ABSTRACT
Cervical cancer is the most common malignant tumor in gynecology with high mortality rate, so novel approaches for cervical cancer treatment are urgently needed. In this study, we analyzed the gene expression data and clinicopathological data of The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression Project (GTEx) downloaded from University of California Santa Cruz (UCSC) Xena database. Chemokine (C-X-C motif) ligand 1 (CXCL1) was screened out as a key prognostic gene for cervical cancer. Revealed by the results of ELISA and Western blot, the expression of CXCL1 and chemokine (C-X-C motif) receptor 2 (CXCR2) in cervical cancer cell lines (HeLa and C33A) was significantly higher than that in the primary cervical epithelial cells. Cellular immunofluorescence was used in this study to observe CXCR2 localization. Through CCK8, clone formation assay, wound healing assay and Annexin V/PI staining, it was found that down-regulation of CXCL1 expression or treatment with CXCR2 antagonist (SB 225002) could reduce the cell viability, affect the proliferation, weaken the migration ability, and promote the apoptosis of cervical cancer cells; however, the effect of CXCR2 antagonist was improved after over-expressed CXCL1. CXCL1/CXCR2 chemokine system regulates the proliferation, migration, and apoptosis of cervical cancer cells in the form of an autocrine loop, thus affecting the development of cervical cancer. This study provides a theoretical basis for researching the molecular mechanism of cervical cancer deterioration and development, and brings forward a new idea for the prevention and treatment of cervical cancer.
GRAPHICAL ABSTRACT

Introduction
Cervical cancer is the most common malignant tumor in gynecology and is the second leading cause of cancer deaths in women [Citation1]. In recent years, the age of cervical cancer patients has a tendency to be younger. It not only seriously threatens women’s health, but also brings heavy economic burdens to the society and the families of patients [Citation2]. The occurrence and development of cervical cancer is a multifactorial and multi-stage process. Many oncogenes, tumor suppressor genes and metastasis-related genes jointly affect the process of cervical cancer [Citation3]. The most common treatments for cervical cancer are surgery and radiotherapy, and comprehensive treatment is considered the new trend for curing the disease. Chemotherapy combined with radiotherapy, chemotherapy combined with surgery, radiotherapy combined with surgery and other treatment modalities are applied increasingly in the clinical practice [Citation4–6]. However, the mortality of cervical cancer is still high. Therefore, it is of great importance to further study the molecular mechanisms of cervical cancer, which will provide a theoretical basis for the treatment of cervical cancer.
Through bioinformatics analysis, we found that the expression of chemokine (C-X-C motif) ligand 1 (CXCL1) was up-regulated in cervical cancer, and significantly correlated with prognosis. CXCL1 is a member of the C-X-C family of chemokines [Citation7]. Studies have found that the expression of CXCL1 in colon cancer is significantly higher than that in normal colon tissues [Citation8,Citation9]; the high expression of CXCL1 is a poor prognostic biomarker in metastatic colorectal cancer (CRC) and stage III CRC [Citation10]. Other studies have found that CXCL1 is related to the pathogenesis of melanoma [Citation11,Citation12], and plays a role in the development of spinal cord by inhibiting the migration of oligodendrocyte precursors, and it participates in the process of angiogenesis, arteriogenesis, inflammation, wound healing and tumorigenesis [Citation13–15]. The chemokine CXCL1 make signals by binding to chemokine (C-X-C motif) receptor 2 (CXCR2) [Citation16], and promote the malignant growth of murine squamous cell carcinoma [Citation8,Citation17]. It has been reported that the introduction of CXCR2 gene into tumor-specific T cells can enhance its tumor localization and improve the anti-tumor immune responses. This strategy helps realize the individualization of cancer treatment through the expression of tumor chemokines [Citation18]. However, there are few studies on CXCL1 in cervical cancer.
We investigated the role of CXCL1/CXCR2 in cervical cancer. CXCL1 was firstly discovered as a key gene for the prognosis of cervical cancer by bioinformatics analysis, and then the effects of CXCL1/CXCR2 in cervical cancer cells were studied with in vitro experiments. This study provides a theoretical basis for the development of cervical cancer and offers a new idea for the treatment of cervical cancer.
Materials and methods
Bioinformatics analysis
The gene expression data and clinicopathological data of The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression Project (GTEx) were downloaded from the University of California Santa Cruz (UCSC) Xena database [Citation19]. Identification (ID) conversion and data merging were performed on the downloaded gene expression data. Data correction was carried out using the Limma package of the R software. To obtain differentially expressed genes (DEGs) in normal vs cervical cancer, differential analysis was conducted on the above grouped sample data using the Limma package (|log2FC|>3, adj.P.Val<0.01) [Citation20]. To clarify the functions of the DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the ClusterProfiler package [Citation21]. To screen out the significant genes, the differential genes were further subjected to Kaplan-Meier (K-M) survival analysis (P < 0.05) and single gene Cox analysis (P < 0.05) [Citation22]. The samples with incomplete clinical information were removed, followed by multivariate independent prognostic analysis, and the genes with P < 0.05 were filtered out for receiver operating characteristic (ROC) analysis [Citation23]. The gene with the largest area under the curve (AUC) was selected for further analysis.
Differential expression analysis of normal vs cervical cancer was performed for CXCL1. Combined with the clinical data, K-M survival analysis and clinical correlation analysis were conducted. After integrating the clinicopathological features with CXCL1 expression using Perl software (v5.30.0), an independent prognostic analysis input file was obtained. The input files were subjected to univariate and multivariate Cox regression analysis using R software (survival, survminer package) and Cox regression results were visualized as forest plots. Perl software was used to prepare expression data set files and phenotype data files for single gene enrichment analysis of CXCL1. Gene Set Enrichment Analysis (GSEA) software was downloaded and installed and set in java8 running environment [Citation24]. KEGG pathway enrichment analysis was performed for CXCL1, and the results were visualized using R software (plyr, ggplot2, grid, gridExtra package).
Cell culture
Primary cervical epithelial cells were cultured and passaged in specific medium. Human cervical cancer cell lines (Hela and C33A) were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) at 37°C in an incubator with 5% CO2 [Citation25]. CXCR2 antagonist SB 225002 (Tocris Bioscience, UK) was treated at a concentration of 200 nM for 48 h.
CCK8
CCK-8 solution (Dojindo, Japan) was added to each well and the cells were incubated for another 2 h in the incubator. The wavelength of 450 nm was selected, and the optical density (OD) value of absorbance in each well was measured by an automated enzyme-linked detection instrument [Citation26].
Clone formation assay
2 × 103 cells HeLa and C33A in different treatment groups (si-NC/siCXCL1, control/SB 225002 treatment, OE-NC/SB 225002+ OE-NC/SB 225002+ CXCL1) were seeded in the culture plate, respectively. Then the cells were cultured in a 37°C cell culture incubator. At the end of study, the cells were carefully washed with phosphate buffered solution (PBS), and fixed with formaldehyde solution. Then the cells were stained with crystal violet [Citation27].
Wound healing assay
Firstly, a marker pen was used to draw horizontal lines evenly behind the 6-well plate, and the line was drawn once about every 0.5–1 cm. The cells were added and cultured overnight to reach a cell density of 90%. In the following day, a scratch was made with the gun tip as perpendicular as possible to the transverse line behind. The cells were washed three times with PBS in order to remove the cells that had been scraped down. Whereafter, we added the serum-free medium, placed the plates into an incubator at 37°C with 5% CO2, and photographed after 24 h incubation [Citation28].
Annexin V/PI staining
HeLa and C33A grown in log phase were seeded in 6-well plates for different treatments. Then, the cells were collected and resuspended with buffer. The cell suspension was added with 5 μl Annexin V-FITC staining solution (vazyme, China) and incubated in the dark at 4°C for 15 min, and then with 10 μl PI staining solution (vazyme, China) and continually incubated for other 5 min. The cell apoptosis was assessed by flow cytometry within 1 h [Citation29].
Enzyme-linked immunosorbent assay (ELISA)
CXCL1 protein levels was detected using CXCL1 ELISA kit (abcam, China). Absorbance was measured at 450 nm, and whereafter, the concentration of CXCL1 protein was calculated according to the manufacturer’s method.
Immunofluorescence
HeLa and C33A were seeded in a 6-well plate. After the cell density rose to 90%, HeLa and C33A were fixed with 4% paraformaldehyde, and then blocked in 5% bovine serum albumin. Subsequently, HeLa and C33A were incubated with CXCR2 antibodies (Invitrogen, USA) overnight at 4°C. In the next day, the cells were incubated with the secondary antibody, and then stained with DAPI. Photos were taken using a fluorescence microscope (Olympus IX51, Japan) [Citation30].
Western blot
HeLa and C33A were collected and lysed on ice. The protein concentration was determined using the BCA kit (abcam, China). The protein was subjected to 10% Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) until the bromine phenol blue reached 1 cm at the bottom of the separation gel, and transferred afterward to PVDF membrane on the ice bath. After blocking the membrane with skim milk, the protein was incubated with the primary antibody overnight. In the following day, the protein was incubated with the secondary antibody at room temperature. After ECL reagent was in full contact with PVDF film, the membranes were scanned with a chemiluminescence imager (Bio-rad, CA, USA). Primary antibodies: CXCR2 (1:2000, Invitrogen, #PA5-100,951); bax (1:2000, Invitrogen, #PA5-11,378); cleaved-caspase 3 (1:1000, Cell Signaling Technology, #9664); pro-caspase 3 (1:1000, Cell Signaling Technology, #9662); bcl2 (1:3000, Invitrogen, #PA5-27,094) [Citation29].
Statistical analysis
Each experiment was repeated 3 times, and the data were presented as ±s. t-test analysis was performed on the data using GraphPad Prism 8.0, and P < 0.05 was considered statistically different.
Results
In this study, CXCL1 was identified as a key prognosis gene for cervical cancer by bioinformatics analysis, and then the effects of CXCL1/CXCR2 in cervical cancer cells (HeLa and C33A) were studied. It was found that CXCL1/CXCR2 chemokine system could regulate the proliferation, migration, and apoptosis of cervical cancer cells in the form of an autocrine loop through ELISA, CCK8, clone formation assay, wound healing assay, Annexin V/PI stainin and Western blot.
Differential genes analysis in cervical cancer
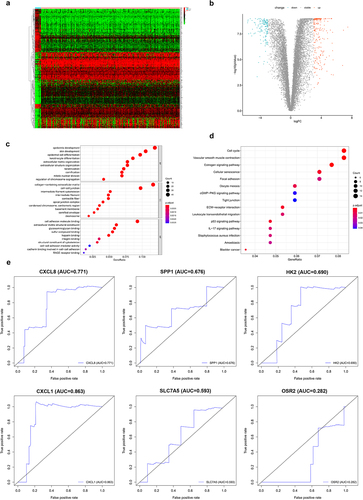
TCGA and GTEX gene expression data and clinicopathological data were downloaded from the UCSC Xena database. After data correction, the sample data were subjected to difference analysis (|log2FC| >3, adj.P.Val <0.01). The results of differential analysis revealed 353 DEGs obtained in normal vs cervical cancer, among which 212 were up-regulated and 141 were down-regulated (,b)). To clarify the functions of the DEGs, GO ()) and KEGG ()) enrichment analyses were performed on the differential genes. The differential genes were subjected to K-M survival analysis and univariate Cox analysis, and significant genes (P < 0.05) were selected (a total of 34). These genes were further combined with clinical data for multivariate Cox analysis, and then significant genes (P < 0.05) were screened out. Finally, it turned out that a total of 6 genes could be used as independent prognostic markers (). ROC analysis was performed on the 6 genes. Seen from the plot, CXCL1 had an AUC area of 0.863, serving as a separate prognostic factor ()).
Table 1. Independent prognostic genes
Clinical sample data analysis of CXCL1 in cervical cancer
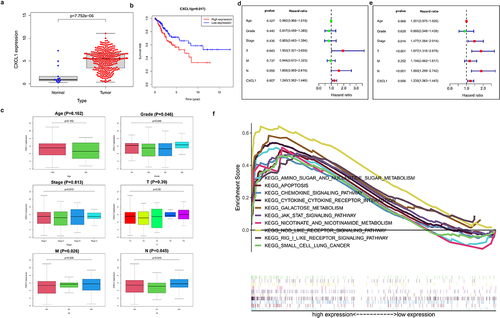
Differential expression analysis was performed on CXCL1, an up-regulated gene in cervical cancer samples ()). The results of K-M survival analysis showed that high expression of CXCL1 was associated with poor patient survival in cervical cancer ()). Further combined with the clinical data (), the clinical correlation analysis (Age, Grade, Stage, T, M, and N) of CXCL1 was performed. CXCL1 expression was proved to be correlated with Grade, M stage, and N stage ()). Immediately afterward, CXCL1 and clinically relevant Age, Grade, Stage, T, M, and N were subjected to univariate and multivariate Cox proportional regression hazard analysis and visualized (). It can be seen that CXCL1 was significantly correlated with the survival status and survival time of patients, thus can be used as a key prognostic factor. Finally, KEGG enrichment analysis of GSEA was performed on CXCL1, from which 10 pathways were selected for multi-GSEA visualization. CXCL1 expression was found to be correlated with pathways such as KEGG apoptosis, and KEGG cytokine-cytokine-receptor-interaction ()).
Table 2. The clinicopathological data of cervical cancer patients
Figure 2. Clinical sample data analysis of CXCL1. A. Differential expression analysis of CXCL1 in normal and tumor; B. K-M survival analysis; C. clinical relevance analysis (age, grade, stage, T, M and N); D. univariate Cox proportional regression hazard analysis; E. multivariate Cox proportional regression hazard analysis; F. KEGG enrichment analysis.
CXCL1/CXCR2 expression in cervical cancer cells
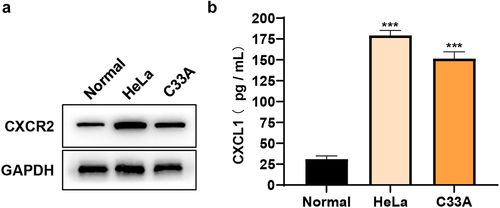
To explore the roles of CXCL1 and its receptor CXCR2 in cervical cancer, HeLa and C33A cells were taken as in vitro cell models in this study. The protein expressions of CXCL1 and CXCR2 were firstly examined in primary cervical epithelial cells and cervical cancer cell lines (HeLa and C33A). The results of Western blot ()) and ELISA ()) showed that both the expressions of CXCL1 and CXCR2 were significantly up-regulated in cervical cancer cell lines and they were expressed at higher levels in HeLa. The above results indicated that CXCL1 and CXCR2 expressions were elevated in cervical cancer cells, which was consistent with the results obtained by bioinformatics analysis.
Effects of down-regulating CXCL1 expression on the proliferation, migration and apoptosis of cervical cancer cells
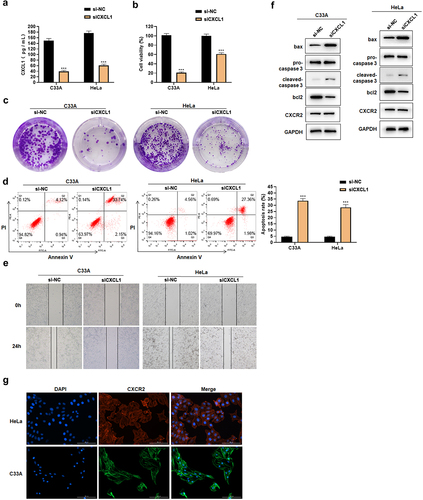
CXCL1 was found highly expressed in cervical cancer, and its expression was correlated with M and N stage. Moreover, it was reported that CXCL1, as a chemokine in cancers such as breast and colorectal cancers, played a crucial role in the process of cell proliferation, migration and apoptosis [Citation31–33]. In this study, we examined whether CXCL1 has an effect on cell proliferation, migration and apoptosis by down-regulating its expression in cervical cancer cell lines. ELISA results showed that CXCL1 secretion decreased after siCXCL1 transfection ()). The assay showed that down-regulating CXCL1 expression could decrease the cell viability and the colony forming ability in HeLa and C33A (). Meanwhile, the down-regulation of CXCL1 induced the cell apoptosis ()), and inhibited the cell migration ability ()). Western blot results indicated that the expression of pro-apoptotic proteins bax and cleaved-caspase 3 increased, while the expression of anti-apoptotic protein bcl2 decreased ()). Cell immunofluorescence results demonstrated that CXCR2 was localized on the cell membrane and cytoplasm ()) to provide the possibility of binding to CXCL1. In addition, through the detection of CXCR2 expression by Western blot ()), a slight down-regulation of CXCR2 expression was found, but no significant changes were observed. The above results revealed that the down-regulation of CXCL1 expression reduced the cell viability, affected the proliferation, attenuated the migratory ability, and promoted the apoptosis of cervical cancer cells.
Figure 4. Effects of siCXCL1 on HeLa and C33A. A. ELISA was used to detect CXCL1 expression; B. CCK8 was used to detect cell viability; C. clone formation assay; D. Annexin V/PI staining; E. wound healing assay; F. Western blot was used to detect CXCR2 and apoptosis-related protein expression; G. cell immunofluorescence of CXCR2. ***P < 0.001.
Effects of CXCR2 antagonist on cervical cancer cell proliferation, migration and apoptosis
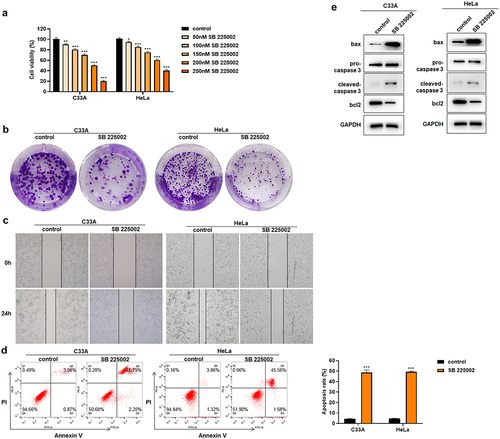
It has been reported that CXCL1/CXCR2 can influence the function of cancer cells in an autocrine loop, with cells secreting CXCL1 factors while CXCL1 acts on cells through its receptor CXCR2 to affect the cell function [Citation8,Citation34,Citation35]. But there has been no related research reported on cervical cancer cells. In this study, the CXCR2 antagonist SB 225002 was used in HeLa and C33A cells to investigate the effects of CXCR2 antagonists on the proliferation, migration and apoptosis of cervical cancer cells [Citation36,Citation37]. First, HeLa and C33A were treated with different concentrations (0, 50, 100, 150, 200, 250 nM) of SB 225002. The results of CCK8 showed that the cell viability continuously decreased with the increased concentration of SB 225002 ()). Finally, 200 nM was selected for the next study. After SB 225002 treatment, the number of colonies decreased in HeLa and C33A cells and the cell migration ability was weakened (). At the same time, the results of Annexin V/PI staining showed that SB 225002 induced the cell apoptosis ()). Western blot indicated that the expression of bax and cleaved-caspase 3 increased and bcl2 decreased ()). The above results indicate that CXCR2 antagonist treatment reduced the cell viability and proliferation, attenuated the migratory ability, but promoted the cell apoptosis of cervical cancer cells.
CXCL1 over-expression ameliorates the effects of CXCR2 antagonist
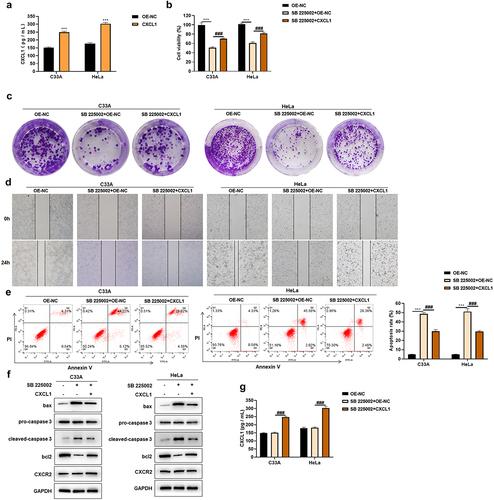
CXCR2 antagonists can prevent CXCL1 and receptor CXCR2 binding, and regulate cellular functions [Citation37]. In this study, after using CXCR2 antagonist SB 225002 (200 nM), CXCL1 was over-expressed and the effects brought by CXCR2 antagonist were found to be improved. First, the effect of CXCL1 over-expression plasmid was examined by ELISA, and it showed that transfection with CXCL1 over-expression plasmid enhanced the secretion of CXCL1 ()). Then, a certain degree of increase in cell viability was revealed by the results of CCK8 assay after the over-expression of CXCL1 with CXCR2 antagonist SB 225002 ()). The reversed effect of over-expression was also indicated in clone formation and wound healing assay (). After CXCL1 over-expressing, a certain decrease in apoptosis rate was demonstrated according to Annexin V/PI staining results ()). The expression of apoptosis-related proteins was consistent with the change of apoptosis rate ()). CXCL1 secretion was examined by ELISA. There was no significant change after SB 225002 treatment, and the secretion of CXCL1 was significantly up-regulated after transfection with the over-expression plasmid ()), whereas CXCR2 expression was not significantly changed ()). The above results indicated that CXCL1 over-expression could ameliorate the effects caused by CXCR2 antagonist.
Figure 6. CXCL1 ameliorates the effects of SB 225002. A. ELISA was used to examine the effect of CXCL1 over-expression plasmid; B. CCK8 was used to detect cell viability; C. clone formation assay; D. wound healing assay; E. Annexin V/PI staining; F. Western blot was used to detect CXCR2 and apoptosis-related protein expression; G. ELISA. OE-NC as control, ***P < 0.001; SB 225002+ OE-NC as control,###P < 0.001.
Discussion
Cervical cancer is a kind of serious and life-threatening malignancy in women, and its incidence and case fatality rate go on rising [Citation38]. Decreased immunity of patients and persistent HPV infection are the main causes of cervical cancer [Citation39]. Although great progress has been made in the clinical treatment of cervical cancer in recent years, the mortality rate of this cancer is still high. How to delay the growth and metastasis of cervical cancer and further improve its therapeutic effect are still difficulties in cervical cancer treatment. In this study, chemokine CXCL1 was identified as a key prognostic gene in cervical cancer through bioinformatics analysis, and CXCL1 expression was found significantly and negatively correlated with patient survival. Meanwhile, CXCL1 was found to be significantly correlated with the clinically relevant Grade, M and N stage. Studies have shown that CXCL1 concentrations were significantly higher in the serum of cervical cancer patients [Citation40,Citation41]. Therefore, we speculated that CXCL1 could affect the biological function of cervical cancer cells.
As a family of small bioactive peptides with various biological functions, chemokines have a relative molecular weight of 8–10 KD. Chemokines are induced by growth factors of tissue cells, cytokines of inflammatory cells and pathogenic stimuli, and are capable of causing directed migration of cells, playing an important part in tumor initiation and progression. Composed of chemokines and chemokine receptors, the chemokine system affects multiple pathways of spontaneous or non-spontaneous cellular progression of tumors, constituting a superfamily of signaling factors capable of predicting unknown domains of tumor evolution. CXCL1 is known to signal through the chemokine receptor CXCR2 to elicit its effects [Citation16]. In this study, cervical cancer cell lines (HeLa and C33A) were selected as cell models. The expression of CXCL1 and CXCR2 were found to be significantly up-regulated in cervical cancer cell lines compared with the primary cervical epithelial cells, and CXCL1/CXCR2 affected cancer cell function in an autocrine loop form. Cervical cancer cells secrete CXCL1 factors, while CXCL1 can act on cervical cancer cells through the cell surface receptor CXCR2 to affect cell function [Citation8,Citation34,Citation35].
As a kind of chemokine in cancers, CXCL1 played a crucial role in the process of cell proliferation, migration, apoptosis, etc [Citation31–33]. The down-regulation of CXCL1 in cervical cancer cells revealed that the cell activity was decreased, the proliferation was affected, and the migration ability was attenuated while the apoptosis was promoted. The CXCR2 antagonist SB 225002 prevents CXCL1 and receptor CXCR2 binding and regulates cell function [Citation36,Citation37]. The use of SB 225002 reduced cervical cancer cell activity, affected proliferation, attenuated migratory ability, but promoted apoptosis. Immediately afterward, CXCL1 was over-expressed after using CXCR2 antagonist SB 225002, and the effects brought by CXCR2 antagonist were found to be ameliorated. We thereby concluded that increasing the concentration of chemokines can lead to an enhanced effect of CXCL1/CXCR2 on cells in the presence of a certain receptor.
This study identified that CXCL1/CXCR2 chemokine system regulated the proliferation, migration and apoptosis of cells in an autocrine loop by in vitro experiments, but to make the research more credible, in vivo experiments were also needed.
Conclusion
This study firstly identified CXCL1 as a key factor in the prognosis of cervical cancer by bioinformatics analysis, and then it was verified by in vitro experiments that the CXCL1/CXCR2 chemokine system regulated the proliferation, migration and apoptosis of cells in an autocrine loop, thus affecting the development of cervical cancer. This study provides a theoretical basis for the study of the molecular mechanism of cervical cancer deterioration and development, and offers new ideas for cervical cancer treatment.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7–16.
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
- Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14(8):3208–3215.
- Siva S, Herschtal A, Thomas JM, et al. Impact of post-therapy positron emission tomography on prognostic stratification and surveillance after chemoradiotherapy for cervical cancer. Cancer. 2011;117(17):3981–3988.
- Cooke SL, Temple J, MacArthur S, et al. Intra-tumour genetic heterogeneity and poor chemoradiotherapy response in cervical cancer. Br J Cancer. 2011;104(2):361–368.
- Lee HJ, Kim YS, Shin SS, et al. Long-term outcomes of concomitant chemoradiotherapy incorporating high-dose-rate brachytherapy to treat locally advanced cervical cancer. Tumori. 2012;98(5):615–621.
- Haskill S, Peace A, Morris J, et al. Identification of three related human GRO genes encoding cytokine functions. Proc Natl Acad Sci U S A. 1990;87(19):7732–7736.
- Wen Y, Giardina SF, Hamming D, et al. GROalpha is highly expressed in adenocarcinoma of the colon and down-regulates fibulin-1. Clin Cancer Res. 2006;12(20 Pt 1):5951–5959.
- Miyake M, Lawton A, Goodison S, et al. Chemokine (C-X-C) ligand 1 (CXCL1) protein expression is increased in aggressive bladder cancers. BMC Cancer. 2013;13:322.
- Oladipo O, Conlon S, O’Grady A, et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. Br J Cancer. 2011;104(3):480–487.
- Richmond A, Thomas HG. Melanoma growth stimulatory activity: isolation from human melanoma tumors and characterization of tissue distribution. J Cell Biochem. 1988;36(2):185–198.
- Anisowicz A, Bardwell L, Sager R. Constitutive overexpression of a growth-regulated gene in transformed Chinese hamster and human cells. Proc Natl Acad Sci U S A. 1987;84(20):7188–7192.
- Vries MH, Wagenaar A, Verbruggen SEL, et al. CXCL1 promotes arteriogenesis through enhanced monocyte recruitment into the peri-collateral space. Angiogenesis. 2015;18(2):163–171.
- Devalaraja RM, Nanney LB, Du J, et al. Delayed wound healing in CXCR2 knockout mice. J Invest Dermatol. 2000;115(2):234–244.
- Owen JD, Strieter R, Burdick M, et al. Enhanced tumor-forming capacity for immortalized melanocytes expressing melanoma growth stimulatory activity/growth-regulated cytokine beta and gamma proteins. Int J Cancer. 1997;73(1):94–103.
- Tsai HH, Frost E, To V, et al. The chemokine receptor CXCR2 controls positioning of oligodendrocyte precursors in developing spinal cord by arresting their migration. Cell. 2002;110(3):373–383.
- Loukinova E, Dong G, Enamorado-Ayalya I, et al. Growth regulated oncogene-alpha expression by murine squamous cell carcinoma promotes tumor growth, metastasis, leukocyte infiltration and angiogenesis by a host CXC receptor-2 dependent mechanism. Oncogene. 2000;19(31):3477–3486.
- Peng W, Ye Y, Rabinovich BA, et al. Transduction of tumor-specific T cells with CXCR2 chemokine receptor improves migration to tumor and antitumor immune responses. Clin Cancer Res. 2010;16(22):5458–5468.
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678.
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
- Yu GC, Wang L-G, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287.
- Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947.
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344.
- Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14(2):482–517.
- Fan YR, Sheng WW, Meng Y, et al. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif Cells Nanomed Biotechnol. 2020;48(1):393–407.
- Jiao X, Zhang SY, Jiao J, et al. Promoter methylation of SEPT9 as a potential biomarker for early detection of cervical cancer and its overexpression predicts radioresistance. Clin Epigenetics. 2019;11(1):120.
- Wu A, Lin L, Li X, et al. Overexpression of ARHGAP30 suppresses growth of cervical cancer cells by downregulating ribosome biogenesis. Cancer Sci. 2021;112(11):4515–4525.
- Liu Y, Li L, Liu Y, et al. RECK inhibits cervical cancer cell migration and invasion by promoting p53 signaling pathway. J Cell Biochem. 2018;119(4):3058–3066.
- Wang XZ, Jia Z, Yang HH, et al. Dibenzoxanthenes induce apoptosis and autophagy in HeLa cells by modeling the PI3K/Akt pathway. J Photochem Photobiol B. 2018;187:76–88.
- Stadler C, Rexhepaj E, Singan VR, et al. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods. 2013;10(4):315–323.
- Wang N, Liu W, Zheng Y, et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-kappaB/SOX4 signaling. Cell Death Dis. 2018;9(9):880.
- Wang D, Sun H, Wei J, et al. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res. 2017;77(13):3655–3665.
- Yang C, Yu H, Chen R, et al. CXCL1 stimulates migration and invasion in ERnegative breast cancer cells via activation of the ERK/MMP2/9 signaling axis. Int J Oncol. 2019;55(3):684–696.
- Luan J, Shattuck-Brandt R, Haghnegahdar H, et al. Mechanism and biological significance of constitutive expression of MGSA/GRO chemokines in malignant melanoma tumor progression. J Leukoc Biol. 1997;62(5):588–597.
- Richmond A, Thomas HG. Purification of melanoma growth stimulatory activity. J Cell Physiol. 1986;129(3):375–384.
- White JR, Lee JM, Young PR, et al. Identification of a potent, selective non-peptide CXCR2 antagonist that inhibits interleukin-8-induced neutrophil migration. J Biol Chem. 1998;273(17):10095–10098.
- Catusse J, Liotard A, Loillier B, et al. Characterization of the molecular interactions of interleukin-8 (CXCL8), growth related oncogen alpha (CXCL1) and a non-peptide antagonist (SB 225002) with the human CXCR2. Biochem Pharmacol. 2003;65(5):813–821.
- Basu P, Mittal S, Bhadra Vale D, et al. Secondary prevention of cervical cancer. Best Pract Res Clin Obstet Gynaecol. 2018;47:73–85.
- Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19.
- Nishikawa R, Suzumori N, Nishiyama T, et al. Clinical significance of serum growth-regulated oncogene alpha (GROalpha) in patients with gynecological cancer. Eur J Gynaecol Oncol. 2012;33(2):138–141.
- Zhi W, Ferris D, Sharma A, et al. Twelve serum proteins progressively increase with disease stage in squamous cell cervical cancer patients. Int J Gynecol Cancer. 2014;24(6):1085–1092.