunction of ASM cells. The main objective of this study was to investigate how miR-15b-5p regulates the proliferation, migration, inflammatory response, and ECM deposition of ASM cells. In our study, we found that miR-15b-5p was inhibited in ASM cells treated by TNF-α, and miR-15b-5p could repress the dysfunction of ASM cells by down-modulating YAP1 expression.
Methods and materials
Cell culture
Human primary ASM cells (ATCC PCS-130-011™, Manassas, VA, USA) were subsequently cultured in Dulbecco’s modified Eagles medium/Ham’s F12 (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA), 100 unit/mL penicillin, and 100 μg/mL streptomycin (Cellgro, Manassas, VA, USA). Human ASM cells were routinely cultured at 37°C in 5% CO2. To establish the cell model of asthma, ASM cells were starved in serum-free medium for 24 h and then subjected to treatment with TNF-α (10 ng/ml; Sigma, St. Louis, MO, USA) for 24 h [Citation19]. ASM cells without TNF-α treatment were set as the blank group.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using the RNAiso kit (Takara, Dalian, China). RNA was treated with DNase (Solarbio, Beijing, China) before cDNA synthesis. RNA was reversely transcribed into cDNA by the miScript reverse transcription kit (Qiagen, Hilden, Germany) and M-MLV reverse transcriptase kit (Promega, Madison, WI, USA). PCR amplification was accomplished on the ABI7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA) with the SYBR Green PCR kit (TaKaRa, Dalian, China), with U6 and β-actin as internal controls. The relative expressions of miR-15b-5p and YAP1 mRNA were calculated with 2−ΔΔCt method [Citation20]. The primer sequences are displayed in .
Table 1. Primer Sequences for RT-qPCR
Cell transfection
GenePharma (Shanghai, China) was the provider of miR-15b-5p mimics, mimics negative control (mimics-NC), pcDNA3.1-YAP1 overexpressing plasmid (YAP1), and empty vector (vector). ASM cells (5 × 105 cells per dish) were transiently transfected with miRNA (final concentration of 50 nM) or plasmid (0.5 μg/mL) by LipofectamineTM 3000 (Invitrogen, Carlsbad, CA, USA). 24 h later, the ASM cells were harvested for further analysis.
Cell proliferation assay
Cell counting kit 8 (CCK-8, Dojindo, Kumamoto, Japan) was adopted to assess the proliferation of ASM cells. ASM cells were seeded into 96-well plates at 2 × 104 cells/well and cultured at 37°C overnight. 24 h later, the cells were followingly treated with 10 ng/ml of TNF-α for 24 h. Next, 10 μL of CCK-8 reagent was loaded into each well at different time (24, 48, 72, and 96 h) and then incubated for 2 h at 37°C. Subsequently, the values of optical density (OD)450 nm were assessed by a Multiscan MS spectrophotometer (Labsystems, Stockholm, Sweden).
5-bromo-2 ′ -deoxyuridine (BrdU) assay
The viability of ASM cells was also detected with a BrdU kit (Invitrogen, Carlsbad, CA, USA). Briefly, the transfected ASM cells were inoculated into 96-well plates at 2 × 103 cells/well and treated with 10 ng/ml of TNF-α for 24 h. Next, the cells were then rinsed with phosphate buffered solution (PBS; Beyotime, Shanghai, China) and 20 μL of BrdU solution was dripped into each well. The cells were followingly incubated for 6 h at 37°C and fixed with 4% paraformaldehyde for 10 min at ambient temperature. After rinsing with PBS, the above cells were incubated with anti-BrdU antibody (Abcam, Cambridge, Massachusetts, USA) for 8 h at 4°C. After that, the nuclei were then stained with 10 μmol/L Hoechst 33342 (Beyotime, Shanghai, China) for 2 h in darkness. Then, cells were then observed under a fluorescence microscope. Cell proliferation rate = numbers of BrdU-stained positive cells/total number of cells × 100%.
Transwell assay
Transwell migration assays were accomplished to test the migration ability of ASM cells with Transwell inserts (8 µm pore size, Corning Incorporated, NY, USA). Briefly, 24 h after cell transfection, the cells were treated with 10 ng/ml of TNF-α for 24 h. Then, the cells were immersed twice in PBS and subsequently resuspended in serum-free medium (1 × 105 cells/mL). Moreover, 200 μl of cell suspension was loaded into the upper compartment of each Transwell insert, and 600 μL of medium with 20% FBS was dripped into the lower compartment, and the cells were cultured at 37°C for 24 h. After that, the cells retained in the upper were removed with a cotton swab and the migrated cells on the lower surface of the filter were fixed with 4% formaldehyde (Sigma-Aldrich, Shanghai, China) for 30 min and subsequently stained with 0.5% crystal violet (Sigma-Aldrich, Shanghai, China) for 20 min. Ultimately, the migrated cells were counted in five randomly selected fields (magnification × 250) under a light microscope (Olympus, Tokyo, Japan).
Enzyme ‐ linked immunosorbent assay (ELISA)
Transfected ASM cells (4 × 105/per well) were inoculated in 12-well plates and treated with 10 ng/ml of TNF-α for 24 h. These cells, together with the medium, were harvested and followingly centrifuged at 1000 × g for 10 min at 4°C. The supernatant was then obtained. Subsequently, IL-6 and IL-8 contents in the samples were measured with the corresponding ELISA kits (TaKaRa, Dalian, China).
Western blot assay
Total proteins were extracted using the RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitor (PMSF), with the protein concentration quantified by the Bicinchoninic Acid (BCA) Protein Assay Kit (Beyotime, Shanghai, China). Equal amounts of protein samples (20 μg per group) were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Then, the membranes were blocked with 5% skimmed milk for 1 h at 37°C and incubated with primary antibodies anti-YAP1 (1:1000, ab52771, Abcam), anti-collagen I (1:1000, ab270993, Abcam), anti-collagen III (1:1000, ab184993, Abcam) and anti-β-actin (1:1000, ab8226, Abcam) overnight at 4°C. The membranes were then washed with Tris Buffered Saline Tween (TBST), and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:5000, ab205718, Abcam) or goat anti-mouse (1:5000, ab205719, Abcam) secondary antibodies for 1 h at 37°C. Ultimately, the protein bands on the membrane were detected with the Pierce ™ ECL Western blotting substrate (Thermo Fisher Scientific, Waltham, MA, USA) and quantified by Image Lab software (Bio-Rad, Hercules, CA, USA).
Bioinformatic analysis
The binding sites between miR-15b-5p and YAP1 3′-UTR were predicted by the StarBase database (http://starbase.sysu.edu.cn/) [Citation21].
Dual-luciferase reporter gene assay
The luciferase reporter vectors carrying the fragments of YAP1 3’-UTR containing wild type (WT) or mutated (MUT) binding sites of miR-15b-5p were available from RiboBio (Guangzhou, China). ASM cells (5 × 105 cells/well) were subsequently inoculated in 24-well plates and cultured for 24 h. Next, the reporter vectors were co-transfected into ASM cells with miR-15b-5p mimics (or mimics-NC) by LipofectamineTM 3000 (Invitrogen, Carlsbad, CA, USA). 48 h later, the luciferase activity was assessed by a dual-luciferase reporter kit (Promega, Madison, WI, USA), with the renilla luciferase reporter vector as an internal control.
Statistical analysis
All data analysis was performed by SPSS 18.0 software. All experiments were independently repeated 3 times. Experimental data were displayed in the form of mean ± standard deviation. The differences between two groups were analyzed by student’s t-test; one-way ANOVA with post-hoc Tukey test, was used to compare the differences among multiple groups. Statistically, P < 0.05 is meaningful.
Results
This work focused on exploring the expression characteristics of miR-15b-5p and investigating its biological function in regulating ASM cell proliferation, migration, inflammatory response, and ECM deposition, with TNF-α-stimulated ASM cells as a model. In addition, the interaction of miR-15b-5p with YAP1 in ASM cells was also investigated. This study showed that miR-15b-5p could inhibit the proliferation, migration, inflammation, and ECM deposition of TNF-α-induced ASM cells by targeting YAP1.
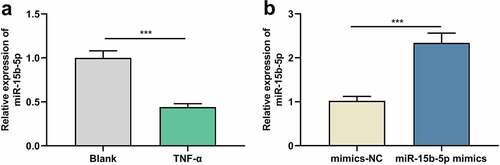
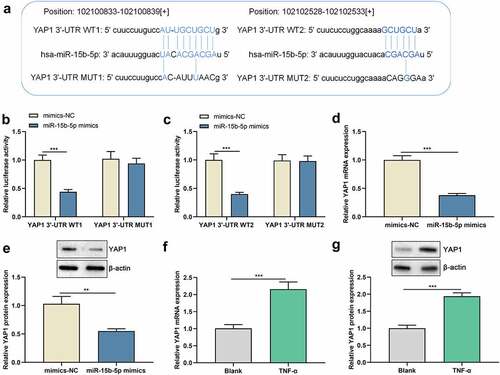
TNF-α suppresses miR-15b-5p expression in ASM cells
We first used TNF-α to treat human ASM cells to establish an in-vitro asthma model and then detected miR-15b-5p expression in ASM cells by RT-qPCR, revealing that miR-15b-5p expression was inhibited in TNF-α-stimulated ASM cells compared with the blank group ()). Thus, it might be involved in the TNF-α-induced dysfunction of ASM cells. To delve into the biological role of miR-15b-5p in ASM cells, we transfected ASM cells with miR-15b-5p mimics, along with mimics-NC as a negative control ()).
Figure 1. The effects of TNF-α on miR-15b-5p expression in ASM cells.
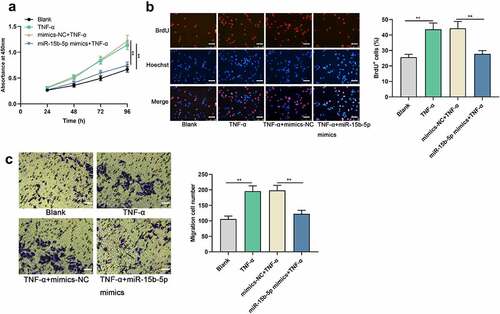
Upregulating miR-15b-5p restrains TNF-α-mediated ASM cell growth and migration
Next, the impact of miR-15b-5p on TNF-α-induced proliferation and migration of ASM cells was investigated. CCK-8, BrdU and Transwell assays suggested that TNF-α stimulation significantly accelerated the proliferation and migration of ASM cell, while miR-15b-5p up-regulation weakened these effects (). Collectively, miR-15b-5p could probably suppress the dysfunction of ASM cells in asthma.
Figure 2. The effects of miR-15b-5p on TNF-α-induced proliferation and migration of ASM cells.
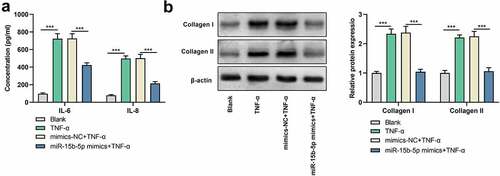
miR-15b-5p up-modulation inhibits TNF-α-induced inflammatory response and ECM deposition
To figure out the impact of miR-15b-5p on the inflammatory response induced by TNF-α, we performed ELISA and observed that IL-6 and IL-8 levels were markedly elevated in ASM cells treated with TNF-α, while those were declined followed by transfecting with miR-15b-5p mimics ()). Additionally, Western blot indicated that the expression levels of collagen I and collagen III proteins were markedly up-regulated in ASM cells in the TNF-α treatment group as against the blank group, while transfecting with miR-15b-5p mimics significantly decreased those levels as against the TNF-α + mimics-NC group ()). Collectively, up-regulation of miR-15b-5p impeded TNF-α-initiated inflammatory response and ECM deposition in ASM cells.
Figure 3. The effects of miR-15b-5p on TNF-α-induced inflammatory response and ECM deposition in ASM cells.
YAP1 is a direct target of miR-15b-5p in ASM cells
As to the hidden mechanism of miR-15b-5p in regulating the phenotypes of ASM cells, we conducted bioinformatics analysis and found that the 3’-UTR of YAP1 harbors two binding sites for miR-15b-5p ()). To clarify whether miR-15b-5p targets YAP1, dual-luciferase reporter gene assay was performed, and it highlighted that miR-15b-5p up-regulation remarkably repressed the luciferase activity of YAP1 3ʹUTR WT1 and WT2, but the luciferase activity of YAP1 3ʹUTR MUT1 and MUT2 were not significantly affected ( and c)). These data suggested that both of the two potential-binding sites were functional. Furthermore, we demonstrated that up-regulation of miR-15b-5p markedly suppressed YAP1 mRNA and protein expression levels ( and e)). In addition, YAP1 mRNA and protein expression levels were dramatically increased in TNF-α-activated ASM cells compared with those of the blank group ( and g)). Collectively, miR-15b-5p could target the YAP1 3 ‘-UTR and inhibit YAP1 expression.
Figure 4. miR-15b-5p directly targets the YAP1 mRNA 3 ′ -UTR.
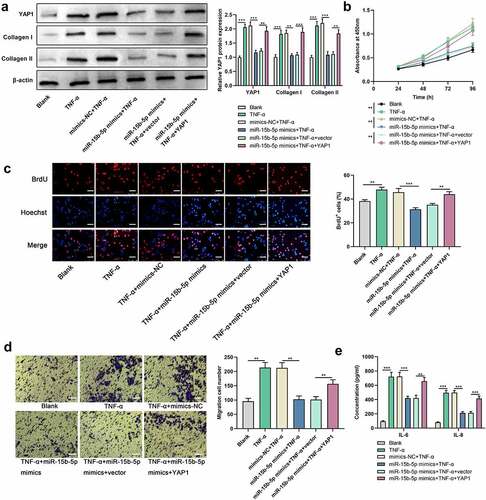
YAP1 restoration abrogates the effect of miR-15b-5p on ASM cells
To investigate whether YAP1 is implicated in miR-15b-5ʹs function in regulating ASM cell proliferation, migration, inflammatory response, and ECM deposition, we co-transfected miR-15b-5p mimics (or mimics-NC) and YAP1 overexpression plasmids (or empty plasmids) into ASM cells. Western blot indicated that the impact of miR-15b-5p overexpression on YAP1, collagen I and collagen III protein expression levels in ASM cells was counteracted by YAP1 overexpression ()). Additionally, YAP1 overexpression mitigated the impact of miR-15b-5p overexpression on TNF-α-mediated ASM cell viability and migration (). ELISA indicated that the effect of transfection of miR-15b-5p mimics on TNF-α-activated IL-6 and IL-8 production in ASM cells were dramatically counteracted by YAP1 overexpression ()). In short, miR-15b-5p impeded TNF-α-induced ASM cell growth, migration, inflammatory response, and ECM deposition by downregulating YAP1 expression.
Figure 5. The restoration of YAP1 expression reverses the effect of the upregulated miR-15b-5p on ASM cells.
Discussion
Airway remodeling in asthma is closely associated with submucosal fibrosis, abnormal proliferation, and migration of ASM cells, and excessive deposition of ECM [Citation22,Citation23]. ASM cell is one of the important sources of inflammatory factors, chemokines, and growth factors [Citation24]. Various cytokines, including TNF-α, transforming growth factor-β (TGF-β), IL-1β, and platelet-derived growth factor (PDGF-BB), have been found to be important mediators of airway remodeling and fibrotic responses, and they can also initiate and drive the inflammatory response in asthma [Citation5,Citation24–26]. As reported, the combined effect of interferon-gamma (IFN-γ), IL-1β, and TNF-α accelerates the release of chemokine CXCL10 in asthmatic ASM cells [Citation27]. Notably, TNF-α stimulation alone can also induce the dysfunction of ASM cells [Citation5,Citation19,Citation28,Citation29]. Here, we found that TNF-α strengthened the proliferative and migrative capabilities of ASM cells; TNF-α could raise the secretion IL-6 and IL-8 of ASM cells, and it increased the expression of ECM-associated proteins (collagen I and collagen III proteins). In other words, TNF-α induces the inflammatory response and ECM deposition in ASM cells, which is consistent with the previous reports [Citation5,Citation19,Citation28,Citation29].
Accumulating research has shown that miRNAs are important modulators in regulating the viability and ECM deposition of asthmatic ASM cells [Citation30,Citation31]. For example, up-regulation of miR-638 targets cyclin D1 and NOR1 and represses PDGF-BB-induced growth and migration of ASM cells [Citation30]. MiR-326 targets TNFSF14 and exacerbates the airway remodeling in asthma by inducing ASM cell proliferation and ECM deposition [Citation31]. MiR-874 inhibits the TNF-α-induced proliferation, migration, inflammation, and ECM deposition in human fetal ASM cells by targeting STAT3 [Citation5]. MiR-15b-5p, as reported, is aberrantly expressed in diverse human cancers [Citation32–34]. Specifically, miR-15b-5p is highly expressed in liver and breast cancer tissues and cell lines, and it can promote the malignant biological processes of tumor cell [Citation32,Citation33]. While in non-small cell lung cancer, miR-15b-5p exerts a tumor-suppressive effect [Citation34]. Here, we discovered that TNF-α stimulation suppressed miR-15b-5p expression in ASM cells. In-vitro experiments showed that miR-15b-5p up-regulation could reverse the impact of TNF-α stimulation on the growth and migration of ASM cells, and restrain TNF-α-induced inflammatory response and ECM deposition. Collectively, miR-15b-5p may be a protective modulator in asthma pathogenesis.
As a vital signal transducer of Hippo pathway, YAP1 is extensively expressed in diverse tissues and organs of mammals, and in normal tissues. YAP1 is regulated by other upstream proteins of Hippo pathway such as Lats1/2, Mst1/2, Sav1, and MOB1/2, leading to the cascade amplification [Citation35]. After entering the nucleus, YAP1 can bind to transcriptional factors such as transcriptional enhancer-related domain (TEAD) to function as a transcriptional enhancer, and thus the transcription of downstream genes is promoted [Citation36]. YAP1, as reported, is highly expressed in tumors such as pancreatic ductal adenocarcinoma, colorectal, and bladder cancer, and it expedites the proliferation, migration, and invasion of cancer cells [Citation37–39]. YAP1 is also widely expressed in respiratory epithelial cells, and the Hippo signal pathway is vital in regulating the growth and differentiation of the epithelial cells [Citation40]. As reported, the dysregulation of YAP1 is pertinent to the pathogenesis of asthma [Citation11,Citation18,Citation41]. YAP1 is observed to be up-regulated in bronchial airway tissue of a mouse model with chronic asthma [Citation41] and it can enhance the viability and migrative potential of ASM cells [Citation11]. In addition, YAP1, reportedly, is modulated by multiple miRNAs, such as miR-375, miR-126-5p, miR-582-3p and miR-27b-3p [Citation11,Citation38,Citation42,Citation43]. Here we found that YAP1 was a downstream target of miR-15b-5p in ASM cells; YAP1 was highly expressed in ASM cells stimulated by TNF-α. Here we proved that the impacts of miR-15b-5p up-regulation on ASM cell growth, migration, inflammatory response and ECM deposition could be rescued by YAP1 overexpression. We therefore summarized that miR-15b-5p/YAP1 axis mediated the TNF-α-initiated ASM cell growth, migration, inflammatory response, and ECM deposition.
Notably, there are some limitations of this study: first, only TNF-α-induced ASM cells were used as a model, and whether miR-15b-5p/YAP1 axis is involved in other pro-inflammatory factors-induced dysfunction of ASM cells remains to be explored in the following work; secondly, in-vivo studies are lacking in the present work; and finally, other downstream targets of miR-15b-5p need to be identified in the future.
Conclusion
On all accounts, miR-15b-5p expression is declined in the TNF-α-initiated ASM cells and miR-15b-5p is involved in repressing the proliferation, migration, inflammatory response as well as ECM deposition of ASM cells by targeting YAP1.
Ethics statement
Our study was approved by the Ethics Review Board of Yantai Yuhuangding Hospital.
Supplemental Material
Download MS Word (6.3 MB)Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Reddel HK, Bateman ED, Becker A, et al. A summary of the new GINA strategy: a roadmap to asthma control. Eur Respir J. 2015 Sep;46(3):622–639.
- Zhu X, Wei Y, Dong J. Long noncoding RNAs in the regulation of asthma: current research and clinical implications. Front Pharmacol. 2020 Sep 11;11:532849.
- Thompson EE, Dang Q, Mitchell-Handley B, et al. Cytokine-induced molecular responses in airway smooth muscle cells inform genome-wide association studies of asthma. Genome Med. 2020 Jul 20;12(1):64.
- Shi ZG, Sun Y, Wang KS, et al. Effects of miR-26a/miR-146a/miR-31 on airway inflammation of asthma mice and asthma children. Eur Rev Med Pharmacol Sci. 2019 Jun;23(12):5432–5440.
- Sun M, Huang Y, Li F, et al. MicroRNA-874 inhibits TNF-α-induced remodeling in human fetal airway smooth muscle cells by targeting STAT3. Respir Physiol Neurobiol. 2018 May;251:34–40.
- PZhu MX, Huang LH, Zhu YK, et al. LncRNA NEAT1 promotes airway smooth muscle cell inflammation by activating the JAK3/STAT5 pathway through targeting of miR-139. Exp Lung Res. 2021 Feb;16:1–13.
- Wang G, Yu X, Xia J, et al. MicroRNA-9 restrains the sharp increase and boost apoptosis of human acute myeloid leukemia cells by adjusting the Hippo/YAP signaling pathway. Bioengineered. 2021 Dec;12(1):2906–2914.
- Yu L, Sun Y, Chu Z. MiR-212-3p promotes proliferation and migration of trophoblast in fetal growth restriction by targeting placental growth factor. Bioengineered. 2021 Dec;12(1):5655–5663.
- Perry MM, Baker JE, Gibeon DS, et al. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am J Respir Cell Mol Biol. 2014 Jan;50(1):7–17.
- Chiba Y, Misawa M. MicroRNAs and their therapeutic potential for human diseases: miR-133a and bronchial smooth muscle hyperresponsiveness in asthma. J Pharmacol Sci. 2010;114(3):264–268.
- Zhao L, Shi X, Wang N, et al. YAP1, targeted by miR-375, enhanced the pro-angiogenesis of airway smooth muscle cells in asthma via STAT3 activation. Cell Cycle. 2020 Jun;19(11):1275–1284.
- Bartel S, Carraro G, Alessandrini F, et al. miR-142-3p is associated with aberrant WNT signaling during airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol. 2018 Aug 1;315(2):L328–L333.
- Dong L, Wang Y, Zheng T, et al. Hypoxic hUCMSC-derived extracellular vesicles attenuate allergic airway inflammation and airway remodeling in chronic asthma mice. Stem Cell Res Ther. 2021 Jan 6;12(1):4.
- Lou L, Tian M, Chang J, et al. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed Pharmacother. 2020 Feb;122:109692.
- Hirai K, Shirai T, Shimoshikiryo T, et al. Circulating microRNA-15b-5p as a biomarker for asthma-COPD overlap. Allergy. 2021 Mar;76(3):766–774.
- Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol. 2016 Jun;13(6):324–337.
- Dong T, Sun X, and Jin H. Role of YAP1 gene in proliferation, osteogenic differentiation, and apoptosis of human periodontal ligament stem cells induced by TNF-α. J Periodontol. 2021 Aug;92(8): 1192-1200.
- Fu J, Zheng M, Zhang X, et al. Fibulin-5 promotes airway smooth muscle cell proliferation and migration via modulating Hippo-YAP/TAZ pathway. Biochem Biophys Res Commun. 2017 Nov 18;493(2):985–991.
- Jude JA, Dileepan M, Subramanian S, et al. miR-140-3p regulation of TNF-α-induced CD38 expression in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2012 Sep;303(5):L460–8.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–408.
- Cheng X, Zhao L, Ke T, et al. Celecoxib ameliorates diabetic neuropathy by decreasing apoptosis and oxidative stress in dorsal root ganglion neurons via the miR-155/COX-2 axis. Exp Ther Med. 2021 Aug;22(2):825.
- Liu G, Cooley MA, Nair PM, et al. Airway remodelling and inflammation in asthma are dependent on the extracellular matrix protein fibulin-1c. J Pathol. 2017 Dec;243(4):510–523.
- Lin J, Feng X, Zhang J, et al. Long noncoding RNA TUG1 promotes airway smooth muscle cells proliferation and migration via sponging miR-590-5p/FGF1 in asthma. Am J Transl Res. 2019 May 15;11(5):3159–3166.
- Kong Y, Ding L, Xu Y, et al. YiQi GuBen formula inhibits PDGF-BB-induced proliferation and migration of airway smooth muscle cells. Pharmacology. 2020;105(7–8):424–433.
- Gao Y, Wang B, Luo H, et al. miR-217 represses TGF-β1-induced airway smooth muscle cell proliferation and migration through targeting ZEB1. Biomed Pharmacother. 2018 Dec;108:27–35.
- Zhang L, Zhang X, Zheng J, et al. Depressive symptom-associated IL-1β and TNF-α release correlates with impaired bronchodilator response and neutrophilic airway inflammation in asthma. Clin Exp Allergy. 2019 Jun;49(6):770–780.
- Alrashdan YA, Alkhouri H, Chen E, et al. Asthmatic airway smooth muscle CXCL10 production: mitogen-activated protein kinase JNK involvement. Am J Physiol Lung Cell Mol Physiol. 2012 May 15;302(10):L1118–27.
- Dang X, He B, Ning Q, et al. Suppression of TRIM8 by microRNA-182-5p restricts tumor necrosis factor-α-induced proliferation and migration of airway smooth muscle cells through inactivation of NF-Κb. Int Immunopharmacol. 2020 Jun;83:106475.
- Wu Y, Zou F, Lu Y, et al. SETD7 promotes TNF-α-induced proliferation and migration of airway smooth muscle cells in vitro through enhancing NF-κB/CD38 signaling. Int Immunopharmacol. 2019 Jul;72:459–466.
- Wang H, Yao H, Yi B, et al. MicroRNA-638 inhibits human airway smooth muscle cell proliferation and migration through targeting cyclin D1 and NOR1. J Cell Physiol. 2018 Jan;234(1):369–381.
- Zhang H, Yan HL, Li XY, et al. TNFSF14, a novel target of miR-326, facilitates airway remodeling in airway smooth muscle cells via inducing extracellular matrix protein deposition and proliferation. Kaohsiung J Med Sci. 2020 Jul;36(7):508–514.
- Dong Y, Zhang N, Zhao S, et al. miR-221-3p and miR-15b-5p promote cell proliferation and invasion by targeting Axin2 in liver cancer. Oncol Lett. 2019 Dec;18(6):6491–6500.
- Wu B, Liu G, Jin Y, et al. miR-15b-5p promotes growth and metastasis in breast cancer by targeting HPSE2. Front Oncol. 2020 Feb 26;10:108.
- Guo K, Qi D, Huang B. LncRNA MEG8 promotes NSCLC progression by modulating the miR-15a-5p-miR-15b-5p/PSAT1 axis. Cancer Cell Int. 2021 Feb 1;21(1):84.
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010 Oct 19;19(4):491–505.
- Yamamoto M, Ohsawa S, Kunimasa K, et al. The ligand Sas and its receptor PTP10D drive tumour-suppressive cell competition. Nature. 2017 Feb 9;542(7640):246–250.
- Xiao Y, Zhang H, Ma Q, et al. YAP1-mediated pancreatic stellate cell activation inhibits pancreatic cancer cell proliferation. Cancer Lett. 2019 Oct 10;462:51–60.
- Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019 Apr;38(14):2627–2644.
- Xu M, Gu M, Zhou J, et al. Interaction of YAP1 and mTOR promotes bladder cancer progression. Int J Oncol. 2020 Jan;56(1):232–242.
- Fodor LE, Gézsi A, Ungvári L, et al. Investigation of the possible role of the Hippo/YAP1 pathway in asthma and allergy. Allergy Asthma Immunol Res. 2017 May;9(3):247–256.
- Zhou J, Xu F, Yu JJ, et al. YAP is up-regulated in the bronchial airway smooth muscle of the chronic asthma mouse model. Int J Clin Exp Pathol. 2015 Sep 1;8(9):11132–11139.
- He J, Su X, Xie W. MiR-582-3p alleviates osteoarthritis progression by targeting YAP1. Mol Immunol. 2020 Dec;128:258–267.
- Miao W, Li N, Gu B, et al. MiR-27b-3p suppresses glioma development via targeting YAP1. Biochem Cell Biol. 2020 Aug;98(4):466–473.
