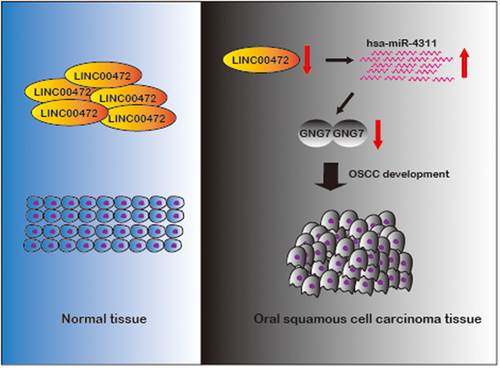
ABSTRACT
Emerging studies indicate that long non-coding RNAs play important roles in oral squamous cell carcinoma (OSCC). However, the function of the majority of long non-coding RNAs is still unclear. Recently, LINC00472 has been reported to play crucial roles in multiple cancers. However, the role of LINC00472 in oral squamous cell carcinoma (OSCC) is still not clear. This study found that LncRNA LINC00472 was significantly down-regulated in several squamous cell carcinoma cancer tissues and OSCC cell lines. Over-expression of LINC00472 in OSCC cells inhibited OSCC progression and alleviated OSCC immune responses. Additionally, we confirmed that LINC00472 functioned as an hsa-miR-4311 sponge and regulated the expression of GNG7 (guanine nucleotide-binding protein, gamma 7). Also, we found that LINC00472 over-expression could suppress xenograft tumor growth in vivo. Our study provides evidence that LINC00472 plays an essential role in inhibiting oral squamous cell carcinoma progression and affecting immune responses by directly binding to hsa-miR-4311 to regulate the expression of GNG7 positively.
Graphical Abstract
KEYWORDS:
Introduction
Oral squamous cell carcinoma (OSCC), one of the most prevalent malignancies across the world, is a subtype of head and neck squamous cell carcinoma (HNSCC) [Citation1,Citation2]. About ninety percent of HNSCC are squamous cell carcinomas developing in mucosal surfaces of the oral cavity and throat [Citation3]. OSCC is the most common HNSCC subtype with increasing disease incidence during recent years [Citation4]. OSCC might be caused by exposure to environmental factors, including tobacco, alcohol, and the most common risk, viruses infection, such as human papillomavirus (HPV)-16 and HPV-18 [Citation5–8]. OSCC lesions caused by multiple environmental factors accumulate genetic alternations, such as chromosomal alternations, genome mutations, and epigenetic alternations. However, a lack of applicable OSCC markers for early diagnosis and understanding underlying molecular mechanisms during cancer development result in poor OSCC treatment outcomes.
Long non-coding RNAs (lncRNAs) are RNA transcripts > 200nt without protein-coding potential, which are uniquely expressed and playimportant roles in specific tissues and cancer types [Citation9–11]. Accumulating evidence suggests that the aberrant expression of lncRNAs in particular cancer types may regulate the cancer progression by acting as a sponge for miRNA inhibition, affecting chromatin methylation, and modulating protein stability [Citation12–14]. For instance, long non-coding RNA 00958, TTN-AS1, and LINC01207 were reported to be involved in regulating OSCC progression [Citation15–17]. LINC00472 (ENSG00000233237), which is also called intergenic non-protein coding RNA 472, located at chromosome 6: 71,343,427–71,420,745, is reported to be involved in the development of multiple cancers, including non-small cell lung cancer, lung adenocarcinoma, osteosarcoma, hepatocellular carcinoma, breast cancer, and bladder carcinoma [Citation18–24]. However, the role of LINC00472 in oral squamous cell carcinoma (OSCC) is still not clear.
Therefore, we hypothesized that LncRNA LINC00472 might be involved in the progression of OSCC in a ceRNA manner. This study aimed to study the role of LINC00472 in OSCC progression using OSCC cell lines and xenografted mice. Here, we found that LINC00472 was down-regulated in OSCC tissues and then found LINC00472 regulated OSCC through the hsa-miR-4311/GNG7 axis. The goal of this study is to identify LINC00472 as an OSCC biomarker and regulator, which may serve as the potential target for the development of OSCC drugs.
Materials and methods
Cell culture and transfection
Normal human oral keratinocytes NHOK cells and 6 human OSCC cancer cell lines, CAL27, FADU, HN12, HSU3, SCC9, and SCC25, were obtained from The Cell Bank Type Culture Collection of the Chinese Academy of Sciences (CBTCCCA, China). All the cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ThermoFisher Scientific, USA) with 10% fetal bovine serum (10100147, ThermoFisher Scientific, USA) and 1% penicillin-streptomycin antibiotics (15140122, ThermoFisher Scientific, USA). All the cell lines were cultured in a 37°C incubator with 5% CO2.
FADU cells were cultured at 1 × 106 density overnight for the cell transfection experiments and transfected with pcDNA3, pcDNA3-LINC00472, hsa-miR-4311 mimics, hsa-miR-4311 inhibitor, pcDNA-GNG7 expression plasmids, and the corresponding control plasmids using Lipofectamine 2000 (11668019, ThermoFisher Scientific, USA).
RNA extraction and quantitative PCR
Total RNA of cells was extracted using TRIzol reagent (15596026, ThermoFisher Scientific, USA) and reverse transcripted using HiScript II 1st Strand cDNA Synthesis Kit (R212-01, Vazyme, China). RT-PCR was performed with SYBR Premix ExTaqTM (RR030A, TaKaRa, China). Relative expression of specific genes was determined using the comparative Ct method [Citation25,Citation26]. To detect hsa-miR-6814-5p, hsa-miR-6868-3p, hsa-miR-660-3p, hsa-miR-4311, and hsa-miR-4496, we performed TaqMan™ MicroRNA Assay (ThermoFisher Scientific, USA).
MTT (3- [4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay
FADU cells transfected with hsa-miR-4311 mimics, GNG7 or hsa-miR-4311 mimics+GNG7 were collected after 24 hours. Then transfected FADU cells were seeded into a 96-well plate at 2 × 103 density. MTT solution was prepared at the concentration of 5 mg/ml. 10 μl of MTT solution was added into each well for 4 hours at 37°C. DMSO dissolved formazan precipitation was measured at 570 nm. We conducted the MTT assay to analyze the proliferation of FADU cells after transfection at the time points of 0 h, 24 h, 48 h, 72 h, and 96 h.
Transwell assays with or without matrigel
FADU cells were collected 24 hours after transfection. 2 × 105 FADU cells suspended in 200 μl of serum-free medium were seeded into the upper compartment of the chamber, and 800 μl of medium with 20% FBS was added to the lower compartment of the chamber (MCEP24H48, Millipore, USA). For the invasion test, the upper chamber was pre-coated with Matrigel Matrix (BD Biosciences, USA). After incubation for 24 hours, the cells were stained with a 0.4% crystal violet solution.
Western blot
Cells cultured in a 6-well plate were collected and lysed with RIPA lysis buffer (P0013B, Beyotime, China). The primary antibodies used in this study were shown as following: CXCL9 (1:200, LS-B13342, LifeSpan BioSciences, USA); CXCL10 (1:100, LS-C104402, LifeSpan BioSciences, USA); GNG7 (1:1000, LS-C807997, LifeSpan BioSciences, USA). Pierce™ ECL Western blotting Substrate was used for the visualization of the blots.
Luciferase activity assay
Luciferase reporter vectors were constructed as illustrated in . FADU cells were seeded in a 24-well plate at 1 × 105 density and cultured for 24 hours. Cells were co-transfected with luciferase reporter vector, miRNA mimics, and Renilla luciferase vectors supplied by Dual-Luciferase® Reporter Assay System (E1910, Promega, USA). 48 hours after transfection, Luciferase and Renilla signals were measured using the Dual-Luciferase® Reporter Assay System (E1910, Promega, USA). The luciferase signal fold change calculation was determined after data normalization to the corresponding Renilla luciferase values.
RNA immune-precipitation (RIP)
In the RNA immune-precipitation (RIP) assay, the anti-AGO2 antibody (MA5-23515, ThermoFisher Scientific, USA) was used. To confirm the direct binding between hsa-miR-4311 and LINC00472, the relative expression level of LINC00472 was examined by quantitative PCR.
Xenograft and in vivo metastasis assay
12 BALB/c nude male mice at 5 weeks were purchased from Shanghai SLAC LABORATORY animal co. Ltd, and divided into 2 groups. The LINC00472 over-expression FADU cells (1 × 106/1 μl DMEM) and negative controls were injected into nude mice subcutaneously described in a previous study [Citation27]. The tumors were collected after 4 weeks of inoculation, and the tumor volume was measured. The xenograft experiment was approved by the animal care and use committee. The lung tissues were collected and subjected to hematoxylin and eosin (HE) staining to analyze tumor nodules [Citation28].
Statistical analysis
Graphpad prism 7.0 was used for statistical analysis in this study. Student’s t tests were conducted to determine the significant differences between any two groups of data. Error bars represented standard deviations (SD). Differences were considered significant when *p < 0.05, **p < 0.01, ***p < 0.001. Data from this study were expressed as the means ± SD from at least three independent repeats.
Results
There are accumulating studies suggesting that long non-coding RNAs participate in regulating multiple cancers, including OSCC. Previous studies have shown that abnormal expression of LINC00472 was correlated with several types of cancers. However, the role of LINC00472 in oral squamous cell carcinoma (OSCC) is still not clear. In this study, we studied the role of LINC00472 in OSCC progression using OSCC cell lines and xenografted mice. We found that LncRNA LINC00472 was significantly down-regulated in several squamous cell carcinoma cancer tissues and OSCC cell lines. Using OSCC cells and xenografted mice, our results suggested activating LINC00472/hsa-miR-4311/GNG7 signaling inhibited OSCC development and affected immune responses, and LINC00472/hsa-miR-4311/GNG7 axis could serve as early diagnostic markers and potential therapeutic targets to improve OSCC prognosis.
LncRNA LINC00472 was down-regulated in several squamous cell carcinomas and OSCC cell lines
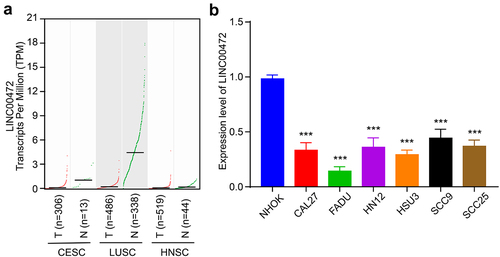
To figure out the role of LINC00472 in squamous cell carcinomas, we conducted LINC00472 expression correlation analysis using GEPIA datasets (Gene Expression Profiling Interactive Analysis, http://gepia2.cancer-pku.cn/#index), including CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), HNSC (head and neck squamous cell carcinoma), and LUSC (lung squamous cell carcinoma) datasets [Citation29]. The results showed that in three types of squamous cell carcinomas, the expression level of LINC00472 was remarkably decreased in the tumor tissue group compared to the normal tissue group (). Next, we examined the expression of LINC00472 in 6 human OSCC cancer cell lines, CAL27, FADU, HN12, HSU3, SCC9, and SCC25, with the normal human oral keratinocytes NHOK cells as control. We found that in all 6 human OSCC cancer cell lines, the LINC00472 level was significantly reduced, especially in the FADU cells (). In summary, the results suggested that LINC00472 was significantly down-regulated in OSCC cells, indicating the potential role of LINC00472 in regulating the progression of OSCC.
Figure 1. LncRNA LINC00472 was down-regulated in several squamous cell carcinomas and OSCC cell lines (a) Transcripts per million (TPM) of LINC00472 detected in CESC, HNSC, and LUSC patients. CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; HNSC, head, and neck squamous cell carcinoma; LUSC, lung squamous cell carcinoma. T represented tumor tissues, and N represented normal tissues. (b) Expression of LINC00472 in normal human oral keratinocytes NHOK cell and 6 human OSCC cancer cell lines, CAL27, FADU, HN12, HSU3, SCC9, and SCC25. Error bars represent standard deviations, n = 3; ***p < 0.001.
LINC00472 over-expression inhibited OSCC progression and affected immune responses in vitro
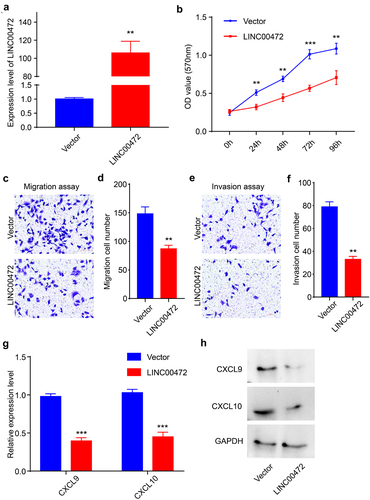
To further explore the role of LINC00472 during the development of OSCC, we selected FADU cells for further functional experiments in vitro. We sub-cloned LINC00472 into the pcDNA3 plasmid and confirmed that the expression level of LINC00472 in FADU cells transfected with LINC00472 plasmids was markedly increased (). First, we conducted an MTT assay to analyze the proliferation of FADU cells after LINC00472 transfection at the time points of 0 h, 24 h, 48 h, 72 h, and 96 h. Over-expression of LINC00472 significantly inhibited the proliferation ability of FADU cells (). Furthermore, we studied the migration and invasion of FADU cells after LINC00472 transfection using Transwell assays. The migration ability of FADU cells after over-expression of LINC00472 was measured by transwell assay without Matrigel, and the results showed that LINC00472 remarkably reduced FADU cells migration (). The results from the transwell assay with Matrigel revealed that LINC00472 also reduced the invasion ability of FADU cells significantly (). In addition, we found that the levels of CXCL9 and CXCL10, the TH1-type chemokines, were decreased after the up-regulation of LINC00472 on both mRNA and protein levels (). The TH1-type chemokines CXCL9 and CXCL10 could be induced by interferon-gamma (IFN-γ) and played essential roles during T-cell trafficking, indicating that LINC00472 may be a mediator during OSCC immune responses. Altogether, the results suggested that LINC00472 inhibited OSCC development and affected immune responses in vitro.
Figure 2. LINC00472 inhibited OSCC progression and affected immune responses in vitro (a) Expression level of LINC00472 after over-expression of LINC00472 quantified using qPCR. (b) Cell proliferation of FADU cells after over-expression of LINC00472 evaluated using MTT assay. (c) The migration ability of FADU cells after over-expression of LINC00472 was measured by transwell experiments (without Matrigel). (d) Quantification result of (C). (e) The invasion ability of FADU cells after over-expression of LINC00472 was measured by transwell experiments (with Matrigel). (f) Quantification result of (E). (g) The expression level of CXCL9 and CXCL10 after over-expression of LINC00472 was quantified using qPCR. (h) CXCL9 and CXCL10 levels after over-expression of LINC00472 were detected using Western blot. Scale bar = 50 μm. Error bars represent standard deviations, n = 3; **p < 0.01, ***p < 0.001.
LINC00472 functioned as the sponge of hsa-miR-4311
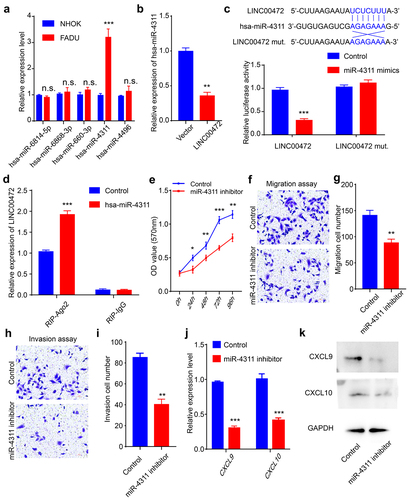
To explore the binding partner of LINC00472, we conducted bioinformatics analysis using LncBase Predicted v.2 and chose the top5 predicted microRNAs for the further experimental test, including hsa-miR-6814-5p, hsa-miR-6868-3p, hsa-miR-660-3p, hsa-miR-4311, and hsa-miR-4496 [Citation30]. By examining the expression levels of the top5 predicted microRNAs, we found that hsa-miR-4311 expression was significantly increased in human OSCC FADU cells compared with normal human oral keratinocytes NHOK cells (). LINC00472 over-expression remarkably decreased the expression level of hsa-miR-4311, indicating that LINC00472 may function as the sponge of hsa-miR-4311 through a ceRNA manner (). Next, we constructed luciferase reporter vectors as illustrated in the upper panel of . The results showed that the relative luciferase activity was significantly reduced in the LINC00472 and hsa-miR-4311 co-transfection group, indicating hsa-miR-4311could directly bind to LINC00472 (). Also, the RIP assay results for AGO2 and IgG showed that hsa-miR-431 could target LINC00472 in the AGO2 dependent manner ().
Figure 3. LINC00472functioned as the sponge of hsa-miR-4311 (a) hsa-miR-4311 was up-regulated in FADU cells. (b) The relative expression level of hsa-miR-4311 after over-expression of LINC00472 was quantified using qPCR. (c) Dual-luciferase assay in FADU cells transfected with either with control or miR-4311 mimics in LINC00472. (d) The correlation of LINC00472 and hsa-miR-4311 was confirmed using RNA immune-precipitation assay with the anti-AGO2 antibody. (e) Cell proliferation of FADU cells after the inhibition of hsa-miR-4311 evaluated using MTT assay. (f) The migration ability of FADU cells after the inhibition of hsa-miR-4311 was measured by transwell experiments (without Matrigel). (g) Quantification result of (F). (h) The invasion ability of FADU cells after the inhibition of hsa-miR-4311 was measured by transwell experiments (with Matrigel). (i) Quantification result of (H). (j) The expression level of CXCL9 and CXCL10 after the inhibition of hsa-miR-4311 was quantified using qPCR. (k) CXCL9 and CXCL10 levels after the inhibition of hsa-miR-4311 were detected using Western blot. Scale bar = 50 μm. Error bars represent standard deviations, n = 3; *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we further explored whether LINC00472 functioned as the sponge of hsa-miR-4311. First, we performed the MTT assay and found that miR-4311 inhibitor could significantly reduce the proliferation of FADU cells (). Also, we utilized transwell assays to confirm that the miR-4311 inhibitor inhibited the migration and invasion of FADU cells (). The expression levels of CXCL9 and CXCL10, the TH1-type chemokines, were significantly decreased after miR-4311 inhibitor transfection (). The results revealed that miR-4311 inhibitor also played an anti-tumor role which is similar to LINC00472. Taken together, the results suggested that LINC00472 functioned as the sponge of hsa-miR-4311.
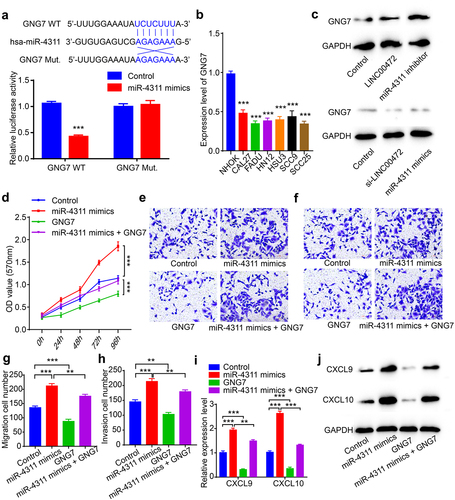
LINC00472 inhibited OSCC progression and affected immune response by regulating the expression of GNG7
To search for the potential protein targets regulated by hsa-miR-4311, we conducted a TargetScan 7.2 bioinformatics analysis [Citation31]. In the gene list given by TargetScan7.2, we screened a potential cancer suppressor gene, guanine nucleotide-binding protein gamma 7 (GNG7). To confirm the correlation between hsa-miR-4311 and GNG7, we conducted a dual-luciferase assay and found that the luciferase activity was significantly decreased in the hsa-miR-4311and GNG7 co-transfection group (). Next, we examined the expression level of GNG7 in several OSCC cancer cell lines and found that GNG7 was remarkably down-regulated in CAL27, FADU, HN12, HSU3, SCC9, and SCC25 cells (). Also, the GNG7 level was negatively correlated with the activity of hsa-miR-4311, indicating that hsa-miR-4311 could regulate the expression of GNG7 (). To verify the function of GNG7, we conducted the MTT assay. The results showed GNG7 reduced the proliferation of OSCC cells, and also inhibited the increased proliferation of miR-4311 mimics transfected OSCC cells (). Next, we utilized transwell assays to confirm whether GNG7 played a role in migration and invasion. We found that GNG7 reduced migration and invasion of FADU cells, and GNG7 partially reversed the cancer-promoting phenotypes induced by miR-4311 mimics (). At last, we test the expression level of CXCL9 and CXCL10, the TH1-type chemokines. The results showed that GNG7 could efficiently decrease the expression of CXCL9 and CXCL10 (). Altogether, our results suggested that LINC00472 inhibited OSCC development and affected immune response by regulating the expression of GNG7.
Figure 4. LINC00472 inhibited OSCC progression and affected immune response by regulating the expression of GNG7 (a) Dual-luciferase assay in FADU cells transfected with either with control or miR-4311 mimics in GNG7. (b) Expression of GNG7 in normal human oral keratinocytes NHOK cell and 6 human OSCC cancer cell lines, CAL27, FADU, HN12, HSU3, SCC9, and SCC25. (c) GNG7 level after over-expression of LINC00472 or down-regulation of hsa-miR-4311; GNG7 level after down-regulation of LINC00472 or up-regulation of hsa-miR-4311. (d) Cell proliferation of FADU cells after up-regulation of hsa-miR-4311, GNG7, or hsa-miR-4311+ GNG7 evaluated using MTT assay. (e) The migration ability of FADU cells after up-regulation of hsa-miR-4311, GNG7, or hsa-miR-4311+ GNG7 measured by transwell experiments (without Matrigel). (f) The invasion ability of FADU cells after up-regulation of hsa-miR-4311, GNG7, or hsa-miR-4311+ GNG7 measured by transwell experiments (with Matrigel). (g) Quantification result of (E). (h) Quantification result of (F). (i) The expression level of CXCL9 and CXCL10 after up-regulation of hsa-miR-4311, GNG7, or hsa-miR-4311+ GNG7 were quantified using qPCR. (j) CXCL9 and CXCL10 levels after up-regulation of hsa-miR-4311, GNG7, or hsa-miR-4311+ GNG7 quantified using Western blot. Scale bar = 50 μm. Error bars represent standard deviations, n = 3; **p < 0.01, ***p < 0.001.
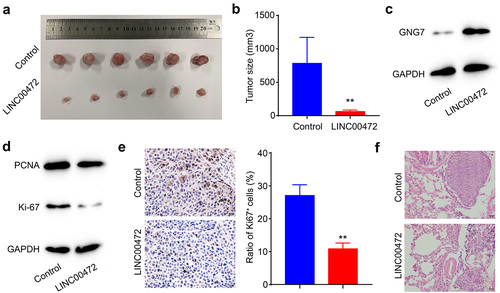
LINC00472 alleviated oral squamous cell carcinoma and inhibited tumor metastasis in vivo
To further determine the effect of over-expression of LINC00472 on tumor growth in vivo, we used a xenograft tumor model. The LINC00472 over-expression FADU cells and negative controls were injected into nude mice subcutaneously. Over-expression of LINC00472 significantly reduced the tumor size of the FADU cells, compared with these of the control group (). Also, we detected that GNG7 was remarkably up-regulated in the LINC00472 group, confirming that GNG7 was the downstream protein effector of LINC00472 (). Next, we examined the levels of two proliferation markers, PCNA and ki-67 in the tumor tissues by Western blot. We found that PCNA and ki-67 levels were significantly decreased in the tumor tissues with up-regulation of LINC00472, indicating that LINC00472 inhibited cell proliferation in vivo (). We then evaluated the protein level of ki-67 in the tumor tissues using immunohistochemistry staining, and results revealed that the ratio of ki-67 positive cells was significantly decreased after overexpression of LINC00472 (). In addition, there were fewer metastatic foci in the lungs of xenografted nude mice in the LINC00472 group than in the negative control group (). In summary, the results suggested that LINC00472 alleviated oral squamous cell carcinoma and inhibited tumor metastasis in vivo.
Figure 5. LINC00472 alleviated oral squamous cell carcinoma and inhibited tumor metastasis in vivo (a) Control or LINC00472-overexpressing FADU cells were injected into the right rear flank of nude mice. The representative pictures of the xenografted tumors. (b) Quantification result of the tumor size of (A). (c) GNG7 level in the OSCC tumor tissues detected by WB. (d) The levels of proliferation markers, PCNA, and ki-67 in the OSCC tumor tissues detected by WB. (e) Ratio of ki-67 positive cells detected by IHC staining. (f) HE staining of metastatic nodules in the lungs of the control or LINC00472-overexpressing group. Scale bar = 20 μm. Error bars represent standard deviations, n = 6; **p < 0.01.
Discussion
The essential roles of lncRNAs in the progression of multiple cancers have been confirmed by emerging studies [Citation32]. The aberrant expression of certain lncRNA is relevant with corresponding cancer progression [Citation33]. Recently, the pathological roles of multiple lncRNAs in the regulation of the progression of OSCC are intensively been identified [Citation34]. For instance, lncRNA Homeobox A11 antisense RNA (HOXA11-AS) promotes proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT), while inhibiting apoptosis of OSCC cells by sponging miR-98-5p to increase YBX2 level [Citation35]. LncRNA CASC9 promotes OSCC progression via inhibiting autophagy-mediated apoptosis through AKT/mTOR pathway [Citation36]. Accumulating evidences suggest that lncRNAs indeed play important roles in the regulation of OSCC development and progression.
LINC00472 was identified to be aberrantly expressed in multiple cancers, including non-small cell lung cancer, lung adenocarcinoma, osteosarcoma, hepatocellular carcinoma, breast cancer, and bladder carcinoma, indicating LINC00472 was correlated with various cancer progression [Citation18–24]. However, the role of LINC00472 in oral squamous cell carcinoma (OSCC) remains unclear. In this study, we found that LINC00472 expression was significantly decreased in CESC, HNSC, and LUSC tissue, and several OSCC cell lines, indicating LINC00472 might play a role in OSCC progression. Next, we further explored the role of LINC00472 in OSCC cells by up-regulating LINC00472 and found that LINC00472 inhibited OSCC progression and alleviated OSCC immune responses in vitro. Also, LINC00472 reduced the levels of TH1-type chemokines CXCL9 and CXCL10, which were induced by interferon-gamma (IFN-γ) and played important roles during T-cell trafficking, indicating that LINC00472 may be a mediator during OSCC immune responses. In addition, we confirmed that LINC00472 significantly reduced the tumor volume of OSCC in vivo using the xenografted OSCC tumors with LINC00472 over-expression.
LncRNAs could interact with microRNAs (miRNAs) to exert their physiological functions [Citation37]. LncRNA competitively binds to particular miRNA to regulate the level of miRNA in a competitive endogenous RNA-related (ceRNA-related) manner, thereby regulating the expression of the downstream protein effector and modulating cancer progression [Citation38]. In our study, we conducted bioinformatics analysis and identified hsa-miR-4311 as the corresponding binding partner of LINC00472 and GNG7 (guanine nucleotide-binding protein, gamma 7) as the downstream protein effector. In addition, we confirmed the roles of hsa-miR-4311 and GNG7 by proliferation, migration, and invasion assays using OSCC cells.
GNG7, a heterotrimeric G protein subunit, is ubiquitously expressed in multiple tissues. However, several studies have found that the expression of GNG7 was decreased in various cancers, including squamous cell carcinoma, esophageal cancer, and pancreatic cancer, indicating the potential tumor suppressor role of GNG7 [Citation39–41]. GNG7 was associated with transmembrane signaling pathways and cell contact-induced growth arrest, thereby inhibiting uncontrolled proliferation in cells [Citation41]. In this study, we found that GNG7 inhibited the progression of GNG7 and was up-regulated in the xenografted tumor tissues with LINC00472 over-expression, suggesting that GNG7 was the downstream protein effector of LINC00472 and hsa-miR-4311. The expression of specific lncRNAs and miRNAs could be considered as the signals of specific cellular states and utilized to identify cellular pathologies such as multiple cancers [Citation42]. LINC00472 and hsa-miR-4311 could serve as early diagnosis markers or potential therapeutic targets to improve OSCC prognosis. Overexpression of LINC00472 could significantly inhibit the progression of OSCC in vitro and in vivo.
However, this study still has some limitations, and numerous questions should be answered in the future. First, for the dissection of the ceRNA mechanism of LINC00472 in vitro, we selected FADU cells because of the low LINC00472 level. Thus, we should further test several other OSCC cell lines in the future. In addition, the role of the LINC00472/hsa-miR-4311/GNG7 axis in OSCC progression should be further tested using patient samples. Hopefully, LINC00472 could serve as an early diagnostical marker and contribute to the therapeutic method.
Conclusions
In conclusion, this study demonstrated that activating LINC00472/hsa-miR-4311/GNG7 signaling inhibited OSCC progression and affected immune responses, and LINC00472/hsa-miR-4311/GNG7 axis could serve as early diagnostic markers and potential therapeutic targets to improve OSCC prognosis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kademani D. Oral Cancer. Mayo Clin Proc. 2007;82(7):878–887.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30.
- Ishida K, Tomita H, Nakashima T, et al. Current mouse models of oral squamous cell carcinoma: genetic and chemically induced models. Oral Oncol. 2017;73:16–20.
- Bagan JV, Scully C. Recent advances in Oral Oncology 2007: epidemiology, aetiopathogenesis, diagnosis and prognostication. Oral Oncol. 2008;44(2):103–108.
- Michaud DS, Langevin SM, Eliot M, et al. High-risk HPV types and head and neck cancer. Int J Cancer. 2014;135(7):1653–1661.
- Stein AP, Saha S, Kraninger JL, et al. Prevalence of Human Papillomavirus in Oropharyngeal Cancer: a Systematic Review. Cancer J. 2015;21(3):138–146.
- Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116–1127.
- Vermorken JB, Trigo J, Hitt R, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol. 2007;25(16):2171–2177.
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504.
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437.
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208.
- Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143.
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669.
- Chen L, Wang W, Cao L, et al. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to regulate cell growth and differentiation in Acute Myeloid Leukemia. Mol Cells. 2016;39:330–336.
- Lu X, Chen L, Li Y, et al. Long non-coding RNA LINC01207 promotes cell proliferation and migration but suppresses apoptosis and autophagy in oral squamous cell carcinoma by the microRNA-1301-3p/lactate dehydrogenase isoform A axis. Bioengineered. 2021;12(1):7780–7793.
- Jin Z, Jiang S. Long non-coding RNA TTN-AS1/microRNA-199a-3p/runt-related transcription factor 1 gene axis regulates the progression of oral squamous cell carcinoma. Bioengineered. 2021;12(1):7724–7736.
- Jiang L, Ge W, Cui Y, et al. The regulation of long non-coding RNA 00958 (LINC00958) for oral squamous cell carcinoma (OSCC) cells death through absent in melanoma 2 (AIM2) depending on microRNA-4306 and Sirtuin1 (SIRT1) in vitro. Bioengineered. 2021;12(1):5085–5098.
- Zou A, Liu X, Mai Z, et al. LINC00472 Acts as a Tumor Suppressor in NSCLC through KLLN-Mediated p53-Signaling Pathway via MicroRNA-149-3p and MicroRNA-4270. Molecular Therapy - Nucleic Acids. 2019;17:563–577.
- Zhang J, Zhang J, Zhang D, et al. Down-regulation of LINC00472 promotes osteosarcoma tumorigenesis by reducing FOXO1 expressions via miR-300. Cancer Cell Int. 2020;20(1):100.
- Su C, Shi K, Cheng X, et al. Long Noncoding RNA LINC00472 Inhibits Proliferation and Promotes Apoptosis of Lung Adenocarcinoma Cells via Regulating miR-24-3p/ DEDD. Technol Cancer Res Treat. 2018;17:1533033818790490.
- Shen Y, Katsaros D, Loo LW, et al. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget. 2015;6(11):8579–8592.
- Li L, Qi F, Wang K. Matrine Restrains Cell Growth and Metastasis by Up-Regulating LINC00472 in Bladder Carcinoma. Cancer Manag Res. 2020;12:7724–7736.
- Deng X, Xiong W, Jiang X, et al. LncRNA LINC00472 regulates cell stiffness and inhibits the migration and invasion of lung adenocarcinoma by binding to YBX1. Cell Death Dis. 2020;11(11):945.
- Chen C, Zheng Q, Kang W, et al. Long non-coding RNA LINC00472 suppresses hepatocellular carcinoma cell proliferation, migration and invasion through miR-93-5p/PDCD4 pathway. Clin Res Hepatol Gastroenterol. 2019;43(4):436–445.
- Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2–3):95–125.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Chai J, Du L, Ju J, et al. Overexpression of KAI1/CD82 suppresses in vitro cell growth, migration, invasion and xenograft growth in oral cancer. Mol Med Rep. 2017;15(4):1527–1532.
- Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26(3):513–523.
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–w60.
- Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D8.
- Agarwal V, Bell GW, Nam J-W, et al. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–463.
- Du Z, Fei T, Verhaak RGW, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913.
- Zhang L, Meng X, Zhu XW, et al. Long non-coding RNAs in Oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol Cancer. 2019;18(1):102.
- Niu X, Yang B, Liu F, et al. LncRNA HOXA11-AS promotes OSCC progression by sponging miR-98-5p to upregulate YBX2 expression. Biomed Pharmacother. 2020;121:109623.
- Yang Y, Chen D, Liu H, et al. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10(2):41.
- Klingenberg M, Matsuda A, Diederichs S, et al. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA Hypothesis: the Rosetta Stone of a Hidden RNA Language? Cell. 2011;146(3):353–358.
- Shibata K, Mori M, Tanaka S, et al. Identification and cloning of human G-protein gamma 7, down-regulated in pancreatic cancer. Biochem Biophys Res Commun. 1998;246(1):205–209.
- Ohta M, Mimori K, Fukuyoshi Y, et al. Clinical significance of the reduced expression of G protein gamma 7 (GNG7) in oesophageal cancer. Br J Cancer. 2008;98(2):410–417.
- Hartmann S, Szaumkessel M, Salaverria I, et al. Loss of protein expression and recurrent DNA hypermethylation of the GNG7 gene in squamous cell carcinoma of the head and neck. J Appl Genet. 2012;53(2):167–174.
- Wang Kevin C, Chang Howard Y. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. 2011;43(6):904–914.
