ABSTRACT
The key role of circular RNA (circRNA) in the malignant progression of cancers has been demonstrated. However, the role of circRNA midline-1 (circMID1) in prostate cancer (PCa) progression has not been clarified. Quantitative real-time PCR was used to measure relative expression. Function analysis was performed using EdU staining, colony formation assay, flow cytometry, wound healing assay, transwell assay and cell glycolysis detection. The protein levels were detected by Western blot analysis. RNA pull-down assay, dual-luciferase reporter assay and RIP assay were performed to verify RNA interaction. Animal experiments were utilized to explore the effects of circMID1 knockdown on PCa tumorigenesis in vivo. Our results showed that circMID1 was upregulated in PCa tissues and cells and its knockdown inhibited PCa cell proliferation, migration, invasion and glycolysis in vitro, as well as PCa tumorigenesis in vivo. IGF1R and YTHDC2 were highly expressed in PCa tissues and cells, and their expression was positively regulated by circMID1. IGF1R and YTHDC2 overexpression reversed the inhibitory effect of circMID1 silencing on PCa cell progression. In terms of mechanism, circMID1 could sponge miR-330-3p and miR-330-3p could target IGF1R and YTHDC2. Functional experiments showed that circMID1 sponged miR-330-3p to regulate PCa progression via the YTHDC2/IGF1R/AKT axis. In conclusion, our data confirmed that circMID1 might play a pro-cancer role in PCa, which promoted PCa progression through regulating the miR-330-3p/YTHDC2/IGF1R/AKT axis.
Graphical abstract
KEYWORDS:
Introduction
Prostate cancer (PCa) is a malignant tumor that occurs in the epithelial cells of the prostate and has become one of the common malignant tumors in urology [Citation1,Citation2]. Because the early symptoms are not obvious, most PCa patients have reached the local advanced stage or metastasis at the time of diagnosis [Citation3,Citation4]. PCa often manifests as symptoms of urinary tract obstruction, and it is accompanied by bone pain and pathological fractures after bone metastasis [Citation5,Citation6]. Therefore, elucidating the underlying molecular mechanism of PCa has important theoretical significance for the treatment of PCa.
Circular RNA (circRNA) is a non-coding RNA formed by back-splicing, which is abundant in the cytoplasm of eukaryotic cells [Citation7,Citation8]. With the development of novel bioinformatics methods, many types and functions of circRNA have been detected and identified, including being an effective microRNA (miRNA) sponge to indirectly regulate downstream gene expression [Citation9,Citation10]. It is worth noting that the high abundance, relative stability and evolutionary conservation of circRNA make circRNA have the potential to become a biomarker for the treatment and diagnosis of many diseases, especially for cancers [Citation11,Citation12]. Many circRNAs have been shown to be associated with the malignant progression of PCa, including circFoxo3 [Citation13], circ_0044516 [Citation14], and circMBOAT2 [Citation15]. Recently study showed that circ_0075542 regulated miR-1197/HOXC11 axis to inhibit PCa proliferation and metastasis [Citation16]. On the contrary, circ_0030586 had been found to enhance the epithelial–mesenchymal transition process of PCa through the regulation of PI3K/AKT pathway [Citation17]. Even so, there are still many new circRNA functions that need to be revealed. In this study, we screened differentially expressed circRNAs in high-grade PCa (H-PCa) tissues and low-grade PCa (L-PCa) tissues using the GEO database and found that hsa_circ_0007933 (derived from midline-1 (MID1) gene, also called circMID1) was significantly overexpressed in H-PCa tissues. However, the role of circMID1 in the progression of PCa remains unclear.
The type 1 insulin-like growth factor receptor (IGF1R) belongs to the receptor tyrosine kinase family and is involved in the regulation of cell growth and differentiation [Citation18]. Studies have shown that IGF1R was significantly highly expressed in PCa cells, and the IGF1R/AKT pathway participated in the regulation of tumor growth, metastasis, and glycolysis [Citation19,Citation20]. As one of the most important mRNA modifications, N6-methyladenosine (m6A) is involved in multiple steps of mRNA processing to achieve the regulation of post-transcriptional gene expression [Citation21]. The YTH domain containing 2 (YTHDC2), an m6A reader, has been shown to bind to IGF1R mRNA, thereby increasing IGF1R mRNA stability and promoting the IGF1R protein translation process [Citation22]. In addition, research had suggested that YTHDC2 was also upregulated in PCa tissues [Citation23].
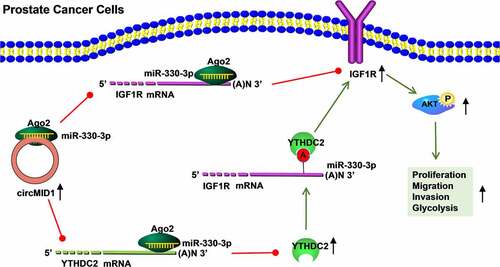
Our study aims to reveal the role and potential molecular mechanism of circMID1 in the progression of PCa. Here, we found that circMID1 could activate the IGF1R/AKT axis, which was achieved by positively regulating the YTHDC2 expression. In addition, in order to reveal the molecular mechanism by which circMID1 regulated the YTHDC2/IGF1R/AKT axis, we explored the miRNAs that could interact with circMID1, YTHDC2 and IGF1R. Therefore, we proposed the hypothesis that circMID1 sponged miR-330-3p to regulate the YTHDC2/IGF1R/AKT axis, thereby regulating PCa progression. Our study is expected to provide new molecular targets for the treatment of PCa.
Materials and methods
Samples collection
In this study, 51 PCa patients with areas of high-grade (Gleason > 8, n = 18) and low-grade (Gleason < 6, n = 33) as well as 27 benign prostatic hyperplasia (BPH) patients were recruited from the First Affiliated Hospital of Zhengzhou University. The clinicopathologic features of PCa patients are shown in . The PCa tissues and the prostatectomy tissues of BPH patients were collected and stored in −80°C. Each patient signed written informed consent. Our research was approved by the Ethics Committee of First Affiliated Hospital of Zhengzhou University and was performed in accordance with the Declaration of Helsinki.
Table 1. Correlation of circMID1 expression and the clinicopathologic features in patients with PCa
Cell culture and transfection
Human PCa cells (PC-3 and DU145) and normal prostate epithelial cells (RWPE-1) were purchased from ATCC (Manassas, VA, USA). PC-3 and DU145 cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA), while RWPE-1 cells were grown in the K-SFM medium (Gibco) at 37°C with 5% CO2. All of the media were supplemented with 10% FBS (Gibco) and 1% penicillin–streptomycin liquid (Beyotime, Shanghai, China). Cell transfection was performed after the cells reached 60% confluences. The lentivirus short hairpin RNA (shRNA) against circMID1 or YTHDC2 (sh-circMID1 or sh-YTHDC2), the pcDNA overexpression vectors of YTHDC2 or IGF1R, miR-330-3p mimic or inhibitor (miR-330-3p or anti-miR-330-3p), and their negative controls were synthesized by Genepharma (Shanghai, China). They were transfected into PCa cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). After transfection for 48 h, cells were collected for function experiments.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using the RNA Isolation Kit (Qiagen, Duesseldorf, Germany). The PrimeScript Reverse Transcriptase Kit (Invitrogen) was used to prepare cDNA. qRT-PCR was performed using PCR System with SYBR Green (Takara, Dalian China). In this, relative expression was analyzed by the 2−∆∆Ct method with β-actin or U6 as internal reference. Primer sequences used for qRT-PCR are shown in .
Table 2. Primer sequences used for qRT-PCR
Identification of circRNA
Universal gDNA Extraction Kit (Takara) was used to collect genomic DNA (gDNA) from PCa cells. After that, the cDNA and gDNA of circMID1 were amplified with convergent primers and divergent primers (Sangon, Shanghai, China), and the PCR products were used for agarose gel electrophoresis.
In the subcellular localization assay, the nucleus and cytoplasm RNA of PCa cells were isolated by PARIS Kit (Invitrogen). Then, circMID1 expression was determined by qRT-PCR. U6 was used as the nucleus control and GAPDH as the cytoplasm control.
EdU staining
As previously described [Citation24], EdU assay was used to evaluate cell proliferation ability by detecting EdU positive (EdU+) cells (%) with BeyoClick™ EdU Cell Proliferation Kit (Beyotime) according to the kit instructions. DAPI solution (Beyotime) was used to stain the cell nucleus.
Colony formation assay
PCa cells were seeded into 6-well plates at a density of 150 cells per dish. Cells were cultured for 2 weeks at 37°C, and then the colony number was counted under microscope after the colonies were fixed with paraformaldehyde and stained with crystal violet according to the previous report [Citation25].
Flow cytometry
According to the previous study [Citation26], the Cell Cycle Analysis Kit (Biovision, Milpitas, CA, USA) was used to measure cell cycle process. Briefly, the transfected PCa cells were harvested and then fixed with 70% cold ethanol. Afterward, the cells were hatched with RNase A for 30 min and then stained with propidium iodide. The distribution of the cell cycle was analyzed using flow cytometry.
Wound healing assay
As previously described [Citation17], PCa cells were seeded onto 6-well plates and cultured until the cells reached 90% confluence. A wound was scraped using the tip of a 200 μL straw in the cell layer. The wound area was photographed at 0 h and 24 h under a microscope, and the wound healing ratio was calculated using ImageJ software.
Transwell assay
According to the previous study [Citation16], Matrigel Invasion Chambers (BD Biosciences, San Jose, CA, USA) were used for detecting cell invasion. PC-3 and DU145 cells were seeded into the upper chambers with RPMI-1640 medium. Serum medium (10%) was used as a chemoattractant in the lower chambers. 24 h later, the bottoms of chambers were stained with crystal violet, and the invaded cell number was counted under a microscope (100 ×).
Cell glycolysis assay
Glucose uptake and lactate products were determined using Glucose Uptake Assay Kit (Abcam, Cambridge, MA, USA) and Lactate Assay Kit (Biovision) according to the kit instructions. Based on the instructions of Glycolysis Rate Determination Kit (Agilent, Santa Clara, CA, USA), the extracellular acidification rate (ECAR) of cells was measured with Seahorse XFe Extracellular Flux Analyzer.
Western blot (WB) analysis
After extracting total proteins with RIPA Lysis Buffer (Beyotime), the same amount of protein was separated by 10% SDS-PAGE gel and then transferred onto PVDF membranes (Invitrogen). The membrane was incubated with anti-IGF1R (1:1,000, ab263907, Abcam), anti-p-AKT (1:25,000, ab81283, Abcam), anti-AKT (1:500, ab8805, Abcam), anti-cyclin D1 (1:200, ab16663, Abcam), anti-GLUT1 (1:2,500, ab14683, Abcam), anti-YTHDC2 (1:1,000, ab220160, Abcam), or anti-β-actin (1:5,000, ab8226, Abcam) followed by hatching with Goat anti-Rabbit (1:50,000, ab205718, Abcam) or Goat-anti-Mouse (1:5,000, ab205719, Abcam). Protein signals were visualized with the BeyoECL Plus Kit (Beyotime).
RIP assay
According to the previous report [Citation24], PC-3 and DU145 cells were lysed with RIP buffer and then hatched with magnetic beads coated with anti-YTHDC2 (1:30, ab220160, Abcam), anti-Ago2 (1:50, ab186733, Abcam) or anti-IgG (1:20, ab6789, Abcam) at 4°C overnight based on the instructions of the Magna RIP Kit (Millipore, Billerica, MA, USA). The enrichment of IGF1R or circMID1 was measured using qRT-PCR.
Actinomycin D (ActD) assay
PC-3 and DU145 cells were transfected with sh-control or sh-YTHDC2 for 48 h. Afterward, the transfected cells were treated with ActD (Millipore) for indicated time points. Then, IGF1R mRNA expression was examined using qRT-PCR.
RNA pull-down assay
Biotin-labeled circMID1 probe, the wild-type (wt)/mutate-type (mut) of miR-330-3p probe (bio-miR-330-3p-wt/mut) and their negative control probes (NC probe and bio-NC) were synthesized by Genepharma. PC-3 and DU145 cells were harvested, and then the cell lysates were incubated with Dynabeads M-280 Streptavidin (Invitrogen) coated circMID1 probe or NC probe. The enrichment of candidate miRNAs was examined by qRT-PCR to screen the targeted miRNA for circMID1. To explore whether miR-330-3p could pull down circMID1, YTHDC2 and IGF1R, PCa cells were transfected with the bio-miR-330-3p-wt/mut probe or bio-NC probe for 24 h, as the previous study [Citation16]. Then, the cell lysates were cultured with the Dynabeads, and the RNA enrichment was analyzed by qRT-PCR.
Dual-luciferase reporter assay
As the previous study [Citation16], the sequences of circMID1, YTHDC2 3ʹUTR and IGF1R 3ʹUTR containing the binding sites or mutate sites with miR-330-3p were cloned into the pmirGLO luciferase vectors, generated the circMID1-wt/mut, YTHDC2 3ʹUTR-wt/mut, IGF1R 3ʹUTR-wt1/mut1 and IGF1R 3ʹUTR-wt2/mut2 vectors. The vectors and miR-330-3p mimic or miR-NC were co-transfected into 293 T cells, 48 h later, Dual-Luciferase Reporter Assay Kit (Vazyme, Nanjing, China) was used to examine luciferase activity.
Xenograft tumor
Ten BALB/c nude mice were purchased from Vital River (Beijing, China). PC-3 cells transfected with sh-circMID1 or sh-control were collected and suspended with PBS (2 × 106 cells/0.2 mL PBS). The cell suspensions were subcutaneously injected into nude mice, and the tumor volume was measured every 4 days starting from day 3. After 27 days, the mice were sacrificed and the tumors were photographed and weighed. The tumor tissue was prepared into paraffin sections, which were then used for H&E staining and YTHDC2 and IGF1R immunohistochemical (IHC) staining according to the instructions of SP Kit (Solarbio, Beijing, China). Animal experiments were approved by the Animal Ethics Committee of First Affiliated Hospital of Zhengzhou University.
Statistical analysis
Statistical analysis was carried out by Graphpad Prism 7.0 software. Data were expressed as means ± standard deviation from three independent experiments. Comparisons were analyzed by Student’s t-test and one-way analysis of variance followed by Tukey’s post-hoc test. P < 0.05 was considered statistically significant.
Results
In our study, we investigated the role and molecular mechanism of circMID1 in PCa progression. We found that circMID1 might promote PCa proliferation, migration, invasion and glycolysis. In terms of mechanism, we found that circMID1 sponged miR-330-3p to regulate the YTHDC2/IGF1R/AKT axis. To sum up, we confirmed that circMID1 promoted PCa progression depending on the regulation of the miR-330-3p/YTHDC2/IGF1R/AKT axis. Our study provided a potential theoretical target for treating PCa.
CircMID1 was upregulated in PCa tissues and cells
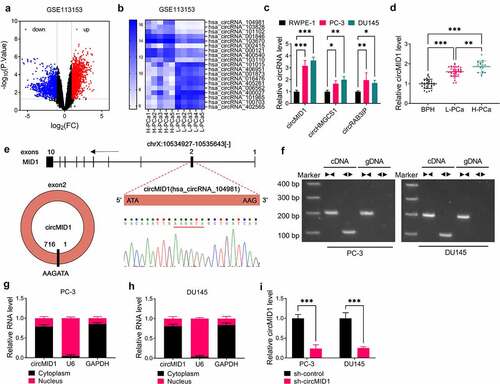
According to the criteria value (|log2FC| > 1, P < 0.05), the differentially expressed circRNAs in five paired H-PCa tissues and L-PCa tissues were screened in GSE113153 database and shown as a volcanic map (). The TOP10 circRNA with the most obvious up-regulated and down-regulated expressions were screened according to the P value for heat map analysis (). By further analyzing the expression of circRNA in PCa cells, we found that circMID1 (probe_hsa_circRNA_104981), circHMGCS1 (probe_hsa_circRNA_103828) and circRAB3IP (probe_hsa_circRNA_101102) were markedly upregulated in PCa cells (PC-3 and DU145) compared to RWPE-1 cells (), and the expression difference of circMID1 was the most significant. Therefore, circMID1 was chosen for the study. Through qRT-PCR, we discovered that circMID1 expression was highly expressed in L-PCa tissues and H-PCa tissues compared to the prostatectomy tissues of BPH patients and was higher in H-PCa tissues than that in L-PCa tissues (). By analyzing the correlation of circMID1 expression and the clinicopathologic features in patients with PCa, we found that the high expression of circMID1 was related to the Gleason score, pathological stage, and whether bone metastasis occurs in PCa patients (). Through analysis, we confirm that circMID1 is derived from the exon 2 of MID1 gene, and the result of Sanger sequencing confirms the head-to-tail splicing of circMID1 (). After amplification by convergent primers and divergent primers, we found that circMID1 only could be amplified by divergent primers in cDNA but not in gDNA, suggesting that circMID1 had circular structure (). Subcellular localization assay results showed that circMID1 is mainly distributed in the cytoplasm of PC-3 and DU145 cells (). To further explore the role of circMID1 in PCa progression, we constructed the sh-circMID1 and confirmed that circMID1 expression indeed could be reduced by sh-circMID1 in PC-3 and DU145 cells ().
Figure 1. CircMID1 was upregulated in PCa tissues and cells. (a) Volcanic map showed the differentially expressed circRNAs in H-PCa tissues and L-PCa tissues screened from GSE113153 database. (b) Heat map showed the TOP10 circRNA with the most obvious up-regulated and down-regulated expressions in H-PCa tissues and L-PCa tissues. (c) The expression of circMID1, circHMGCS1 and circRAB3IP in RWPE-1, PC-3 and DU145 cells was measured by qRT-PCR. (d) CircMID1 expression in L-PCa tissues, H-PCa tissues and the prostatectomy tissues of BPH patients was detected by qRT-PCR. (e) The basic information of circMID1 was shown. (f) After the cDNA and gDNA of circMID1 were amplified by divergent primers and convergent primers, the PCR products were used in agarose gel electrophoresis analysis. (g-h) Subcellular localization assay was used to confirm the distribution of circMID1 in cytoplasm and nucleus of PC-3 and DU145 cells. (i) The transfection efficiency of sh-circMID1 was confirmed by detecting circMID1 expression using qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001.
Downregulation of circMID1 inhibited the proliferation, migration, invasion and glycolysis of PCa cells
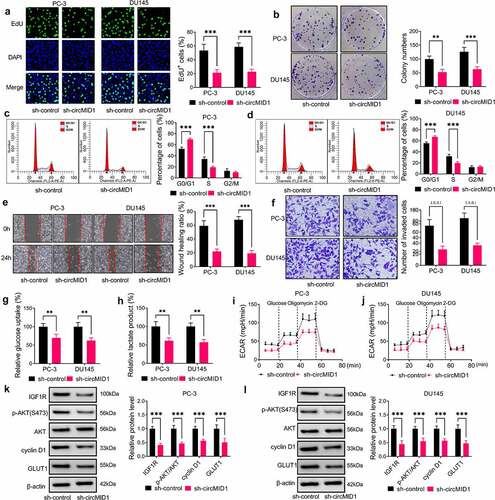
Then, we measured the progression of PCa cells transfected with sh-circMID1. Our results showed that circMID1 silencing repressed the EdU+ cell rate, the colony numbers and the cell numbers at the S phase in PC-3 and DU145 cells (), showing that circMID1 might promote PCa cell proliferation. In addition, circMID1 knockdown inhibited the wound healing ratio and the invaded cell number in PC-3 and DU145 cells (), confirming that circMID1 could also increase PCa cell migration and invasion. Moreover, the glucose uptake, lactate product and ECAR in PC-3 and DU145 cells also were reduced by silencing circMID1 (). Then, we examined the protein expression of IGF1R and p-AKT/AKT and found that circMID1 silencing could decrease the protein levels of IGF1R and p-AKT/AKT in PC-3 and DU145 cells (). In addition, we also discovered that the proliferation marker cyclin D1 and glycolysis marker GLUT1 were also markedly reduced by circMID1 knockdown (). These data revealed that circMID1 might play an oncogenic role in PCa.
Figure 2. Downregulation of circMID1 inhibited the proliferation, migration, invasion and glycolysis of PCa cells. PC-3 and DU145 cells were transfected with sh-control or sh-circMID1. EdU staining (a), colony formation assay (b) and flow cytometry (c-d) were used to assess cell proliferation. Wound healing assay (e) and transwell assay (f) were performed to measure cell migration and invasion. Glucose uptake (g), lactate product (h) and ECAR (i-j) were determined to evaluate cell glycolysis. (k-l) The protein levels of IGF1R, p-AKT/AKT, cyclin D1 and GLUT1 were evaluated using WB analysis. **P < 0.01, ***P < 0.001.
CircMID1 positively regulated IGF1R expression via YTHDC2
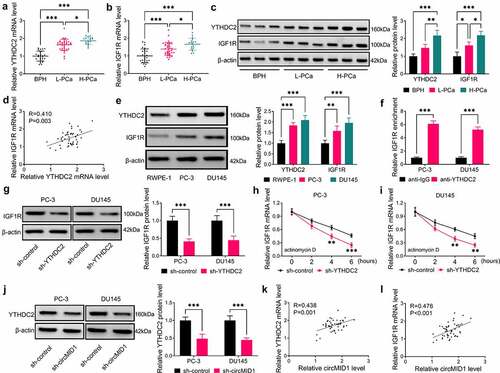
In this, we measured YTHDC2 and IGF1R expression and found that the mRNA and protein expression levels of YTHDC2 and IGF1R were significantly upregulated in L-PCa and H-PCa tissues and were much higher in H-PCa tissues than those in L-PCa tissues (). Further analysis revealed that YTHDC2 expression was positively correlated with IGF1R expression in PCa tissues (). Besides, the protein expression levels of YTHDC2 and IGF1R were also increased in PC-3 and DU145 cells compared to RWPE-1 cells (). To further confirm the interaction between YTHDC2 and IGF1R, we performed RIP analysis, and the results showed that the enrichment of IGF1R in anti-YTHDC2 was significantly higher than that in anti-IgG (). After decreasing the YTHDC2 protein expression using sh-YTHDC2 in PC-3 and DU145 cells (Supplementary Fig. 1A-B), we found that IGF1R protein expression was remarkably decreased by sh-YTHDC2 (), and the mRNA stability of IGF1R mRNA was also significantly reduced under the treatment of ActD (). In addition, our data revealed that circMID1 knockdown significantly inhibited the protein expression of YTHDC2 (), which was consistent with its inhibitory effect on IGF1R expression. Correlation analysis results further suggested that circMID1 expression was positively correlated with YTHDC2 and IGF1R mRNA expression in PCa tissues (). Therefore, our results proposed that circMID1 might positively regulate IGF1R expression by upregulating YTHDC2.
Figure 3. CircMID1 positively regulated IGF1R expression via YTHDC2. (a-b) The expression of YTHDC2 and IGF1R in L-PCa tissues, H-PCa tissues and the prostatectomy tissues of BPH patients was measured by qRT-PCR. (c) The protein expression of YTHDC2 and IGF1R in L-PCa tissues, H-PCa tissues and the prostatectomy tissues of BPH patients was determined using WB analysis. Three lanes represent three different tissue samples. (d) Pearson correlation analysis was used to assess the correlation between YTHDC2 and IGF1R in PCa tissues. (e) WB analysis was used to detect the protein expression of YTHDC2 and IGF1R in RWPE-1, PC-3 and DU145 cells. (f) RIP assay was used to confirm the interaction between YTHDC2 and IGF1R. (g) After transfected with sh-control or sh-YTHDC2 into PC-3 and DU145 cells, the protein expression of IGF1R was measured by WB analysis. (h-i) ActD assay was performed to evaluate the effect of sh-YTHDC2 on the stability of IGF1R. (j) WB analysis was used to measure IGF1R protein expression in PC-3 and DU145 cells transfected with sh-control or sh-circMID1. (k-l) Pearson correlation analysis was used to evaluate the correlation between circMID1 and YTHDC2 or IGF1R in PCa tissues. *P < 0.05, **P < 0.01, ***P < 0.001.
Overexpression of YTHDC2 and IGF1R reversed the effects of circMID1 knockdown on PCa progression
Then, the YTHDC2 or IGF1R overexpression vector was constructed, and it was confirmed that YTHDC2 or IGF1R protein expression was markedly increased after transfection (Supplementary Fig. 1A-C). To further confirm that circMID1 mediated PCa progression by regulated YTHDC2/IGF1R axis, sh-circMID1 and pcDNA YTHDC2 or IGF1R overexpression vector were co-transfected into PC-3 and DU145 cells. The results indicated that the suppressive effects of circMID1 knockdown on the EdU+ cell rate, the colony numbers and the cell cycle process in PC-3 and DU145 cells could be overturned by YTHDC2 or IGF1R overexpression (). Also, overexpressed YTHDC2 or IGF1R also reversed the inhibition effects of circMID1 silencing on the wound healing ratio, the invaded cell numbers, glucose uptake, lactate product, and ECAR in PC-3 and DU145 cells (). WB analysis results suggested that the negative regulation of circMID1 knockdown on the protein levels of IGF1R, p-AKT/AKT, cyclin D1 and GLUT1 in PC-3 and DU145 cells also could be abolished by the overexpression of YTHDC2 or IGF1R (). These data illuminated that circMID1 might promote PCa progression by upregulating the YTHDC2/IGF1R axis.
Figure 4. Overexpression of YTHDC2 and IGF1R reversed the effects of circMID1 knockdown on PCa progression. PC-3 and DU145 cells were transfected with sh-control + pcDNA, sh-circMID1 + pcDNA, sh-circMID1 + YTHDC2 or sh-circMID1 + IGF1R. Cell proliferation was determined using EdU staining (a), colony formation assay (b) and flow cytometry (c-d). Cell migration and invasion were measured by wound healing assay (e) and transwell assay (f). Cell glycolysis was evaluated by detecting glucose uptake (g), lactate product (h) and ECAR (i-j). (k-l) WB analysis was performed to test the protein levels of IGF1R, p-AKT/AKT, cyclin D1 and GLUT1. *P < 0.05, **P < 0.01, ***P < 0.001.
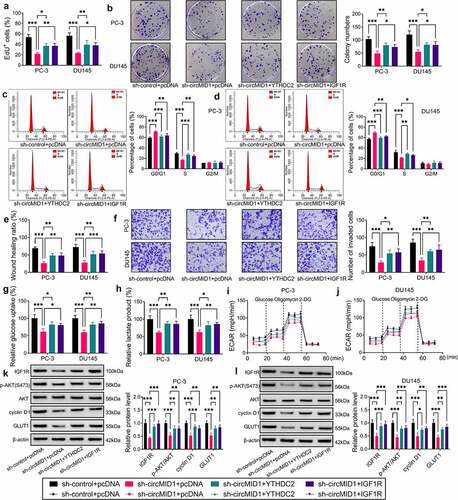
CircMID1 interacted with miR-330-3p to modulate YTHDC2/IGF1R axis
Using the RIP assay, we found that circMID1 could combine with Ago2, while its knockdown could reduce the enrichment level of circMID1 in Ago2 (). To confirm that the miRNA could interact with circMID1, YTHDC2 and IGF1R, we performed bioinformatics analysis using Circinteractome and starBase software. The Venn diagram showed that a total of three miRNAs could interact with circMID1, YTHDC2 and IGF1R (). RNA pull-down assays showed that only miR-330-3p could be pulled down by the circMID1 probe (), so miR-330-3p was selected as a candidate miRNA for circMID1. The binding sites between miR-330-3p and circMID1, YTHDC2 or IGF1R are shown in , where miR-330-3p and IGF1R shared two binding sites. After confirming that the miR-330-3p mimic indeed increased miR-330-3p expression (Supplementary Fig. 1D), we performed the dual-luciferase reporter assay. The results suggested that miR-330-3p mimic could reduce the luciferase activities of circMID1-wt, YTHDC2 3ʹUTR-wt, IGF1R 3ʹUTR-wt1 and IGF1R 3ʹUTR-wt2 vectors, while having no effect on that of the corresponding mut vectors (). Moreover, circMID1, YTHDC2 and IGF1R could also be significantly enriched in the bio-miR-330-3p-wt probe rather than the bio-miR-330-3p-mut probe (). After being transfected with miR-330-3p mimic or inhibitor into PC-3 and DU145 cells, we confirmed that miR-330-3p expression was markedly increased or decreased, respectively (Supplementary ). By detecting YTHDC2 and IGF1R protein expression, we discovered that YTHDC2 and IGF1R protein expression levels could be reduced by miR-330-3p overexpression, while enhanced by miR-330-3p inhibition (Supplementary ). Furthermore, we analyzed the correlation between miR-330-3p and YTHDC2 or IGF1R in PCa tissues. The results showed that miR-330-3p expression was negatively correlated with YTHDC2 or IGF1R expression in PCa tissues (Supplementary Fig. 2A-B). In addition, miR-330-3p inhibitor could also reverse the decreasing effect of circMID1 knockdown on the protein levels of YTHDC2 and IGF1R in PC-3 and DU145 cells (). These results revealed that circMID1 sponged miR-330-3p to positively regulate the YTHDC2/IGF1R axis.
Figure 5. CircMID1 interacted with miR-330-3p to modulate YTHDC2/IGF1R axis. (a-b) RIP assay was used to confirm that circMID1 could combine with Ago2. (c) Venn diagram showed the targeted miRNAs that could interact with circMID1, YTHDC2 and IGF1R. (d-e) RNA pull-down assay was used to confirm the interaction between circMID1 and candidate miRNAs. (f) The binding sites and mutate sites between miR-330-3p and circMID1, YTHDC2 or IGF1R were exhibited. Dual-luciferase reporter assay (g-j) and RNA pull-down assay (k-l) were used to confirm the interaction between miR-330-3p and circMID1, YTHDC2 or IGF1R. (m-o) The protein levels of YTHDC2 and IGF1R were measured by WB analysis in PC-3 and DU145 cells transfected with sh-control + anti-NC, sh-circMID1 + anti-NC, or sh-circMID1 + anti-miR-330-3p. *P < 0.05, **P < 0.01, ***P < 0.001.
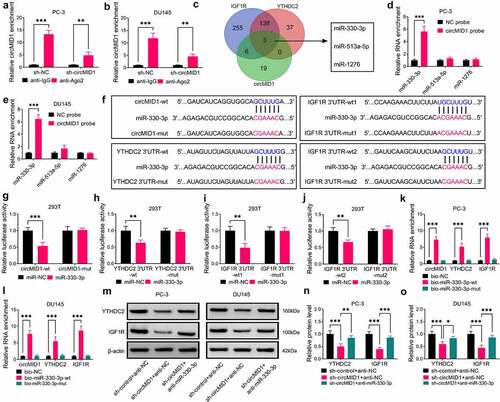
CircMID1 promoted PCa progression via regulating the miR-330-3p/YTHDC2/IGF1R/AKT axis
After that, sh-circMID1, anti-miR-330-3p and YTHDC2 overexpression vectors were co-transfected into PC-3 and DU145 cells to perform the rescue experiments. Function experiments results showed that the miR-330-3p inhibitor partially reversed the negative regulation of circMID1 silencing on the EdU+ cell rate, the colony numbers, the cell cycle process, the wound healing ratio, the invaded cell numbers, glucose uptake, lactate product and ECAR in PC-3 and DU145 cells (). In addition, the combined action of anti-miR-330-3p and YTHDC2 overexpression vectors could completely reverse the effect of circMID1 silencing on PCa cell proliferation, migration, invasion and glycolysis (). Also, the decreasing effect of circMID1 knockdown on the protein levels of IGF1R, p-AKT/AKT, cyclin D1 and GLUT1 in PC-3 and DU145 cells also could be partially reversed by miR-330-3p inhibitor and completely reversed by miR-330-3p inhibitor + YTHDC2 overexpression vector (). Therefore, we confirmed that circMID1 mediated PCa progression by regulating the YTHDC2/IGF1R/AKT axis through sponging miR-330-3p.
Figure 6. CircMID1 promoted PCa progression via regulating the miR-330-3p/YTHDC2/IGF1R axis. PC-3 and DU145 cells were transfected with sh-control + anti-NC + pcDNA, sh-circMID1 + anti-NC + pcDNA, sh-circMID1 + anti-miR-330-3p + pcDNA or sh-circMID1 + anti-miR-330-3p + YTHDC2. EdU staining (a), colony formation assay (b) and flow cytometry (c-d) were performed to detect cell proliferation. Cell migration and invasion were determined using wound healing assay (e) and transwell assay (f). Glucose uptake (g), lactate product (h) and ECAR (i-j) were measured to assess cell glycolysis. (k-l) The protein levels of IGF1R, p-AKT/AKT, cyclin D1 and GLUT1 were examined by WB analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
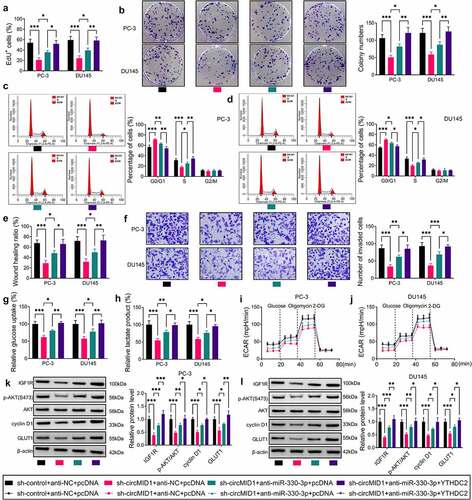
CircMID1 interference inhibited PCa tumor growth in vivo
To further verify the role of circMID1 in PCa progression, the xenograft tumor was constructed using PC-3 cells transfected with sh-control or sh-circMID1. Our data suggested that the tumor volume and weight were markedly reduced in the sh-circMID1 group compared to the sh-control group (). The detection of circMID1 expression confirmed that circMID1 expression was indeed decreased in the sh-circMID1 group (). shows the H&E staining results and IHC staining results. IHC staining showed that compared to the sh-control group, YTHDC2 and IGF1R positive cells were significantly reduced in the sh-circMID1 group (). Also, the protein levels of YTHDC2, IGF1R, p-AKT/AKT, cyclin D1 and GLUT1 were obviously suppressed in the sh-circMID1 group (). All data confirmed that circMID1 played a pro-tumor role in PCa.
Figure 7. CircMID1 interference inhibited PCa tumor growth in vivo. PC-3 cells transfected with sh-control or sh-circMID1 were injected into nude mice. (a) Tumor volume was measured every 4 days starting from day 3. (b) Tumor weight in each group was detected after 27 days. (c) The circMID1 level in the tumor tissues of each group was tested by qRT-PCR. (d) H&E staining results and IHC staining results were shown in the tumor tissues of each group. (e) WB analysis was used to determine the protein levels of YTHDC2, IGF1R, p-AKT/AKT, cyclin D1 and GLUT1 in the tumor tissues of each group. **P < 0.01, ***P < 0.001.
Discussion
Currently, the role of many circRNAs in the progression of PCa has been clarified. For example, circFOXO3 was uncovered to be overexpressed in PCa tissues, which could enhance cancer cell proliferation and inhibit apoptosis via regulating the miR-29a-3p/SLC25A15 network [Citation27]. On the contrary, circUCK2 had been discovered to suppress PCa proliferation and invasion through upregulating TET1 by sponging miR-767-5p, so it might act as a tumor suppressor in PCa [Citation28]. Here, we selected a circRNA, circMID1, which was highly expressed in H-PCa tissues, to explore its role in the progression of PCa. Through experimental verification, we confirmed that circMID1 did have a circular structure. Loss-of-function experiments revealed that circMID1 knockdown had an inhibition effect on PCa cell proliferation, migration, invasion and glycolysis. Additionally, our data also indicated that downregulated circMID1 reduced PCa tumorigenesis in vivo. These data confirmed that circMID1 acted as a tumor promoter in PCa.
Many studies have pointed out that IGF1R, as an oncogene, is involved in the regulation of cancer malignant progression, and its mediated AKT signaling pathway is activated in many cancers [Citation29,Citation30]. In this, we found that circMID1 could decrease IGF1R expression and p-AKT/AKT expression, showing that circMID1 could activate the IGF1R/AKT axis. Previous studies have shown that the differential expression of YTHDC2 mediates the malignant progression of a variety of cancers [Citation31,Citation32]. Overexpressed YTHDC2 was discovered to promote the migration and invasion of colon cancer [Citation33] and was confirmed to be positively correlated with the Gleason grades of PCa [Citation23]. In this, we determined that the expression of YTHDC2 in H-PCa tissues was significantly higher than that in L-PCa tissues and found that its expression was positively regulated by circMID1. Importantly, our study indicated that YTHDC2 could positively regulate IGF1R protein expression, which was consistent with previous research results [Citation22]. The reversal experimental results further suggested that circMID1 accelerated PCa proliferation, migration, invasion and glycolysis through the YTHDC2/IGF1R/AKT axis. This is a groundbreaking discovery for us.
The presentation of the circRNA/miRNA/mRNA axis shows that circRNA can act as a miRNA sponge to mediate downstream gene expression [Citation9,Citation10]. To search for the miRNA that could be sponged by circMID1 and could target YTHDC2 and IGF1R, we performed bioinformatics analysis and confirmed that miR-330-3p could interact with circMID1, YTHDC2 and IGF1R. MiR-330-3p played an anti-tumor function in many cancers, such as liver cancer [Citation34], colorectal cancer [Citation35] and gastric cancer [Citation36]. In PCa-related studies, miR-330-3p was confirmed to be downregulated in PCa tissues [Citation37], and Li et al. reported that circ-0016068 could accelerate PCa cell growth and metastasis via decreasing miR-330-3p [Citation38]. Here, our results revealed that the miR-330-3p inhibitor partially reversed the suppressive effect of circMID1 knockdown on PCa progression and IGF1R/AKT expression, and pcDNA YTHDC2 overexpression vector completely reversed the regulation of sh-circMID1-mediated PCa progression and IGF1R/AKT expression. These data confirmed that circMID1 regulated PCa progression by regulating the YTHDC2/IGF1R/AKT axis through sponging miR-330-3p.
Conclusions
In summary, our study revealed a novel circRNA that regulated PCa progression. Our research showed that circMID1 facilitated PCa proliferation, migration, invasion and glycolysis to promote cancer progression, which was achieved by regulating the miR-330-3p/YTHDC2/IGF1R/AKT axis. Our findings provided a new potential therapeutic target for PCa, suggesting that targeted inhibition of circMID1 might be an effective approach for treating PCa.
Highlights:
1. CircMID1 silencing inhibits PCa cell progression and tumorigenesis.
2. CircMID1 positively regulates YTHDC2/IGF1R/AKT axis.
3. CircMID1 sponges miR-330-3p.
4. MiR-330-3p targets YTHDC2 and IGF1R.
Supplemental Material
Download Zip (5.8 MB)Acknowledgements
None
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Grozescu T, Popa F. Prostate cancer between prognosis and adequate/proper therapy. J Med Life. 2017;10(1):5–12.
- Rebbeck TR. Prostate cancer genetics: variation by race, ethnicity, and geography. Semin Radiat Oncol. 2017;27(1):3–10.
- Chang AJ, Autio KA, Roach M, et al. High-risk prostate cancer-classification and therapy. Nat Rev Clin Oncol. 2014;11(6):308–323.
- Wang G, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. Genes Dev. 2018;32(17–18):1105–1140.
- Komura K, Sweeney CJ, Inamoto T, et al. Current treatment strategies for advanced prostate cancer. Int J Urol. 2018;25(3):220–231.
- Holm HV, Dahl AA, Klepp OH, et al. Modern treatment of metastatic prostate cancer. Tidsskr Nor Laegeforen. 2017;137(11):803–805.
- Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics. 2020;112(2):1680–1685.
- Zhang HD, Jiang L-H, Sun D-W, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7.
- Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301.
- Shen Z, Zhou L, Zhang C, et al. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101.
- Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121(3):2118–2126.
- Shi J, Liu C, Chen C, et al. Circular RNA circMBOAT2 promotes prostate cancer progression via a miR-1271-5p/mTOR axis. Aging (Albany NY). 2020;12(13):13255–13280.
- Han Y, Wen X, Li X, et al. Circular RNA hsa_circ_0075542 acts as a sponge for microRNA-1197 to suppress malignant characteristics and promote apoptosis in prostate cancer cells. Bioengineered. 2021;12(1):5620–5631.
- Luo GC, Chen L, Fang J, et al. Hsa_circ_0030586 promotes epithelial-mesenchymal transition in prostate cancer via PI3K-AKT signaling. Bioengineered. 2021;12(2):11089–11107.
- Li J, Choi E, Yu H, et al. Structural basis of the activation of type 1 insulin-like growth factor receptor. Nat Commun. 2019;10(1):4567.
- Tognon CE, Sorensen PH. Targeting the insulin-like growth factor 1 receptor (IGF1R) signaling pathway for cancer therapy. Expert Opin Ther Targets. 2012;16(1):33–48.
- Xiong W, Huang C, Deng H, et al. Oncogenic non-coding RNA NEAT1 promotes the prostate cancer cell growth through the SRC3/IGF1R/AKT pathway. Int J Biochem Cell Biol. 2018;94:125–132.
- Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–1355.
- He JJ, Li Z, Rong ZX, et al. m(6)A Reader YTHDC2 promotes radiotherapy resistance of nasopharyngeal carcinoma via activating IGF1R/AKT/S6 signaling axis. Front Oncol. 2020;10:1166.
- Wu Q, Xie X, Huang Y, et al. N6-methyladenosine RNA methylation regulators contribute to the progression of prostate cancer. J Cancer. 2021;12(3):682–692.
- Gao C, Wen Y, Jiang F, et al. Circular RNA circ_0008274 upregulates granulin to promote the progression of hepatocellular carcinoma via sponging microRNA −140-3p. Bioengineered. 2021;12(1):1890–1901.
- Ji F, Du R, Chen T, et al. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 Axis. Mol Ther Nucleic Acids. 2020;19:413–420.
- Li J, Fan R, Xiao H. Circ_ZFR contributes to the paclitaxel resistance and progression of non-small cell lung cancer by upregulating KPNA4 through sponging miR-195-5p. Cancer Cell Int. 2021;21(1):15.
- Kong Z, Wan X, Lu Y, et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J Cell Mol Med. 2020;24(1):799–813.
- Xiang Z, Xu C, Wu G, et al. CircRNA-UCK2 Increased TET1 inhibits proliferation and invasion of prostate cancer cells via sponge MiRNA-767-5p. Open Med (Wars). 2019;14(1):833–842.
- Du P, Liu F, Liu Y, et al. Linc00210 enhances the malignancy of thyroid cancer cells by modulating miR-195-5p/IGF1R/Akt axis. J Cell Physiol. 2020;235(2):1001–1012.
- Li XX, Huang L-Y, Peng -J-J, et al. Klotho suppresses growth and invasion of colon cancer cells through inhibition of IGF1R-mediated PI3K/AKT pathway. Int J Oncol. 2014;45(2):611–618.
- Li Y, Zheng J-N, Wang E-H, et al. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ. 2020;8:e10385.
- Zhuang J, Lin C, Ye J. m(6) A RNA methylation regulators contribute to malignant progression in rectal cancer. J Cell Physiol. 2020;235(9):6300–6306.
- Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett. 2016;376(1):34–42.
- Jin Z, Jia B, Tan L, et al. miR-330-3p suppresses liver cancer cell migration by targeting MAP2K1. Oncol Lett. 2019;18(1):314–320.
- Huang Y, Sun H, Ma X, et al. HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 2020;254:117180.
- Guan A, Wang H, Li X, et al. MiR-330-3p inhibits gastric cancer progression through targeting MSI1. Am J Transl Res. 2016;8(11):4802–4811.
- Wu YP, Lin X-D, Chen S-H, et al. Identification of prostate cancer-related circular RNA through bioinformatics analysis. Front Genet. 2020;11:892.
- Li Q, Wang W, Zhang M, et al. Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:827.
