ABSTRACT
Cisplatin (DDP) therapy is widely used for the treatment of non-small cell lung cancer (NSCLC), but the curative effect is limited by chemoresistance. This study was designed to explore circ_0020123 function in DDP resistance of NSCLCDDP. Expression detection for circ_0020123, microRNA-140-3p (miR-140-3p) and homeobox B5 (HOXB5) was performed by real-time polymerase chain reaction (qRT-PCR). Half inhibitory concentration (IC50) of DDP and cell proliferation was detected by Cell Counting Kit-8 (CCK-8) assay. Colony formation ability was assessed using colony formation assay. Cell migration and invasion were evaluated via transwell assay. Cell apoptosis was examined by flow cytometry. Protein analysis was conducted by Western blot. Dual-luciferase reporter assay was used to affirm target interaction. Circ_0020123 expression was upregulated in DDP-resistant NSCLC cells. DDP resistance was reduced by downregulation of circ_0020123 in NSCLC cells. Circ_0020123 was identified as a miR-140-3p sponge. The effect of si-circ_0020123 on DDP resistance was partly associated with miR-140-3p upregulation. HOXB5 was a downstream target for miR-140-3p. Overexpression of HOXB5 mitigated miR-140-3p-induced inhibition of DDP resistance in NSCLC cells. Circ_0020123 upregulated the level of HOXB5 partly via sponging miR-140-3p. Also, circ_0020123 promoted tumor growth in NSCLC/DDP xenografts by regulating miR-140-3p and HOXB5 levels at least in part. These results revealed that circ_0020123 promoted DDP resistance in NSCLC cells partly by targeting miR-140-3p/HOXB5 axis, indicating that circ_0020123 might be used as a molecular target in DDP treatment for NSCLC.
Graphical Abstract
Introduction
Non-small cell lung cancer (NSCLC) is the most common pathological subtype of lung cancer, and it accounts for more than 80% of cases in lung cancer [Citation1]. NSCLC is a heterogeneous disease, and prognosis is poor in metastatic NSCLC patients [Citation2]. Drug resistance has become a major barrier for the treatment of various malignant tumors. Cisplatin (DDP) is a first-line chemotherapy for the advanced NSCLC patients. Exploring the molecular mechanism of DDP resistance and discovering more therapeutic targets are essential to improve DDP therapy for NSCLC [Citation3,Citation4].
Noncoding circular RNAs (circRNAs) and microRNAs (miRNAs) are pivotal regulators in cancer progression and treatment [Citation5]. CircRNAs can affect gene expression by sequestering miRNA expression, which is termed as ‘miRNA sponge’ mechanism [Citation6]. Huang et al. found that circRNA AK strain thymoma serine/threonine kinase 3 (circAKT3) facilitated DDP resistance in gastric cancer by suppressing miR-198 to upregulate the level of phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) [Citation7] and Sang et al. reported that circ_0025202 regulated the Forkhead Box-class O3a (FOXO3a) expression to inhibit tamoxifen resistance by acting as a miR-182-5p sponge in breast cancer [Citation8]. Circ_0020123 has been confirmed to be an oncogenic factor in NSCLC [Citation9,Citation10], while circ_0020123 function in DDP resistance is unclear. In addition, microRNA-140-3p (miR-140-3p) induced anti-tumor response and promoted DDP sensitivity in NSCLC [Citation11,Citation12]. Whether circ_0020123 can act as a miR-140-3p sponge in NSCLC remains unknown.
Homeobox B5 (HOXB5) is a member of HOX family with carcinogenic regulation in different kinds of cancers [Citation13–15]. NSCLC cell growth and metastasis were impeded after the knockdown of HOXB5 [Citation16], and HOXB5 was confirmed as a downstream target of miR-625 or miR-455-3p in NSCLC [Citation17,Citation18]. Herein, we will explore the potential of HOXB5 as a target of miR-140-3p.
In this research, circ_0020123 was assumed to regulate HOXB5 level via targeting miR-140-5p. This study aimed to investigate the biological role and functional mechanism of circ_0020123 in DDP resistance in NSCLC.
Materials and methods
DDP-resistant cell lines
Human bronchial epithelial cell line 16 HBE was bought from Sigma-Aldrich (St. Louis, MO, USA), and NSCLC cell lines (A549 and H1299) were purchased from BEINUO BIOLOGY (Shanghai, China). Cell culture was performed in a 37°C incubator with 5% CO2 using Roswell Park Memorial Institute 1640 (RPMI 1640; Sigma-Aldrich) containing 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% Antibiotic Antimycotic Solution (100 ×, Sigma-Aldrich). A549 and H1299 cell lines were treated with cisplatin (DDP; Sigma-Aldrich) to construct DDP-resistant cell lines [Citation19], then A549/DDP and H1299/DDP cell lines were cultivated under 40 µM DDP for 3 months to maintain DDP resistance.
Cell transfection
Transfection of oligonucleotides or vectors was carried out using Lipofectamine™ 3000 Reagent (Invitrogen, Carlsbad, CA, USA) as per the user’s manuals. The used oligonucleotides (RIBOBIO, Guangzhou, China) were shown as follows: small interfering RNA targeting circ_0020123 (si-circ_0020123), miR-140-3p mimic (miR-140-3p), miR-140-3p inhibitor (anti-miR-140-3p) and the matched negative controls (si-NC, miR-NC and anti-miR-NC). The overexpression vector pcDNA-HOXB5 (HOXB5) was constructed using control vector pcDNA (pcDNA-con, Invitrogen). Lentiviral vector containing short hairpin RNA targeting circ_0020123 (sh-circ_0020123) was obtained from RIBOBIO, with sh-NC as the negative control.
The quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA isolation and reverse transcription were performed by Trizol reagent (Invitrogen) and ReverTra Ace® qPCR RT Kit (Toyobo, Kita-Ku, Osaka, Japan). The expression quantification was completed via THUNDERBIRD® SYBR® qPCR Mix (Toyobo) following the manufacturer’s direction. The relative expression level of each molecule was calculated using the comparative cycle threshold (2−∆∆Ct) method [Citation20]. Forward (F) and Reverse (R) primer sequences were presented in . The circ_0020123 and HOXB5 levels were standardized by glyceraldehyde-phosphate dehydrogenase (GAPDH), while miR-140-3p expression was normalized by U6.
Table 1. Primer sequences used for qRT-PCR
Cell counting Kit-8 (CCK-8) assay
Cells in the log phase were treated with different DDP concentrations (0 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 40 μM, 80 μM) for 48 h and added with 10 μL CCK-8 solution (Dojindo, Kumamoto, Japan) for 2 h, followed by the determination of the optical density (OD) value at 450 nm under a microplate reader. The half inhibitory concentration (IC50) of DDP was defined as the DDP concentration when cell viability was reduced to 50%. In addition, the 96-well plates were planted with A549/DDP and H1299/DDP at the density of 5 × 103 cells/well overnight. Then, cells in the log phase were transfected for different times (0 h, 24 h, 48 h, 72 h). The OD value at each time point was measured, and cell proliferation curve was plotted. The detection was repeated for three times with three parallel samples.
Colony formation assay
Two hundred transfected A549/DDP and H1299/DDP cells were seeded into each well of the 6-well plates and cultured at 37°C for 14 days. The cell colonies were then fixated with 4% paraformaldehyde (Sigma-Aldrich) and stained with 0.1% crystal violet (Sigma-Aldrich). Subsequently, the stained colonies were manually counted. The experiment was repeated for three times with three parallel samples.
Transwell assay
Cell motility was assessed through a transwell assay [Citation21]. Cell migration was detected in transwell chamber (Corning Inc., Corning, NY, USA). 2 × 104 A549/DDP and H1299/DDP cells were resuspended in a serum-free RPMI 1640 medium, and then cell suspension was pipetted into the upper chamber. The lower chamber was added with 10% FBS+RPMI 1640 medium. Cell invasion was examined after the upper chamber was coated with matrigel (Corning Inc.). 2 × 104 cells and RPMI 1640 medium containing 10% FBS were seeded into the upper and lower chambers, respectively. 24 h later, migrated and invaded cells into the lower chamber were fastened and stained by 4% paraformaldehyde and crystal violet (Sigma-Aldrich). Cell images were acquired under 100 × magnification using the inverted microscope (Olympus, Tokyo, Japan) and cell number of migration or invasion was counted. The assay was repeated for three times with three parallel samples.
Flow cytometry
A549/DDP and H1299/DDP cells were transfected for 72 h, and then the harvested cells were stained with Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and Propidium Iodide (PI) in accordance with the operating procedures of Annexin-V-FITC Apoptosis Kit (BD Biosciences, San Diego, CA, USA). In brief, cells were washed in cold Phosphate Buffer solution (PBS; Sigma-Aldrich) and resuspended in 1 × Binding Buffer. 1 × 105 cells were transferred into a 2 mL culture tube, followed by the incubation of 5 µL Annexin FITC and 5 µL PI for 15 min. Instantly, the apoptotic cells in each tube were analyzed through the flow cytometer (BD Biosciences). The examination was repeated for three times with three parallel samples.
Western blot
Western blot was performed as previously described [Citation22]. Protein extraction was conducted by Radioimmunoprecipitation assay (RIPA; Millipore, Billerica, MA, USA). The concentrations of proteins were determined by BCA Protein quantification Kit (Sigma-Aldrich), and 40 µg proteins were loaded on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Whereafter, proteins in the gels were transferred to polyvinylidene fluoride membranes (Millipore) and membranes were immersed in 5% skim milk (Invitrogen). Then, the membranes were incubated with the primary antibodies targeting P-glycoprotein (P-gp; Abcam, Cambridge, UK, ab129450, 1:1000), lung resistance-related protein (LRP; Abcam, ab227734, 1:1000), HOXB5 (Abcam, ab229345, 1:1000) or GAPDH (Abcam, ab9485, 1:3000). After the incubation with the secondary antibody (Abcam, ab205718, 1:5000), protein blots were presented using ECL Ultra Western HRP Substrate (Millipore). The analysis of gray level was performed using Image Lab software (Bio-Rad, Hercules, CA, USA), and protein expression was normalized to GAPDH.
Bioinformatics analysis
Online circinteractome (https://circinteractome.nia.nih.gov/) was used to predict the potential miRNA targets (miR-95, miR-944, miR-384, miR-876-3p, miR-140-3p, miR-607, miR-600, miR-587). Cyclin-dependent kinase 6 (CDK6), mitogen-activated protein kinase 1 (MAPK1), fibroblast growth factor 9 (FGF9), homeobox B5 (HOXB5), p21 (RAC1) activated kinase 2 (PAK2), eukaryotic translation initiation factor 5A2 (EIF5A2), NOVA alternative splicing regulator 1 (NOVA1), jagged canonical Notch ligand 1 (JAG1) were predicted as potential targets of miR-140-3p by Targetscan (http://www.targetscan.org). Then, the target was further selected using qRT-PCR.
Dual-luciferase reporter assay
Target interaction was carried out by a dual-luciferase reporter assay [Citation23]. Circ_0020123 sequence contained the binding sites of miR-140-3p was defined as a wild-type (WT) sequence, while circ_0020123 sequence after mutation of miR-140-3p binding sites was considered as the mutant-type (MUT) sequence. WT sequence or MUT sequence of circ_0020123 was cloned into the luciferase vector psiCHECK-2 (Promega, Madison, WI, USA) to obtain the recombinant luciferase vectors circ_0020123 WT and circ_0020123 MUT. Then, circ_0020123 WT and circ_0020123 MUT were, respectively, transfected into A549/DDP and H1299/DDP cells with miR-NC or miR-140-3p, followed by cell incubation for 48 h and luciferase signal detection using the dual-luciferase reporter system (Promega). Also, HOXB5 luciferase vectors (HOXB5 3ʹUTR WT and HOXB5 3ʹUTR MUT) were constructed, and luciferase activity was measured after co-transfection with miR-NC or miR-140-3p.
Tumor xenograft assay
BALB/c nude mice (Vital River Laboratory Animal Technology Co. Ltd., Beijing, China) were subcutaneously injected with 1 × 106 transfected A549/DDP cells with 12 mice per group (sh-circ_0020123 or sh-NC). Subsequently, mice were intraperitoneally injected with 2 mg/kg DDP or PBS (Sigma-Aldrich) twice a week. Tumor volume (length × width2/2) of each group (sh-NC+PBS, sh-NC+DDP, sh-circ_0020123+ PBS, sh-circ_0020123+ DDP) was measured after cell injection for each week. Four weeks later, all mice were euthanized and tumors were weighed. Total RNA and protein were extracted from tissues, then the expression levels (circ_0020123, miR-140-3p and HOXB5) were detected using qRT-PCR and Western blot. Our animal experiment was ratified by the Animal Ethical Committee of Xiantao First People’s Hospital Affiliated to Yangtze University.
Statistical analysis
Each assay was independently repeated for three times with three technical parallels. Data were revealed as the mean ± standard deviation (SD) and analyzed by SPSS 24.0. Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for analysis of statistical difference. P < 0.05 indicated a significant difference.
Results
Circ_0020123 level was increased in DDP-resistant NSCLC cells
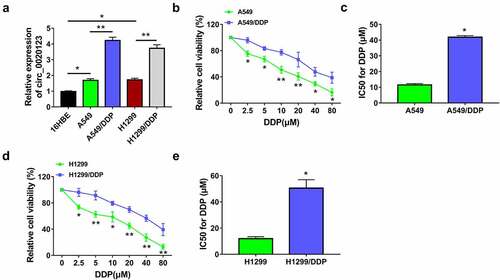
CircRNAs have essential roles in regulating drug resistance in cancers, and circRNA/miRNA/mRNA axis is an important mechanism in cancer biology. The aims of this study were to investigate regulatory role and molecular mechanism of circ_0020123 in DDP resistance. Circ_0020123 was hypothesized to sponge miR-140-3p to regulate HOXB5. To study the role of circ_0020123 in DDP resistance of NSCLC, we first detected the level of circ_0020123 in DDP-resistant NSCLC cells. Circ_0020123 was highly expressed in NSCLC cells (A549 and H1299) relative to normal 16HBE cells, and this upregulation was also detected in DDP-resistant cells (A549/DDP and H1299/DDP) relative to the parental NSCLC cells (). Cell viability detection showed that IC50 of DDP was elevated in A549/DDP cells () and H1299/DDP cells () contrasted with A549 and H1299 cells, suggesting the generation of DDP resistance in A549/DDP and H1299/DDP cells. Thus, the high expression of circ_0020123 was found in DDP-resistant NSCLC cells.
Figure 1. Circ_0020123 level was increased in DDP-resistant NSCLC cells. (a) The qRT-PCR was used to measure circ_0020123 expression in 16HBE cells, NSCLC cells (A549 and H1299) and DDP-resistant NSCLC cells (A549/DDP and H1299/DDP). (b-e) IC50 of DDP was detected by CCK-8 assay in A549 and A549/DDP cells (b-c) or H1299 and H1299/DDP cells (d-e). Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test was used for statistical analysis. *P < 0.05, **P < 0.01.
Knockdown of circ_0020123 repressed DDP resistance in NSCLC cells
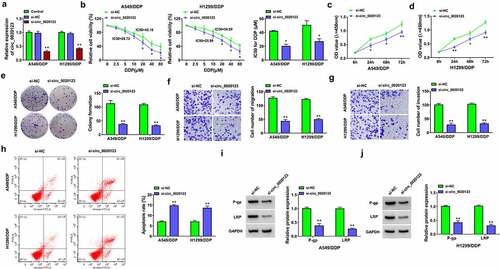
Knockdown of circ_0020123 was achieved by siRNA transfection. The results revealed that circ_0020123 expression of si-circ_0020123 group was significantly downregulated compared with si-NC and control groups in A549/DDP and H1299/DDP cells (). Cell viability detection suggested that IC50 value of DDP was decreased in si-circ_0020123 group of 10 µM DDP-treated A549/DDP and H1299/DDP cells relative to si-NC group (). By performing CCK-8 assay () and colony formation assay (), cell proliferation was inhibited after circ_0020123 downregulation in 10 µM DDP-treated A549/DDP and H1299/DDP cells. Transwell assay and flow cytometry suggested that silencing circ_0020123 suppressed cell migration or invasion () while increasing the apoptotic rate () in 10 µM DDP-treated A549/DDP and H1299/DDP cells. Drug resistance can be assessed by detection of resistance-related proteins, such as p-gp and LRP. Western blot manifested that si-circ_0020123 transfection downregulated the protein levels of p-gp and LRP compared to si-NC transfection in 10 µM DDP-treated A549/DDP and H1299/DDP cells, showing that DDP resistance was reduced by circ_0020123 knockdown (). Taken together, the expression inhibition of circ_0020123 could sensitize NSCLC cells to DDP.
Figure 2. Knockdown of circ_0020123 repressed DDP resistance in NSCLC cells. Transfection of si-NC or si-circ_0020123 was performed in A549/DDP and H1299/DDP cells. (a) Relative expression of circ_0020123 was determined using qRT-PCR. (b) IC50 for DDP was analyzed by CCK-8 assay after different concentrations of DDP treatment. (c-e) Cell proliferation was assessed through CCk-8 assay (c-d) and colony formation assay (e) after 10 µM DDP treatment. (f-g) Cell migration and invasion were examined by transwell assay after 10 µM DDP treatment. (h) Flow cytometry was exploited for detecting cell apoptosis after 10 µM DDP treatment. (i-j) The levels of resistance-related proteins were measured by Western blot after 10 µM DDP treatment. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test was used for statistical analysis. *P < 0.05, **P < 0.01.
Circ_0020123 acted as a sponge of miR-140-3p
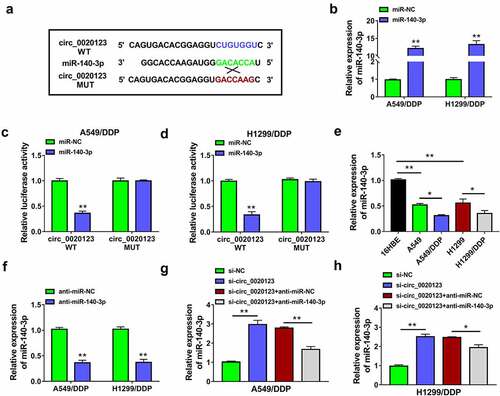
Online circinteractome prediction indicated that many miRNAs might be the targets of circ_0020123, and some downregulated miRNAs in lung cancer were selected as candidate targets. The qRT-PCR further demonstrated that the promoting effect of circ_0020123 downregulation on miR-140-3p level was the most significant among 8 candidate miRNAs (miR-95, miR-944, miR-384, miR-876-3p, miR-140-3p, miR-607, miR-600, miR-587). Thus, miR-140-3p was used for further target research of circ_0020123 in NSCLC. The complementary binding site of miR-140-3p in circ_0020123 sequence was exhibited in . The overexpression efficiency of miR-140-3p mimic was great in A549/DDP and H1299/DDP cells (). The interaction between circ_0020123 and miR-140-3p was analyzed by dual-luciferase reporter assay. As shown in , overexpression of miR-140-3p suppressed luciferase activity of circ_0020123 WT plasmid, while it did not affect that of circ_0020123 MUT plasmid in A549/DDP and H1299/DDP cells. Compared with A549 and H1299 cells, miR-140-3p level was evidently downregulated in A549/DDP and H1299/DDP cells (). The miR-140-3p expression was inhibited after transfection of anti-miR-140-3p by contrast with transfection of anti-miR-NC (), and miR-140-3p inhibition promoted DDP resistance in DDP-resistant NSCLC cells. The introduction of anti-miR-140-3p counteracted si-circ_0020123-mediated upregulation of miR-140-3p (), which exhibited that circ_0020123 induced the negative regulation of miR-140-3p. The qRT-PCR data also showed that circ_0020123 overexpression directly downregulated miR-140-3p expression in A549/DDP dan H1299/DDP cells. Circ_0020123 was a molecular sponge of miR-140-3p.
Figure 3. Circ_0020123 acted as a sponge of miR-140-3p. (a) The binding sites between circ_0020123 and miR-140-3p were shown by circinteractome. (b) The expression of miR-140-3p was assayed by qRT-PCR in miR-NC or miR-140-3p transfected A549/DDP and H1299/DDP cells. (c-d) The binding between circ_0020123 and miR-140-3p was validated using dual-luciferase reporter assay. (e) The expression analysis of miR-140-3p was performed by qRT-PCR in DDP-sensitive and resistant cells. (f) The inhibition of anti-miR-140-3p on miR-140-3p level was analyzed via qRT-PCR. (g-h) The qRT-PCR was conducted for miR-140-3p detection after transfection of si-circ_0020123, si-circ_0020123+ anti-miR-140-3p or negative controls. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for statistical analysis. *P < 0.05, **P < 0.01.
Inhibition of miR-140-3p partly rescued the effect of si-circ_0020123 on DDP resistance in NSCLC cells
To explore the relation between circ_0020123 and miR-140-3p in the regulation of DDP resistance in NSCLC, A549/DDP and H1299/DDP cells were transfected with si-circ_0020123, si-circ_0020123+ anti-miR-140-3p or the corresponding negative controls. CCK-8 assay indicated that si-circ_0020123-induced repressive effect on DDP resistance was attenuated by anti-miR-140-3p in A549/DDP and H1299/DDP cells (). Cell proliferation inhibition by si-circ_0010123 was also countervailed by miR-140-3p inhibitor in 10 µM DDP-treated A549/DDP and H1299/DDP cells (). Colony formation ability (), cell migration () and invasion () of 10 µM DDP-treated A549/DDP and H1299/DDP cells were promoted in si-circ_0020123+ anti-miR-140-3p group contrasted to si-circ_0020123+ anti-miR-NC group. Transfection of anti-miR-140-3p also attenuated si-circ_0020123-induced promotion of cell apoptosis () and downregulation of P-gp or LRP protein level () in 10 µM DDP-treated A549/DDP and H1299/DDP cells. These results suggested that the regulation of circ_0020123 in DDP resistance was related to the sponge effect on miR-140-3p.
Figure 4. Inhibition of miR-140-3p partly rescued the effect of si-circ_0020123 on DDP resistance in NSCLC cells. Transfection of si-NC, si-circ_0020123, si-circ_0020123+ anti-miR-NC or si-circ_0020123+ anti-miR-140-3p was performed in A549/DDP and H1299/DDP cells. (a-b) CCK-8 assay was carried out to analyze the IC50 for DDP after different concentrations of DDP treatment. (c-e) CCK-8 assay (c-d) and colony formation assay (e) were used for proliferation analysis under 10 µM DDP treatment. (f-g) Transwell assay was applied to evaluate cell migratory or invasive ability under 10 µM DDP treatment. (h) Cell apoptosis was measured by flow cytometry under 10 µM DDP treatment. (i-j) Western blot was implemented for the protein expression detection of P-gp and LRP under 10 µM DDP treatment. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for statistical analysis. *P < 0.05, **P < 0.01.
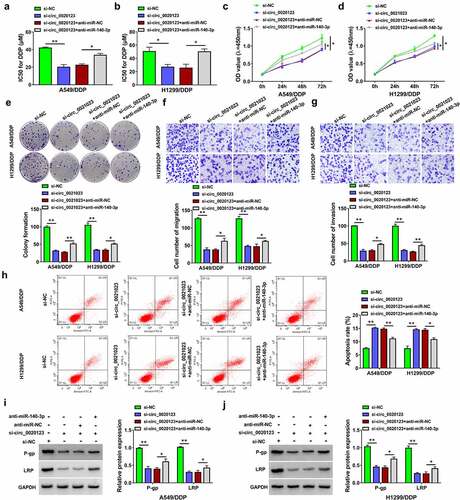
HOXB5 was a target of miR-140-3p
Through Targetscan prediction and literature screening, eight genes (CDK6, MAPK1, FGF9, HOXB5, PAK2, EIF5A2, NOVA1, JAG1) were selected as candidate targets for miR-140-3p. Subsequently, we observed that HOXB5 was downregulated by miR-140-3p overexpression with the most significant change in A549/DDP and H1299/DDP cells. Then, target relation between miR-140-5p and HOXB5 was further researched. The binding sites between the sequences of miR-140-3p and HOXB5 3ʹUTR are displayed in . In A549/DDP and H1299/DDP cells, luciferase activity of HOXB5 3ʹUTR WT group was reduced by miR-140-3p upregulation but that of HOXB5 3ʹUTR MUT group was not changed (). The qRT-PCR and Western blot revealed that HOXB5 mRNA and protein levels were upregulated in A549/DDP and H1299/DDP cells compared with the parental cells (). The promoting effect of pcDNA-HOXB5 on the level of HOXB5 was conspicuous by performing Western blot assay (). Moreover, miR-140-3p mimic triggered the inhibitory influence of HOXB5 protein level and this inhibition was lightened by pcDNA-HOXB5 (). The above data exhibited that miR-140-5p could directly downregulate the expression of HOXB5.
Figure 5. HOXB5 was a target of miR-140-3p. (a) Targetscan was used to predict the binding between miR-140-3p and HOXB5 3ʹUTR. (b-c) Dual-luciferase reporter assay was performed for verifying the interaction between miR-140-3p and HOXB5 3ʹUTR. (d-e) Western blot was applied to examine the protein expression of HOXB5 in 16HBE cells or DDP-sensitive/resistant NSCLC cells (d) and pcDNA or pcDNA-HOXB5 transfected cells (e). (f-g) The protein level of HOXB5 was analyzed by Western blot after transfection of miR-140-3p, miR-140-3p+pcDNA-HOXB5. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for statistical analysis. *P < 0.05, **P < 0.01.
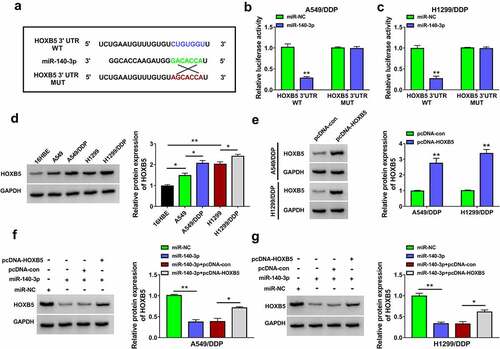
HOXB5 overexpression partly alleviated the inhibition of miR-140-3p in DDP resistance of NSCLC cells
The function of miR-140-3p and its regulatory mechanism were further researched through the reverted experiments. IC50 of DDP was reduced in miR-140-3p-transfected A549/DDP and H1299/DDP cells, while this effect was weakened after the introduction of pcDNA-HOXB5 (). The miR-140-3p-induced inhibitory effects on cell proliferation (), migration and invasion () but the stimulative effect on cell apoptosis () in 10 µM DDP-treated A549/DDP and H1299/DDP cells were all partly relieved by upregulation of HOXB5. The protein levels of P-gp and LRP were increased after co-transfection of miR-140-3p and pcDNA-HOXB5 contrasted with along miR-140-3p transfection in 10 µM DDP-treated A549/DDP and H1299/DDP cells (). All these findings demonstrated that miR-140-3p suppressed DDP resistance partly by inhibiting the expression of HOXB5.
Figure 6. HOXB5 overexpression partly alleviated the inhibition of miR-140-3p in DDP resistance of NSCLC cells. Transfection of miR-NC, miR-140-3p, miR-140-3p+pcDNA-con or miR-140-3p+pcDNA-HOXB5 was performed in A549/DDP and H1299/DDP cells. (a-b) The detection of IC50 for DDP was performed using CCK-8 assay after different concentrations of DDP treatment. (c-e) Cell proliferation ability was analyzed using CCK-8 assay (c-d) and colony formation assay (e) following 10 µM DDP treatment. (f-g) Transwell assay was applied to evaluate cell migration or invasion ability following 10 µM DDP treatment. (h) The assessment of cell apoptosis was completed via flow cytometry following 10 µM DDP treatment. (i-j) The levels of P-gp and LRP were determined through Western blot following 10 µM DDP treatment. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for statistical analysis. *P < 0.05, **P < 0.01.
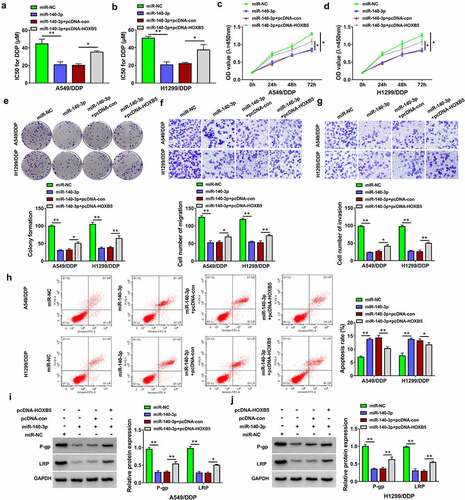
Circ_0020123 regulated the HOXB5 level partly via targeting miR-140-3p
To analyze whether HOXB5 was regulated by circ_0020123 via miR-140-3p, we determined the effect of circ_0020123/miR-140-3p on HOXB5 expression. The results of Western blot presented that knockdown of circ_0020123 led to protein downregulation of HOXB5, whereas miR-140-3p inhibitor eliminated this regulation in part (). In addition, the silence of HOXB5 reduced drug resistance and cell proliferation in A549/DDP and H1299/DDP cells. Hence, circ_0020123 function was achieved partly by regulating miR-140-3p-related HOXB5 expression.
Figure 7. Circ_0020123 regulated the HOXB5 level partly via targeting miR-140-3p. (a-b) Western blot was performed for the measurement of HOXB5 protein level in si-NC, si-circ_0020123, si-circ_0020123+ anti-miR-NC or si-circ_0020123+ anti-miR-140-3p transfection group in A549/DDP (a) and H1299/DDP (b) cells. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for statistical analysis. *P < 0.05, **P < 0.01.
Circ_0020123 inhibition reduced tumor growth of NSCLC in A549/DDP xenograft model partly through the regulation of miR-140-3p/HOXB5 axis
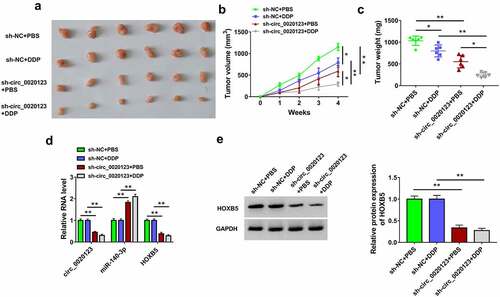
A549/DDP xenograft models were established in mice, followed by DDP or PBS treatment. After injection for 4 weeks, tumors were excised and photographed (). Data indicated that tumor volume and weight were reduced in sh-circ_0020123+ PBS group relative to sh-NC+PBS group in A549/DDP xenografts, and knockdown of circ_0020123 also inhibited tumor growth under the treatment of DDP in mice (). The qRT-PCR detection revealed the downregulation of circ_0020123 and HOXB5 but the upregulation of miR-140-3p in sh-circ_0020123+ PBS or sh-circ_0020123+ DDP group compared to sh-NC+PBS or sh-NC+DDP group (). The protein expression of HOXB5 was reduced after circ_0020123 silence in vivo (). These findings in vivo elucidated that circ_0020123 downregulation decreased tumor growth partly by regulating the miR-140-3p/HOXB5 axis in A549/DDP xenografts.
Figure 8. Circ_0020123 inhibition reduced tumor growth of NSCLC in A549/DDP xenograft model partly through the regulation of miR-140-3p/HOXB5 axis. (a) The tumors of sh-NC+PBS, sh-NC+DDP, sh-circ_0020123+ PBS or sh-circ_0020123+ DDP group were excised from mice. (b-c) The measured tumor volume (b) and weight (c) of each group. (C) The expression levels of circ_0020123, miR-140-3p and HOXB5 mRNA were assayed by qRT-PCR. (e) HOXB5 protein level was analyzed using Western blot. Each experiment was performed for three times with three parallels each time. Data were exhibited as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were used for statistical analysis. *P < 0.05, **P < 0.01.
Discussion
Drug resistance has limited the chemotherapeutic effects on human cancers, and the radiosensitization of cancer cells to drug is crucial for cancer treatment [Citation24]. In this report, we validated that circ_0020123 downregulation reduced DDP resistance in NSCLC cells via the regulation of miR-140-3p/HOXB5 axis. It is of great importance to improve DDP therapy for NSCLC patients.
A handful of circRNAs have been reported to regulate drug resistance in NSCLC. For example, circ_0002483 inhibited taxol resistance, while circ_0004015 increased gefitinib resistance in NSCLC cells [Citation25,Citation26]. Circ_0002130 and circ_0011292 promoted cancer progression in osimertinib-resistant and paclitaxel-resistant NSCLC cells [Citation27,Citation28]. Huang et al. asserted that circ_0001946 retarded cancer development and decreased the resistance of DDP in NSCLC cells [Citation29]. Circ-Smarca5 was shown to enhance the chemosensitivity of NSCLC cells to DDP [Citation30]. Additionally, circ_0096157 and circ_0085131 contributed to DDP resistance in NSCLC cells [Citation31,Citation32]. Our expression analysis revealed that circ_0020123 was overexpressed in DDP-resistant NSCLC cells, implying that high circ_0020123 expression might be correlated to DDP resistance. By knocking down the level of circ_0020123, we found that DDP resistance was repressed in NSCLC cells and anti-tumor effects on DDP-resistant NSLCC cells were facilitated. Hence, circ_0020123 was involved in the generation of DDP resistance, and circ_0020123 downregulated could enhance DDP sensitivity.
Hsa_circRNA_102336 elevated DDP resistance via sponging miR-515-5p in bladder cancer [Citation33]. Inhibition of circCDR1as conferred chemosensitivity of breast cancer cells to 5-fluorouracil by increasing the level of miR-7 [Citation34]. By predicting the circinteractome and the expression effects of circ_0020123 on different candidate miRNAs, miR-140-3p was chosen as a probable target for circ_0020123. Dual-luciferase reporter assay further proved that circ_0020123 could directly bind to miR-140-3p. More interestingly, si-circ_0020123-mediated suppression of DDP resistance was partly attributed to the upregulation of miR-140-3p. A previous study manifested that miR-140-3p regulated the sensitivity of sorafenib in hepatocellular carcinoma via targeting pregnane x receptor (PXR [Citation35]. DDP sensitivity in NSCLC could be promoted by miR-140-3p [Citation12], while it has not been discovered whether the effect of miR-140-3p was associated with its downstream targets. Herein, target relation between miR-140-3p and HOXB5 was confirmed in NSCLC cells. The miR-140-3p-induced reduction of DDP resistance was achieved through the downregulation of HOXB5 in NSCLC cells.
Furthermore, circ_0020123 regulated the expression of HOXB5 via inhibiting miR-140-3p. CircRNA/miRNA/mRNA axes have been found in the regulation of DDP resistance in NSCLC, such as circ_0076305/miR-296-5p/Signal transducer and activator of transcription 3 (STAT3) axis [Citation36], circRNA carboxypeptidase A4/let-7 miRNA/programmed cell death-Ligand (PD-L1) axis [Citation37] and circRNA Protein arginine N-methyltransferase 5/miR-4458/Protein reversion less 3-like (REV3L) axis [Citation38]. Also, previous reports in Bioengineered indicated that circRNAs were involved in resistance regulation by mediating miRNA/mRNA axis. For instance, circRNA SRY-Box 13 increased DDP resistance in NSCLC via miR-3194-3p-related upregulation of microtubule-associated protein RP/EB family member 1 (MAPRE1) [Citation39]. Zheng et al. found that circ_0074027 enhanced DDP resistance by miR-379-5p/IGF1 axis in NSCLC [Citation40]. No research of circ_0020123 was disclosed in Bioengineered, and circ_0020123/miR-140-3p/HOXB5 axis was first reported in chemoresistance of NSCLC. Also, in vivo assay suggested that knockdown of circ_0020123 inhibited tumor growth inhibition of NSCLC.
However, this study still has some limitations. For example, there is a lack of clinical data. It is necessary to provide more data of clinical samples. In addition, the result did not support the effect of circ_0020123 on DDP resistance in vivo. Animal assay should be modified to better affirm the current conclusion.
Conclusion
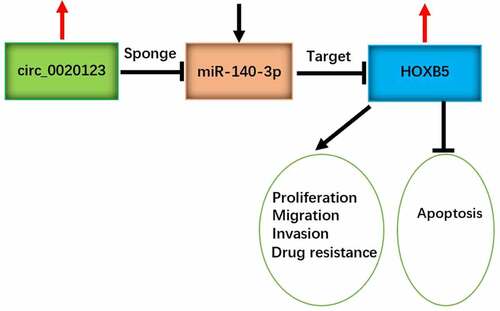
Summarily, we have provided the first-hand information about the promoting influence of circ_0020123 on DDP resistance in NSCLC and the discovery of circ_0020123/miR-140-3p/HOXB5 signal axis (Graphical Abstract). This study emphasized that circ_0020123 acted as a biomolecular target for chemoresistance in DDP therapy. Knockdown of circ_0020123 can be used for sensitization of DDP in treatment of NSCLC patients.
Highlights
Circ_0020123 downregulation inhibits cisplatin resistance in NSCLC cells.
Circ_0020123 targets miR-140-3p to upregulate HOXB5 level.
Circ_0020123 contributes to tumor growth in vivo by miR-140-3p/HOXB5 axis.
Supplemental Material
Download Zip (31.8 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Wood K, Hensing T, Malik R, et al. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol. 2016;2(6):805–812.
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. 2019;322(8):764–774.
- Fennell DA, Summers Y, Cadranel J, et al. Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat Rev. 2016;44:42–50.
- Liao M. Non-surgical therapy for patients with advanced non-small cell lung cancer. Respirology. 1998;3(3):151–157.
- Wang J, Zhu S, Meng N, et al. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27(10):1718–1725.
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141.
- Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71.
- Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–1652.
- Qu D, Yan B, Xin R, et al. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8(8):1387–1402.
- Bi R, Wei W, Lu Y, et al. High hsa_circ_0020123 expression indicates poor progression to non-small cell lung cancer by regulating the miR-495/HOXC9 axis. Aging (Albany NY). 2020;12(17):17343–17352.
- Hu C, Zou Y, Jing LL. miR-140-3p inhibits progression of non-small cell lung cancer by targeting Janus kinase 1. J Biosci. 2020;45. DOI:10.1007/s12038-020-0003-3
- Wu S, Wang H, Pan Y, et al. miR-140-3p enhances cisplatin sensitivity and attenuates stem cell-like properties through repressing Wnt/beta-catenin signaling in lung adenocarcinoma cells. Exp Ther Med. 2020;20(2):1664–1674.
- Gao Y, Fei X, Kong L, et al. HOXB5 promotes proliferation, migration, and invasion of pancreatic cancer cell through the activation of the GSK3beta/beta-catenin pathway. Anticancer Drugs. 2020;31(8):828–835.
- Lee K, Chang JW, Oh C, et al. HOXB5 acts as an oncogenic driver in head and neck squamous cell carcinoma via EGFR/Akt/Wnt/beta-catenin signaling axis. Eur J Surg Oncol. 2020;46(6):1066–1073.
- Su B, Zhang X, Luo G. Homeobox B5 suppression attenuates proliferation and elevates apoptosis in hepatoma cell lines through ERK/MDM2 signalling. Clin Exp Pharmacol Physiol. 2020;47(6):1058–1066.
- Zhang B, Li N, Zhang H. Knockdown of homeobox B5 (HOXB5) inhibits cell proliferation, migration, and invasion in non-small cell lung cancer cells through inactivation of the Wnt/beta-catenin pathway. Oncol Res. 2018;26(1):37–44.
- Tan X, Jiang L, Wu X, et al. MicroRNA-625 inhibits the progression of nonsmall cell lung cancer by directly targeting HOXB5 and deactivating the Wnt/beta-catenin pathway. Int J Mol Med. 2019;44(1):346–356.
- Gao X, Zhao H, Diao C, et al. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495(1):1074–1080.
- Liu ZL, Jin BJ, Cheng CG, et al. Apatinib resensitizes cisplatin-resistant non-small cell lung carcinoma A549 cell through reversing multidrug resistance and suppressing ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(23):5370–5377.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Zhang W, Mao S, Shi D, et al. MicroRNA-153 decreases tryptophan catabolism and inhibits angiogenesis in bladder cancer by targeting indoleamine 2,3-dioxygenase 1. Front Oncol. 2019;9:619.
- Sun J, Xin K, Leng C, et al. Down-regulation of SNHG16 alleviates the acute lung injury in sepsis rats through miR-128-3p/HMGB3 axis. BMC Pulm Med. 2021;21(1):191.
- Liu Y, Yang C, Zhao Y, et al. Overexpressed methyltransferase-like 1 (METTL1) increased chemosensitivity of colon cancer cells to cisplatin by regulating miR-149-3p/S100A4/p53 axis. Aging (Albany NY). 2019;11(24):12328–12344.
- Panda M, Biswal BK. Cell signaling and cancer: a mechanistic insight into drug resistance. Mol Biol Rep. 2019;46(5):5645–5659.
- Li X, Yang B, Ren H, et al. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10(12):953.
- Zhou Y, Zheng X, Xu B, et al. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun. 2019;508(2):527–535.
- Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307.
- Guo C, Wang H, Jiang H, et al. Circ_0011292 enhances paclitaxel resistance in non-small cell lung cancer by regulating miR-379-5p/TRIM65 axis. Cancer Biother Radiopharm. 2020. DOI:10.1089/cbr.2019.3546
- Huang MS, Liu JY, Xia XB, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508.
- Tong S. Circular RNA SMARCA5 may serve as a tumor suppressor in non-small cell lung cancer. J Clin Lab Anal. 2020;34(5):e23195.
- Lu H, Xie X, Wang K, et al. Circular RNA hsa_circ_0096157 contributes to cisplatin resistance by proliferation, cell cycle progression, and suppressing apoptosis of non-small-cell lung carcinoma cells. Mol Cell Biochem. 2020. DOI:10.1007/s11010-020-03860-1
- Kong R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol Int. 2020;44(9):1945–1956.
- Gong P, Xu R, Zhuang Q, et al. A novel circular RNA (hsa_circRNA_102336), a plausible biomarker, promotes the tumorigenesis by sponging miR-515-5p in human bladder cancer. Biomed Pharmacother. 2020;126:110059.
- Yang W, Gu J, Wang X, et al. Inhibition of circular RNA CDR1as increases chemosensitivity of 5-FU-resistant BC cells through up-regulating miR-7. J Cell Mol Med. 2019;23(5):3166–3177.
- Li J, Zhao J, Wang H, et al. MicroRNA-140-3p enhances the sensitivity of hepatocellular carcinoma cells to sorafenib by targeting pregnenolone X receptor. Onco Targets Ther. 2018;11:5885–5894.
- Dong Y, Xu T, Zhong S, et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984.
- Hong W, Xue M, Jiang J, et al. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39(1):149.
- Pang J, Ye L, Zhao D, et al. Circular RNA PRMT5 confers cisplatin-resistance via miR-4458/REV3L axis in non-small-cell lung cancer. Cell Biol Int. 2020;44:2416–2426.
- Zhang L, Peng H, Xu Z, et al. Circular RNA SOX13 promotes malignant behavior and cisplatin resistance in non-small cell lung cancer through targeting microRNA-3194-3p/microtubule-associated protein RP/EB family member 1. Bioengineered. 2022;13(1):1814–1827.
- Zheng S, Wang C, Yan H, et al. Blocking hsa_circ_0074027 suppressed non-small cell lung cancer chemoresistance via the miR-379-5p/IGF1 axis. Bioengineered. 2021;12(1):8347–8357.
