ABSTRACT
Biofilm formation on biotic or abiotic surfaces is caused by microbial cells of a single or heterogeneous species. Biofilm protects microbes from stressful environmental conditions, toxic action of chemicals, and antimicrobial substances. Quorum sensing (QS) is the generation of autoinducers (AIs) by bacteria in a biofilm to communicate with one other. QS is responsible for the growth of biofilm, synthesis of exopolysaccharides (EPS), and bioremediation of environmental pollutants. EPS is used for wastewater treatment due to its three-dimensional matrix which is composed of proteins, polysaccharides, humic-like substances, and nucleic acids. Autoinducers mediate significantly the degradation of environmental pollutants. Acyl-homoserine lactone (AHL) producing bacteria as well as quorum quenching enzyme or bacteria can effectively improve the performance of wastewater treatment. Biofilms-based reactors due to their economic and ecofriendly nature are used for the treatment of industrial wastewaters. Electrodes coated with electro-active biofilm (EAB) which are obtained from sewage sludge, activated sludge, or industrial and domestic effluents are getting popularity in bioremediation. Microbial fuel cells are involved in wastewater treatment and production of energy from wastewater. Synthetic biological systems such as genome editing by CRISPR-Cas can be used for the advanced bioremediation process through modification of metabolic pathways in quorum sensing within microbial communities. This narrative review discusses the impacts of QS regulatory approaches on biofilm formation, extracellular polymeric substance synthesis, and role of microbial community in bioremediation of pollutants from industrial effluents.
Graphical Abstract
1. Introduction
Biofilm is an aggregation of single or multiple microbes that are adhered to biotic or abiotic surfaces irreversibly and are covered with a self-produced extracellular polymeric substance (EPS) [Citation1]. The term ‘biofilm’ was introduced by Costerton et al. in 1978 [Citation2]. The development of biofilm includes several steps such as adherence of microbiome to the surface, synthesis of EPS, the interaction between microbes through signaling molecules, and dissemination of microbial cells into planktonic form [Citation3]. Biofilm protects microbes from the stressful environmental conditions, toxic action of chemicals, and antimicrobial substances [Citation4]. The microbial cell population in biofilm ranges from 108 to 1011 per g wet weight [Citation5]. Biofilm formation by bacteria can occur in natural or anthropogenic environments. EPS of biofilm can hinder the action of pesticides, hydrocarbons, and heavy metals which are present in the close environment of biofilm [Citation6]. Quorum sensing (QS) is defined as a communication procedure between bacterial cells in biofilm through chemical mediators which are known as autoinducers (AIs). The concentration of AI determines the expression of genes that control the population of a microbial cell [Citation7]. QS contributes significantly to the evolution of biofilm and secretion of exopolysaccharides [Citation8]. In gram-negative bacteria, the QS system is LuxI/LuxR type whereas QS in gram-positive bacteria is oligopeptide/two-component-type sensor histidine kinases [Citation9]. Gram-negative bacteria use acylated homoserine lactone (AHL) as an auto-inducer whereas gram-positive bacteria use autoinducer peptide (AIP). Autoinducer-2 (AI-2) is used by both gram-negative and gram-positive bacteria in QS systems. Ligands such as AI-1, AI-2, AHL, and AIP-1 bind with their respective receptors such as LuxN, LuxP, LuxR, and AgrC to determine communication between bacterial species at the molecular level [Citation10]. Different types of AIs from a variety of bacterial species are involved in environmental bioremediation such as remediation of toxic pollutants in soil and wastewater [Citation11, Citation12]. In biological wastewater treatments, bacterial biofilm is predominated by gram-negative bacteria that use AHLs for communication. In activated sludges of wastewater treatment plants (WWTPs), AHLs-producing QS and AHLs-degrading QQ (quorum quenching) bacteria contribute significantly to controlling the development of biofilm in biological wastewater treatments [Citation13]. Biofilm based environmental bioremediation is more eco-friendly and cost-effective as compared to other chemicals, physical, and thermal approaches. Microbial cells within biofilm provide resistance against the action of xenobiotic substances [Citation14]. Biofilm is used for the treatment of wastewater where it is connected with biological oxygen demand (BOD), ammonia, nitrogen, nitrate, and dissolved O2. Nutrients of wastewater induce the growth of microbes and microbe derived metabolites which are used to remove the contaminants from the wastewater. Biofilm reactors such as biological contact oxidation tank, biological rotating disc, biological aerated filter, biofilm fluidized bed (BFB), moving bed biofilm reactor (MBBR) and integrated fixed-film activated sludge reactor are used to remove pollutants from domestic sewage and a variety of industrial wastewater. Biofilm reactors are capable to remove organic pollutants and nitrogenous substances at a high rate and they produce the least amount of sludge in wastewater treatment [Citation15,Citation16]. Microorganisms in biofilms on the surface of hydrocarbons are preferred for the removal of slowly degraded pollutants due to their high cell density, debilitate pollutants through biosorption, bioaccumulation, and biomineralization [Citation17]. The utilization of biofilm in bioremediation depends on the interaction of microorganisms with xenobiotic substances in the environment. Immobilized cells in the biofilm are involved in the synthesis of cofactors and enzymes which are contributed significantly to bioremediation. Due to horizontal gene transfer (HGT) between the bacterial cells in biofilm, bacterial biofilms are more efficient for enhanced bioremediation as compared to planktonic cells [Citation18].
Bibliometric analysis based on scientific search engines like PubMed revealed the applications of biofilm in bioremediation and wastewater treatments. The objective of this narrative review is to attempt to underline the current stage of characteristics of biofilm, the role of QS in biofilm development and their microbiological niches, and their role in bioremediation and wastewater treatment. The approaches for the characterization of biofilm using available analytical and molecular techniques are also briefly summarized. Role of exo-electrogens in biofilm growth in MFCs and biofilm associated EPS in bioremediation have been highlighted. Furthermore, this review also explores the need for genome editing technology in bioremediation.
2. Characteristics features of biofilm
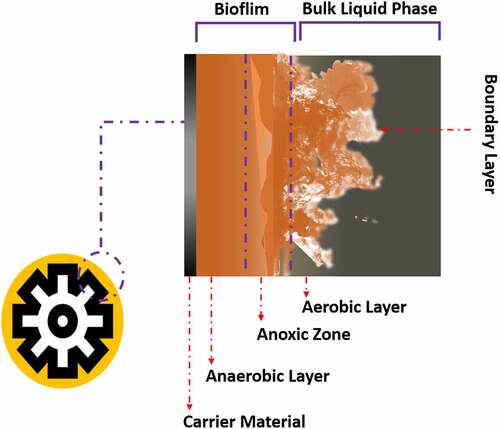
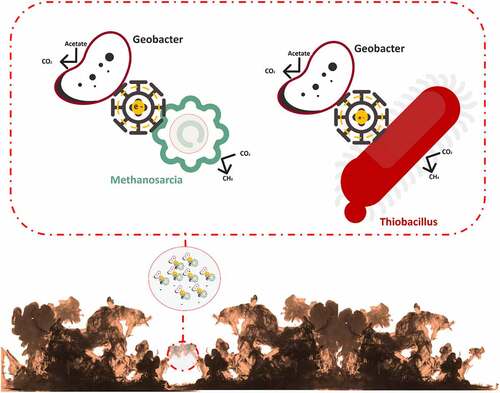
In the natural environment, biofilms formation occurs between species belonging to algae, bacteria, fungi, and protozoa. The matrix of biofilm consists of either water or solvent. Free-living bacteria that do not adhere to surfaces are considered planktonic bacteria. The abundance of bacteria in planktonic cells becomes lower than bacteria in biofilms. Bacteria in biofilms can survive in adverse environmental conditions such as alterations of pH, presence of toxic substances and free radicals, and low amount of nutrient availability. The surfactants of EPS are used in solubilizing non-degradable compounds such as organic pollutants [Citation19–21]. EPS matrix which is 0.2–1.0 mm thick shares 50% of total biofilm whereas microorganisms constitute the rest of the portion of biofilm [Citation22]. Biosurfactants are involved in the manufacture of biofilm [Citation23]. Water channels in a biofilm are involved in the diffusion of antimicrobial substances, oxygen, and nutrients [Citation24]. Exopolysaccharide, cell population, pH, metabolites, oxygen level, and gene expression pattern are varied in different niches of biofilm [Citation10]. The formation of biofilm is a dynamic process [Citation25]. The evolution of biofilm has several steps such as reversible adherence of planktonic bacteria to the biotic or abiotic surface through Van der Waals interactions, irreversible adherence of bacteria through their flagella, fimbriae, and pilli, the proliferation of bacterial cells, secretion of EPS, QS, maturation of biofilm and dissemination of planktonic bacteria [Citation26] (). The water-binding capacity and mobility of the biofilm are responsible for the diffusion within the biofilm matrix which is composed of nutrients, polymers, and metabolites. D-glucuronic, D-galacturonic, and mannuronic acids contribute anionic property of biofilms. The presence of peptidoglycan, phospholipids, proteins, polysaccharides, DNA, and RNA is reported in biofilm [Citation27]. Inorganic and organic substances which are secreted by the microbes provide a favorable condition for the adherence of microbial cells to the substratum [Citation28]. Several saccharolytic enzymes are produced from microbial communities of biofilm. These enzymes are involved in the detachment of microbes from surfaces to colonize in a new location. Autoinducers are involved in the coordination of bacterial species in the biofilm [Citation11]. HGT is responsible for the enhancement of genetic diversity such as genes responsible for the development of antibiotic and heavy-metal resistance and genes involved in the degeneration of environmental pollutants in a biofilm. Microbes in biofilms are communicated with each other through QS which regulates the expression of genes that drive biofilm formation [Citation29]. Syntrophic interaction between bacteria is a well-documented phenomenon in biofilms. It occurs in between fermentative bacteria and methanogenic archaea during anaerobic wastewater treatment. Interspecies electron transfer (IET) is a crucial phase in syntrophic interactions. Syntrophic genera such as Clostridium, Geobacter, Pelobacter, and Smithella have highly abundant genes which are involved in the synthesis of diffusible signal factor (DSF) and c-di-GMP. Type IV pili are produced by c-di-GMP, which also regulates flagellar movement and biofilm matrix components that contribute to biofilm development and dissemination [Citation30] (). Heterotrophic bacteria such as Alcaligenes, Bacillus, Escherichia coli, Nocardias, Pseudomonas, Streptococcus faecalis, Sphaerotilus, Thiobacillus, and Zoogleas predominate in the biofilm [Citation31].
Figure 1. Stages of bacterial biofilm development along with factors involved in controlling biofilm formation.
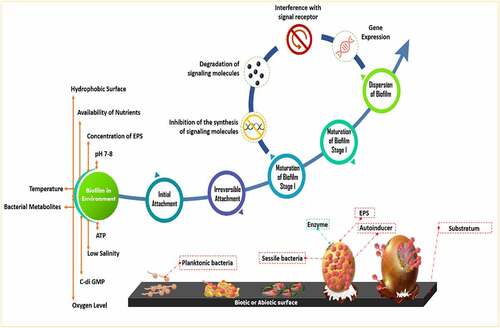
Figure 2. Syntrophic interaction in biofilms between fermentative bacteria and methanogenic archaea during anaerobic wastewater treatment.
2.1 Factors influencing morphology and behavior of biofilm
Environmental factors such as bacterial metabolites, oxygen level, pH, and nutrients determine the formation of biofilm [Citation27]. Hydrophobic surface, low salinity, low temperature, and pH of 7–8 are favorable parameters for biofilm formation [Citation16] (). Dissolved oxygen content, temperature, and nutrients in wastewater are responsible to determine the bacterial abundance in the biofilm. The activity of biofilm is regulated by the level of adenosine triphosphate, dehydrogenase, deoxyribonucleic acid, and solid and volatile solid [Citation16]. The activity of microbial communities in the biofilms is determined by the level of ATP. The aging of biofilm reduced the capability of biofilm in wastewater treatment [Citation32]. pH alters the adhesion property of bacteria through alterations of its surface electrical charges. Repulsion occurs when the distance between the bacteria and the surface is 10–20 nm. EPS prevents microbes from the toxic effect of heavy metal and organic pollutants [Citation33]. Biofilm formation is influenced by the concentration of EPS [Citation34].
Escherichia coli K12 induced biofilm formation is regulated by aerobic granular sludge producing N-acyl-homoserine lactones (AHLs) [Citation35]. The intracellular concentration of c-di-GMP is also involved in the formation of biofilm. A higher concentration of c-di-GMP enhances the adherence of bacteria to the surface and induces the formation of biofilm. Reduced concentration of intracellular c-di-GMP induces the dissemination of biofilm [Citation36]. c-di-GMP regulated biofilm formation contributes significantly to wastewater treatment. Overexpression of c-di-GMP prevents biofilm formation caused by Staphylococcus aureus. Overexpression of the c-di-GMP regulated BalA protein promotes the spread of dissemination of Pseudomonas aeruginosa induced biofilm [Citation37]. The transcription factor RpoS is involved in the formation and maturation of biofilm in Escherichia coli and Pseudomonas aeruginosa [Citation38]. Overexpression of genes that encode adhesion and ribosomal proteins and downregulation of flagellar genes contribute significantly to biofilm formation [Citation39].
2.2 Physiochemical properties of biofilm matrix
The biofilm matrix is also named EPS. EPS regulates the structure and stability of biofilm through the presence of Ca+2. The molecular weight of EPS is 500 to 2000 kDa. Exopolysaccharides contain sucrose-derived glucans, fructans, and cellulose. They are either polyanionic or polycationic. In Pseudomonas aeruginosa, exopolysaccharide such as alginate, Pel, and Psl37 are reported. Alginate contributes significantly in determining the formation of biofilm [Citation40]. Amyloids are predominant components in the biofilms of wastewater treatment plants [Citation41]. Extracellular DNA (eDNA) of the biofilm matrix provides the mechanical strength of biofilms and ensures the transfer of genetic information between bacterial cells [Citation42]. The protein in EPS provides protection to biofilms against environmental stress [Citation43].
3. Role of QS in biofilm development
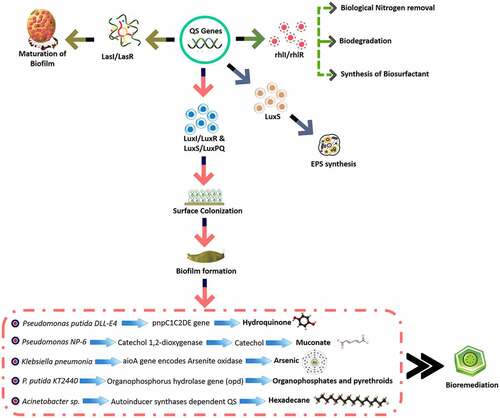
QS depends on the population of microbial cells in the biofilm. QS also regulates the expression of genes that are involved in bioremediation, bioluminescence, production of antibiotics, and virulence factors [Citation44]. AIs are produced by the bacteria as a signaling molecule during QS [Citation45]. Boron-containing QS molecule (AI-2) is considered a common mode of the signal during communication. AI-2-mediated QS has been reported in Deinococcus radiodurans, Escherichia coli, Salmonella typhimurium, and Vibrio cholerae. The marine bacterium Vibrio harveyi uses LuxLM driven AI-1 (N-(3-hydroxybutanoyl)-homoserine lactone) for intraspecies communication and LuxS driven AI-2 (furanosyl borate diester) for interspecies communication [Citation10]. In Streptococcus mutans, comC gene encodes competence-stimulating peptide (CSP) pheromone which induces biofilm formation [Citation46]. Quorum sensing genes such as lasI and rhlI which encode AHL synthase contribute significantly to the growth of biofilm in Pseudomonas aeruginosa N6P6 which are involved in the degradation of polycyclic aromatic hydrocarbon (PAH) [Citation47]. The AHL regulatory locus swr in S. liquefaciens and S. marcescens are involved in the manufacturer of biofilm [Citation6]. In P. aeruginosa, lasI/lasR QS system is involved in the maturation of biofilm whereas Rhl (rhlI/rhlR) QS system is associated with the synthesis of biosurfactant [Citation48].
3.1 QS in gram-negative bacteria
In Gram-negative bacteria, LuxI (autoinducer synthase) gene encodes acyl-homoserine lactone (AHL) autoinducer, and LuxR is the cytoplasmic AI receptor/DNA-binding transcriptional activator. The concentration of AHL enhances due to increased cell density. Due to its lipophilic nature, AHL can easily pass through the cell due to which intraspecies communication, biofilm formation, and EPS synthesis occur. The synthesis of AHL is species specific [Citation49]. Acyl-homoserine lactone (AHL)-based quorum sensing has been reported in Serratia liquefaciens and Rhodobacter sphaeroides [Citation27,Citation50]. Acinetobacter, Aeromonas, Burkholderia cepacian, Chitinimonas, Citrobacter, Enterobacter, Klebsiella, Pseudomonas aeruginosa, Pseudomonas putida, and Serratia liquefaciens use AHLs for communication [Citation51,Citation52]. Homoserine lactone ring is a common feature of all AHLs [Citation53]. Bacteria belong to Proteobacteria are commonly involved in the synthesis of AHLs. LuxI, LuxM, and HdtS protein families are involved in the synthesis of AHLs. The LuxI is mainly involved in the synthesis of AHL synthases. AHL-LuxR protein complex has been activated due to interaction between AHL signals and LuxR receptor and this complex binds to QS promoters to induce the transcription of genes that are regulated by the QS system. In Pseudomonas aeruginosa, AHL signals such as C4-HSL and 3-oxo-C12-HSL interact with RhlR and LasR for QS. Quorum quenching enzymes such as AHL-acylases, AHL-lactonases, and oxidoreductases are involved in the inhibition of AHL activity. Pseudomonas strain PAI-A and Variovorax paradoxus use AHL signals for their development. Anabaena sp. PCC7120, Pseudomonas aeruginosa, and Ralstonia strains are involved in the production of AHL-acylases. The presence of AHL biosensors has been reported in Agrobacterium tumefaciens NT1 plasmid pZLR4, A. tumefaciens A 136 (pCF218) (pCF372), Chromoterbacium violaceum CV026, and E. coli plasmid pSB401 [Citation13,Citation54]. Acidobacteria sp., A. tumefaciens C58, and Bacillus sp. strain 240B1 are involved in the synthesis of AHL-lactonases which are involved in the cleavage of the bond between homoserine lactone moiety and the acyl chain [Citation55].
3.2 QS in gram‑positive bacteria
In gram-positive bacteria, AIP is used as an autoinducer which induces the structural diversity of biofilm. Membrane-associated ATP-binding cassette (ABC) transporter induces the secretion of AIP. The gene agrD encodes autoinducing peptides [Citation56]. The Gram-positive bacterium Exiguobacterium sp is involved in inducing the AHL bioreporters, namely Chromobacterium violaceum CV026, Agrobacterium tumefaceins A136, and E. coli JM 109(psb1075) [Citation57]. Gram-positive bacteria such as Clostridium botulinum, C. perfringens, C. difficile, Enterococcus faecalis, Lactobacillus plantarum, Listeria monocytogenes, and S. aureus are involved in the production of AIPs [Citation18].
3.3 Mode of gene transfers in biofilm
Transformation, transduction, and conjugation are responsible for the transfer of genes among bacterial species in biofilms [Citation27]. Horizontal transfers of plasmids carrying catabolic genes in biofilms modulate bacterial populations to enhance the degradation of xenobiotic compounds [Citation58]. Genes responsible for the degradation of xenobiotic compounds such as PAHs are located in the bacterial plasmids or transposons. Goris et al. reported the effect of a transfer of plasmid pC1 of Delftia acidovorans tagged with a mini-Tn5 transposon encoding the gene for the oxidative deamination of 3-chloroaniline into Pseudomonas putida on activated sludge bacteria. Biofilm showed the highest frequency of gene transfer due to high microbial density [Citation59]. Springael et al. reported transposon mediated 3-chlorobenzoate-degradative genes such as clc-element transfer from P. putida BN210 into other 3-chlorobenzoate mineralizing bacteria. HGT through conjugation and transformation is a common phenomenon in biofilm [Citation60]. HGT of AHLs genes in Novosphingobium enhances the degradation of PAHs and pesticides [Citation52]. Horizontal transfers of catabolic plasmids in biofilms result in a changed microbial population capable of degrading a variety of pollutants. Plasmids in the biofilm are immobile due to the quiescent nature of microbial cells in the biofilm. QS-mediated transformation, conjugative transfer, and prophage induction mechanisms have been reported in B. subtilis, Rhizobium leguminosarum, and Rhodobacter capsulatus. QS-mediated transformation enhances the catabolic potential of bacterial strains. Novosphingobium has aromatic ring hydroxylating dioxygenases and luxR homologous. Multiple mobile genetic elements which are present upstream of the luxR homologous region enhanced the HGT of AHLs genes [Citation18].
4. Electro-active biofilm formation
Electrically potent microorganisms are involved in the formation of Electrochemically active biofilms (EABs) in wastewaters. They are involved in the electrochemical reactions in bioremediation. EABs are employed in bioelectrochemical systems (BESs) such as microbial fuel cells (MFC) and microbial electrolysis cells (MEC). Electrons are transported via pili/nanowires of some EABs. Bio-electrochemical treatment systems (BET) are used for wastewater treatment [Citation61–63]. Metal-reducing bacteria use iron or manganese oxides as electron acceptors for the respiratory mechanism [Citation64]. The development of biofilm in MFCs is determined by the bacterial species, temperature, pH, the pattern of substrate, and composition of electrode. Mixed culture biofilm generates more energy and shows higher potential for electron transfer as compared to pure culture biofilm [Citation65]. The pH factor is involved in the redox potential in MFCs. At neutral pH, microbial enzymes are performed well for biofilm formation [Citation66]. In the anaerobic anode compartment of MFCs, biofilm performs as a biocatalyst to synthesize protons and electrons through hydrolysis of the substrate such as ferricyanide, humic acid, thionin, and methylene blue [Citation67]. In MFCs, microorganisms are involved in the formation of electricity through the transfer of electrons from bacterial cells toward the anode surface [Citation68]. Exo-electrogens are involved in the formation of electro-active biofilms through the transmission of electrons through c-Cyts or pili. The exo-electrogenic potential of MFCs has been reported in β-Proteobacteria (Rhodoferax), γ-Proteobacteria (Shewanella and Pseudomonas), δ-Proteobacteria (Aeromonas, Desulfuromonas, Desulfobulbus, Geobacter, and Geopsychrobacter), Acidobacteria (Geothrix) and Firmicutes (Clostridium). C-type cytochromes (c-Cyts) and conductive pili (nanowires) which are located on the outer membrane-bound of Geobacter and Shewanella species involved in electron transmission. Type IV pili, c-Cyts pili, omcZ, and pilA are involved in the biofilm formation in Aeromonas spp., Geobacter spp., and G. sulfurreducens [Citation65,Citation69] (). Bacteria belonging to phylum Proteobacteria, Acidobacteria, and Firmicutes are involved in the generation of electricity. Aeromonas hydrophila, Clostridium butyricum, Enterococcus gallinarum, Geobacter spp., Pseudomonas aeruginosa, Rhodoferax ferrireducens, Rhodobacter sphaeroide, Shewanella spp., and Shewadella oneidensis MR-1 are involved in the generation of electricity in MFCs. Clostridium beijerinckii, Clostridium butyricum, and Escherichia coli K12 contributed production of hydrogen electrochemically on the anode [Citation65] [Citation70].,reported that the anodic bacterial community in MFCs fed domestic wastewater was overrepresented by Geobacter metallireducens, G. sulfurreducens, G. lovleyi, and G. uraniireducens. Proteobacteria, Firmicutes, and Bacteroidetes were highly abundant on the anode surface [Citation71]. Spirochaetaceae (Spirochaeta spp.) and Methanobacteriaceae showed higher abundance in acetate accustomed biofilm whereas butyrate accustomed biofilm showed a higher abundance of bacterial communities such as Syngistaceae, Geobacteraeceae and Syntrophomonadaceae and Archeal communities such as Methanosarcinaceae [Citation72]. Gram-positive bacteria showed a significant contribution to the EAB formation. Thermincola spp. of thermophilic MFCs is involved in the transmission of electrons through c-type cytochromes of its cell envelope. Thick layer anodic biofilms generate more electrical energy as compared to thin layer anodic biofilm [Citation73].
Table 1. Bacteria involved in electrochemically active biofilms [EABs) formation in wastewater
5. Application of biofilm-associated extracellular polymeric substances (EPS) and biosurfactants in bioremediation
The EPS ensures the binding of a bacterial cell to the surface of xenobiotics substances for the degradation of xenobiotics substances [Citation74–76] [Citation77].,reported that a higher amount of calcium dependent EPS augments P. mendocina NR802 induced degradation of PAHs. EPSs are hydrophobic in nature due to the presence of surfactants and lipids. Biosurfactants such as glycolipids, lipopetides, ionic lipids, and neutral lipids which are nontoxic and biodegradable are used in the enhancement of bioremediation of organic and inorganic pollutants. Biosurfactants are also involved in the development of biofilm through nutrient transport in biofilms via water channels [Citation66, Citation78]. Biosurfactants such as rhamnolipids are reported in the biofilm matrix of P. aeruginosa. rhl QS system is involved in the synthesis of rhamnolipids in this bacterium. Biosurfactants are used in the bioremediation of polycyclic aromatic hydrocarbons (PAHs) [Citation79–82]. The architecture of EPS in bacterial genera such as Aeromonas, Bacillus, Burkholderia, Pseudomonas, and Xanthomonas are involved in bioremediation and wastewater treatment through QS [Citation76]. Bacteria such as Enterobacter cloacear, Gordonia alkanivorans and Halomonas eurihalina synthesize EPS having emulsifiers such as polysaccharides, proteins, and lipids which are involved in the degradation of hydrocarbons [Citation83]. In a biofilm, oxidoreductase enzymes (laccase, polyphenoloxidase, and catalase) and EPS hydrolyase are involved in pyrene breakdown [Citation6]. EPS played significantly in the wastewater treatment and bioremediation in soil [Citation84].
6. Techniques applied for analysis and characterization of biofilm communities
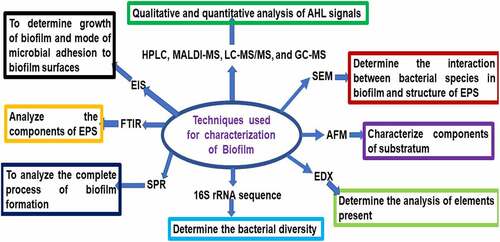
Scanning electron microscope (SEM), confocal laser scanning microscopy (CLSM), and atomic force microscopy (AFM) are used to determine the morphology of mature biofilms such as roughness, topography, and stiffness. AFM is used to characterize components of the substratum in the biofilm. SEM is also used to determine the interaction between bacterial species in biofilm and the structure of EPS. Energy Dispersive X-ray (EDX) spectroscopy is used to determine the analysis of elements present in biofilm. Fourier Transform Infrared (FTIR) spectroscopy is used to analyze the components of EPS in biofilm. Surface Plasmon Resonance (SPR) spectroscopy is used to analyze the complete process of biofilm formation. Electrical Impedance Spectroscopy (EIS) is used to determine the growth of biofilm and mode of microbial adhesion to biofilm surfaces. 16S rRNA sequence analysis is used to determine the bacterial diversity in the biofilm. Crystal violet assay is used to estimate the growth of biofilm [Citation16]. High-performance liquid chromatography (HPLC), MALDI-MS, LC-MS/MS, and GC-MS have been used for qualitative and quantitative analysis of AHL signals [Citation13] (). 16S rRNA gene sequencing of the anodic microbial community in MFCs fed domestic wastewater determined the presence of Geobacter metallireducens, G. sulfurreducens, G. lovleyi and G. uraniireducens [Citation65]. High performance liquid chromatography–mass spectrometry (HPLC–MS) analyses are used to determine AHL in activated sludge [Citation85].
7. Role biofilms in bioremediation of organic compounds
Aerobic bacteria are involved in the biodegradation of polycyclic aromatic hydrocarbons (PAHs) through oxidation of the benzene ring by dioxygenase enzymes into dihydrodiols [Citation86,Citation87]. Several bacteria are involved in the degradation of naphthalene, anthracene, benzo (b) fluoranthene, dibenzo (a,h) and indeno (1,2,3-c,d) pyrene, and phenanthrene (). Biosurfactants such as surfactin, rhamnolipid, and sophorolipid are produced by B. subtilis, P. aeruginosa and Torulopsis bombicola which are involved in the bioremediation of PAH [Citation21, Citation88, Citation89]. Glycolipids biosurfactants and polymeric biosurfactants are involved in the degradation of organic pollutants whereas biosurfactants of ionic lipids and lipopetides are involved in the insulation of pollutants [Citation90–92]. Sphingomonas sp. augments the degradation of phenanthrene in the existence of biosurfactants [Citation93]. Chryseobacterium sp., Ochrobactrum intermedium, Klebsiella oxytoca, Pseudomonas alcaligenes, P. putida, Sphingobacterium sp., and S. maltophilia are involved in the biodegradation of diesel in the presence of rhamnolipid [Citation94]. Synthesis of biosurfactants may be augmented by the genetic modification of QS systems. Sulfate-reducing bacteria belong to Desulfobacteriaceae in a natural biofilm are used in the oxidation–reduction of sulfur [Citation95]. Chlorinated aromatic compounds such as 2, 4-dichlorophenol (DCP) are present in the effluents of the chemical industry. Rotating perforated tube biofilm reactor which contains activated sludge culture enriched with P. putida containing microbial biomass is used to degrade DCP [Citation96]. Microbial exopolymers in biofilms are used in the process of adsorption of chlorinated herbicide such as diclofop-methyl, and methyl 2-[4-(2,4-dichlorophenoxy) phenoxy] pyruvate [Citation97]. Pseudoalteromonas haloplanktis TAC125 is involved in the bioremediation of aromatic compounds in marine environments into catechols [Citation98]. Cycloclasticus zancles 78-ME, C. sp. DSM 27168, C. pugetii PS-1, C. sp. P1, and C. sp. PY97M are involved in the aerobic degradation of naphthalene, phenanthrene, and pyrene which are present in seawater and marine sediments [Citation99]. Dehalococcoide is responsible for removing chlorinated ethenes [Citation100]. Biofilms of Burkholderia sp. NK8 and P. aeruginosa PA01 are involved in the degradation of chlorinated benzoates [Citation101]. Biofilms of Pseudomonas stutzeri T102 having naphthalene-degrading genes are involved in the bioremediation of naphthalene [Citation102]. Pseudomonas sp. AKS2 is also involved in the degradation of low-density polyethylene (LDPE) through the formation of Biofilm on the surface of LDPE [Citation103]. S. marcescens and S. liquefaciens are involved in the degradation of phenanthrene, diazinon, and catechol [Citation104]. Burkholderia cepacian is also involved in the degradation of PAH [Citation105]. QS regulates biofilm formation in Pseudomonas aeruginosa and Pseudomonas putida, both of which belong to the γ-proteobacteria. In P. putida, QS is controlled by ppuI and ppuR. Biofilm of P. putida is involved in the degradation of 3-chlorobenzoate due to the presence of 3-chlorobenzoate degradation cluster of plasmid within biofilms [Citation60] (). EPS of Enterobacter cloacear, Halomonas eurihalina, and Gordonia alkanivorans are involved in the degradation of hydrocarbons [Citation83].
Table 2. Bacteria involved in bioremediation of environmental pollutants
Biofilms manufactured by Alcaligenes, Methylosinus Pseudomonas, Rhodococcus, and Sphingomonas are involved in the degradation of organic compounds [Citation27]. Bacterial strains such as Azospirillum, Burkholderia, Brachymonas, Cupriavidus, Moraxella, Pseudomonas, Sphingomonas, and Variovorax are involved in the biodegradation of hydroquinone. Nine genes of Pseudomonas putida DLL-E4 such as pnpA, pnpR, and pnpC1C2DECX1X2 are involved in the metabolism of 4-nitrophenol. Gene pnpC which encodes hydroxyquinol 1, 2-dioxygenase is involved in the conversation of hydroxyquinol to maleylacetic acid. Gene such as pnpC1C2DE of Pseudomonas putida DLL-E4 is involved in the transformation of hydroquinone into metabolites of tricarboxylic acid cycle [Citation106]. Sphingomonas sp. Ibu-2 strain of wastewater treatment plant is involved in the biodegradation of ibuprofen under aerobic conditions [Citation107]. Variovorax sp. Ibu-1 strain of activated sludge is involved in the transformation of ibuprofen to trihydroxyibuprofen [Citation108]. Bacillus thuringiensis B1 is also involved in the degradation of ibuprofen by using their ipfABDEF gene cluster in their genomes [Citation109]. Pseudomonas putida F1 strain having cmt operon, Micrococcus lysodeikticus and Bacillus sp having homoprotocatechuate pathway, Flavobacterium strain having homogentisate degradation pathway, Streptococcus rimosus and Pseudomonas cepacia are involved in the degradation of ibuprofen. Gram-negative bacteria are involved in the aerobic degradation of phenylacetic acid by using (phenylacetyl)-coenzyme A ligase pathway. Corynebacterium glutamicum and Pseudomonas sp. KT2440 are involved in the degradation of benzoate by inducing the over expression of ABC transporter, cytochromes and NADH-dehydrogenases proteins [Citation107]. Biofilm-forming soil bacteria such as Azospirillum, Bradyrhizobium, Pseudomonas, and Rhizobium are attracted to benzoate, benzoylformate, mandelate, β-phenylpyruvate, and salicylate through several QS genes such as rhlI [Citation110] (). Understanding the interactions between microorganisms and chemicals is essential for successful bioremediation.
The biodegradation of plastics is involved in several steps such as bio-deterioration, bio-fragmentation, assimilation, and mineralization. In bio-deterioration, biofilm induces physical and chemical deterioration of plastic. In bio-fragmentation, enzymes such as oxygenases, lipases, and esterases which are secreted by the bacteria in biofilm are involved in the fragmentation of polymers in plastic into oligomers and monomers. In assimilation, oligomers that are generated due to bio-fragmentation are assembled in bacterial cells of biofilm as a carbon source. During mineralization, bacteria are involved in the oxidation of oligomers into CO2, N2, CH4, and H2O [Citation111]. Wastewater treatment plants (WWTPs) are the main source of microplastics which are mostly present in sewage sludge [Citation112]. Arthrobacter, Aspergillus, Micrococcus, Pseudomonas, and Rhodococcus are mainly responsible for the biodegradation of plastics in seawater, and plastic dumping sites [Citation113] [Citation114].,reported that Enterobacter asburiae YT1 and Bacillus sp. YP1 which are isolated from the gut of waxworms are involved in the degradation of polyethylene [Citation115].,reported that Acinetobacter calcoaceticus, Burkholderia cepacian, and Escherichia coli are involved in the bioremediation of microplastics. Cyclobacteriaceae, Pirellulaceae, Phycisphaerales, and Roseococcus showed a higher abundance in biofilms on microplastics. Microbial enzymes are involved in the depolymerization of microplastics. Predominant microbial species of cold environments such as Arthrobacter, Corynebacterium, Micrococcus, Pseudomonas, Rhodococcus, and Streptomyces are also involved in biodegradation [Citation116]. Bacterial genera such as Alcanivorax, Moritella, Pseudomonas, Psychrobacter, Shewanella, and Tenacibaculum from deep-sea sediment are involved in the biodegradation of poly (ε-caprolactone) [Citation117]. Organic carbon which is derived from plastic of seawater induces the growth of heterotrophic microbes [Citation118] (). Polyethylene terephthalate (PET) is commonly used in bottles and synthetic fibers. Due to its long carbon chains bearing aromatic rings, PET is more stable in the environment and it is difficult to biodegrade. Pseudomonas mendocina, Ideonella sakaiensis, Nocardia sp., and Thermobifida fusca are involved in the degermation of PET by using enzymes such as lipases and esterases [Citation119]. Rhodococcus ruber is involved in the metabolism of polyethylene by using laccase enzyme [Citation120]. Algal derivatives such as polyhydroxyalkanoates (PHAs) from cyanobacteria, and starch from microalgae are involved in the synthesis of bioplastic [Citation121].
8. Role of quorum sensing in biological wastewater treatments
QS signals such as AHLs of Agrobacterium, Aeromonas, and Pseudomonas contribute significantly to wastewater treatment [Citation11]. AHLs enhance the degradation of phenol in wastewater treatment [Citation122]. QS signals of Acinetobacter sp. DR1 is involved in the degradation of hexadecane [Citation123]. The rhl QS system and expression of catechol 2, 3-dioxygenase in P. aeruginosa is involved in the degradation of benzoate, phenanthrene, and phenol [Citation124]. QS induces the development of aerobic granule (AG) through aggregation in wastewater treatment [Citation11]. AHL producing bacterial genera such as Aeromonas and Pseudomonas showed a higher abundance in Activated sludge (AS) which are commonly used for the purification of domestic and industrial wastewater Citation6. The structure and metabolic activity of activated sludge in wastewater treatment are controlled by N-heptanoyl-L-homoserine lactone [Citation125]. AHLs enhance the expression of amoA genes of ammonia-oxidizing archaea and bacteria in wastewater treatment [Citation126]. Biofilms have both favorable roles such as granular sludge, and moving bed biofilm reactors, and the adverse role such as membrane biofouling in wastewater treatment [Citation51]. AI-2 is involved in the development of biofilm by Escherichia coli through the synthesis of EPS [Citation127]. AI-2 impacts aerobic granulation through the synthesis of EPS [Citation128]. Quorum quenching (QQ) activity is reduced by the granulation [Citation129]. AHL regulates the synthesis of EPS by P. aeruginosa through LuxS and Esal/EsaR systems [Citation130]. Bacteria belonging to Flavobacterium and Xanthomonadaceae showed higher abundance in activated sludge with an upsurge of AHLs concentrations [Citation51]. The abundance of bacteria belong to Anaerolinea, Bacteroidetes, Proteiniphilum, and Syntrophobacter become elevated in the presence of short-chain AHLs [Citation131]. The RhlI/RhlR system of P. aeruginosa is involved in the treatment of industrial and municipal wastewater particularly in the degradation of phenol. Acinetobacter sp. is involved in the metabolism of hexadecane through autoinducer synthases dependent QS. AHL is used for the removal of pollutants from wastewater through the synthesis of biofilm and biosurfactant, horizontal gene transfer, and expression of genes that are involved in the xenobiotic metabolism. QQ enzymes such as lactonases (quorum-quenching N-acyl-homoserine lactonases), acylase amidases such as amidohydrolases or acylases, reductases, and cytochrome oxidases are involved in the degradation of AHLs. Management of microbiome at the community and molecular level through QS contributes significantly to the bioremediation of organic pollutants [Citation51]. Pseudomonas sp. 1A1, Variovorax paradoxus VA1-C and Rhodococcus sp. BH4 showed inhibitory activity against QS signals in the presence gamma-caprolactone (GCL) of activated sludge [Citation132] [Citation129].,reported the presence of QS and QQ genes in activated sludge of wastewater treatment facilities by using metagenomic approaches. AHL-quenching bacteria were present in a higher proportion as compared to AHL-producing bacteria in aerobic granules [Citation133]. Overgrowth of biofilm is responsible for the poor performance of membrane biofilm reactors (MBfR) and moving bed biofilm reactors (MBBR). QQ-bacteria such as Acinetobacter sp.,Afipia sp.,Microbacterium sp.,Micrococcus sp.,Pseudomonas sp., and Rhodococcus sp are responsible for the prevention of the growth of biofilm [Citation134]. Quorum quenching activity was also exhibited by Brevundimonas, Comamonas, Mesorhizobium, Pedobacter, and Variovorox [Citation135]. Candida albicans synthesize QQ agents such as farnesol which inhibits the growth of biofilm on MBR [Citation136] (). Geobacter metallireducens is involved in the reduction of Mn (IV) to Mn (II) whereas G. sulfurreducens and G. metallireducens are involved in the reduction of U(VI) to U(IV) and Cr (VI) to less toxic Cr (III) [Citation137]. Acetonitrile which is commonly discharged through industrial wastewater becomes hazardous to aquatic organisms. B. subtilis E2, E3, and N4 as well as Rhodococcus rhodochrous BX2, are used to degrade acetonitrile [Citation138a]. B. subtilis N4-pHT01-nit is involved in the degradation of acetonitrile along with Moving bed biofilm reactor (MBBR) [Citation139]. Arthrobacter sp., Pseudochrobactrum saccharolyticum LY10 sp., Aspergillus carbonarius and P. mendocina NR802 are involved in the elimination of xenobiotic substances and toxic heavy metals [Citation140,Citation141]. P. aeruginosa CGMCC1.860 is involved in the synthesis of AHL molecules such as C4HSL and C6HSL which are involved in the biodegradation of aromatic compounds [Citation142]. Sulfate reducing bacteria (SRB) such as Desulfovibrio are involved in the bioremediation of wastewater containing Cu+2 [Citation143]. Biodegradation of bisphenol A (BPA) from wastewater is aided by Acidovorax sp., Luteimonas sp., and Pseudomonas sp [Citation144]. Mezorhizobium, Devosia, Pseudoxanthomonas, Bosea, and Paracocci have been found in wastewater treatment systems that use nitrogen metabolism [Citation145]. QS and QQ have been found to influence the synthesis of EPS, biofilm formation, and biodegradation of organic pollutants and biofouling management in wastewater treatment.
Table 3. Quorum sensing (QS] and quorum quenching [QQ) bacteria involved in treatment of municipal or industrial wastewater
9. Application of moving bed biofilm reactor (MBBR) in waste water treatment plant (WWTP)
Nutrients of wastewater are a major source of contamination in the environment [Citation146–148]. Biological Nutrient Removal (BNR) is not efficient to remove the nutrients from wastewater due to the presence of microorganisms [Citation149]. MBBR has been considered an efficient system for introducing biomass as biofilm [Citation150,Citation151]. Moving bed biofilm technology has several advantages such as reduced obstruction, resistance against pH, temperature, and toxic substances, and maintains of biomass as a biofilm on carriers [Citation152,Citation153]. The carrier materials such as polyethylene, polypropylene, and polyvinyl chloride are responsible for the construction of biofilm and microbial diversity in the biofilm. The amino functional group (-NH2) into polyethylene and polypropylene increases the thickness and density of biofilm [Citation154] (). Membrane bioreactors (MBRs) are considered the best wastewater treatment approach. Membrane biofouling is one of the most serious threats in MBR applications. In membrane biofouling, unwanted microorganisms become assemble and form biofilm on the membrane surface through attachment and expansion [Citation155]. Bacteria such as Aeromonas, Citrobacter, Enterobacter, Klebsiella, Pseudomonas, and Serratia are involved in the growth of biofilm on the membrane surface through AHL-based QS system [Citation156, Citation157]. Enzymes such as acylases or lactonases which are involved in the quenching of AHL activity showed a significant role in controlling the membrane biofouling in MBRs. Bacteria such as Anabaena sp. PCC7120, A. tumefaciens C58, Bacillus sp. strain 240B1, P. aeruginosa PAO1, Rhodococcus erythropolis strain W2, and Ralstonia sp. XJ12B is responsible for the synthesis of produce quorum quenching enzymes [Citation156]. QQ technology is an appealing and cost-effective method of biofouling control in MBRs.
Biofilms based WWT technologies are used to remove contaminants such as organic and nitrogenous substances from wastewater. In the sludge bed of the bioreactor, the microbiota adheres to the sludge granules. Anaerobic bioreactors are involved in the generation of methane as a byproduct through the metabolism of organic substances [Citation158]. Prokaryotes belong to Phylum Euryarchaeota and six orders such as Methanobacteriales, Methanococcales, Methanomicrobiales, Methanosarcinales, Methanopyrales, and Methanocellales are involved in methane synthesis [Citation159]. Methanobacterium, Methanosaeta, and Methanosarcina showed higher abundance in the fixed-bed reactor and seven fluidized-bed reactors that are involved in the treatment of industrial wastewaters. Methanobacteriales and Methanomicrobiales showed co-existence in fixed-film anaerobic reactors [Citation160]. Methanobacteriales, Methanomicrobiales, and Methanospirillum showed abundance in fixed-bed anaerobic baffled reactors (FABRs) [Citation161] [Citation162].,reported that Methanomethylovorans hollandica, Methanobacterium aarhusense, Methanobacterium subterraneum and Methanolinea tarda were highly abundant in the granular bioreactor. Methanomicrobiales showed higher abundance in anaerobic fixed-bed biofilm systems at low temperatures [Citation163]. Methanobacteriaceae, Methanomicrobiaceae, and Methanosarcinaceae showed higher abundance in the packed-bed biofilm reactors [Citation164]. Methanogenic archaea play a critical role in anaerobic wastewater treatment.
Genome editing tools such as Clustered regularly interspaced short palindromic repeats (CRISPR-Cas), Transcription-activators like effector nucleases (TALEN), and zinc finger nucleases (ZFNs) are presently involved in the enhancement of bioremediation. Among them, the CRISPR-Cas system which consists of guide RNA (gRNA) linked crisper derived RNA (crRNA) and transacting antisense RNA (trcRNA) is widely used in Pseudomonas and Escherichia coli. The CRISPR system is also used to express genes involved for bioremediation in Achromobacter sp. HZ01, Comamonas testosterone, and Rhodococcus ruber TH. Genome editing enhances the survival of bacteria in a toxic environment. Metabolic engineering is involved in the modification of microbial enzymes such as esterases, monooxygenases, oxidases, oxidoreductases, and phenoloxidases which are involved in bioremediation. Enzyme-based bioremediation is compatible with our ecological condition [Citation165]. In Klebsiella pneumonia, aioA gene encodes arsenite oxidase which is involved in the bioremediation of arsenic [Citation166]. Catechol 1,2-dioxygenase of Pseudomonas NP-6 is involved in the degradation of catechol into muconate compounds [Citation167]. Insecticides used in agricultural fields such as organophosphates (OP) and organochlorines (OC) entered into the water. Genetically modified P. putida KT2440 has been used in the bioremediation of organophosphates and pyrethroids [Citation168]. Enzymes (haloalkane dehalogenase, wild-type haloalcohol dehalogenase, and wild-type epoxide hydrolase) of genetically modified E. coli strain have been utilized in the degradation of 1, 2, 3-trichloropropane [Citation169].
Insertion of organophosphorus hydrolase gene (opd) and pnp operon encoding enzymes in P. putida is involved in the bioremediation of organophosphorus and paraoxon [Citation170]. In Rhodococcus opacus R7, pobA and chcpca gene clusters are involved in the bioremediation of naphthenic acid. Expression of the aliA1 gene which encodes fatty acid CoA ligase is involved in the degradation of alicyclic naphthenic acid [Citation171]. The enzyme oxygenase is involved in the degradation of organic compounds. OxDBase provides information about the enzyme oxygenases which are involved in bioremediation [Citation172–174]. Bionemo (Biodegradation Network Molecular Biology) database provides information about the expression of genes involved in biodegradation [Citation175]. EAWAGBBD PPS database provides information about the pathways involved in the biodegradation of 1-naphthyl-N-methyl carbamate [Citation176]. Alcaligenes xylosoxidans and Pseudomonas stutzeri showed enhancement of biodegradation of polychlorinated biphenyls in the presence of biphenyl [Citation177]. Squamocin induces biofilm formation in Bacillus atrophaeus CN4 which is involved in the bioremediation of naphthalene [Citation178]. Cyanobacterium, Phormidium autumnale UTEX1580 has been involved in the metabolism of indigo dye which is released from textile industries [Citation179]. Cloning of flavin-diffusible monooxygenase encoding genes such as cphC-I and cphB of Arthrobacter chlorophenolicus in E. coli enhances bioremediation of 4-chlorophenol [Citation180]. The application of synthetic biology approaches in microbes improves the effectiveness of microbial bioremediation processes for specific contamination [Citation181,Citation182,Citation183].
10. Future prospects and conclusions
Aggregation of toxic xenobiotic substances in the environment is increasing, therefore it is essential to reduce concentration of xenobiotic substances through different approaches. Microbes based removal of environmental pollutants is ecofriendly. Biofilm based bioremediation approach is one of the potential approaches for reducing the level of pollutants in the environment. Biofilms are natural habitats where bacterial cells exchange genetic material, signaling molecules, and metabolites. QS is responsible for the interaction between bacterial cells of different species. To understand bioremediation of environmental contaminated sites such as wastewaters, it is essential to understand bacterial diversity in biofilm by using metagenomic approaches such as 16s rRNA sequencing. Microbiome based bioremediation process has now emerged as an essential approach in the global scenario. The development of genetically engineered biofilm formation enhances bioremediation of pollutants. Synthetic biology approaches are made to increase the efficiency of microbial enzymes in bioremediation. QS technology enhances bioremediation process in industrial wastewater treatment plants. QS regulates the development of biofilm, synthesis of EPS, and biosurfactant which contribute significantly to the removal of heavy metals and organic substances from the ecosystem. In WWTPs, QS and QQ are responsible for aggregation, colonization, granulation, removal of nutrients, metabolism of organic substances, and biofouling. It is essential to understand the role of AHL-based QS and QQ systems in wastewater treatments. Genome editing, omic based approaches, and metabolic engineering along with in silico databases and bioinformatics tools may open a new avenue for biofilm based bioremediation research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Costerton JW, Lewandowski Z, Caldwell DE, et al. Microbial biofilms. Annu Rev Microbiol. 1995;49(1):711–745.
- *Costerton JW, Geesey GG, Cheng KJ. How bacteria stick. Sci Am. 1978;238(1):86–95.
- Azeredo J, Azevedo NF, Briandet R, et al. Critical review on biofilm methods. Crit Rev Microbiol. 2017;43(3):313–351.
- Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867.
- Flemming HC, Wingender J, Szewzyk U, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575.
- *Mangwani N, Shukla SK, Kumari S, et al. Effect of biofilm parameters and extracellular polymeric substance composition on polycyclic aromatic hydrocarbon degradation. RSC Adv. 2016;6(62):57540–57551.
- Rajamanikandan S, Jeyakanthan J, Srinivasan P. Molecular docking, molecular dynamics simulations, computational screening to design quorum sensing inhibitors targeting LuxP of Vibrio harveyi and its biological evaluation. Appl Biochem Biotechnol. 2017;181(1):192–218.
- Li YH, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors. 2012;12(3):2519–2538.
- Swem LR, Swem DL, Wingreen NS, et al. Deducing receptor signaling parameters from in vivo analysis: luxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134(3):461–473.
- Balan B, Dhaulaniya AS, Varma DA, et al. Microbial biofilm ecology, in silico study of quorum sensing receptor-ligand interactions and biofilm mediated bioremediation. Arch Microbiol. 2020;Aug 12. DOI:10.1007/s00203-020-02012-9.
- Feng L, Wu Z, Yu X. Quorum sensing in water and wastewater treatment biofilms. J Environ Biol. 2013;34(2 Spec No):437–444.
- *Varjani S, Rakholiya P, Ng HY, et al. Microbial degradation of dyes: an overview. Bioresour Technol. 2020;314:123728.
- Huang J, Shi Y, Zeng G, et al. Acyl-homoserine lactone-based quorum sensing and quorum quenching hold promise to determine the performance of biological wastewater treatments: an overview. Chemosphere. 2016;157:137–151.
- Halan B, Buehler K, Schmid A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012;30(9):453–465.
- Escudie R, Cresson R, Delgenes J-P, et al. Control of start-up and operation of anaerobic biofilm reactors: an overview of 15 years of research. Water Res. 2011;45(1):1–10.
- Huang H, Peng C, Peng P, et al. Towards the biofilm characterization and regulation in biological wastewater treatment. Appl Microbiol Biotechnol. 2019;103(3):1115–1129.
- Barkay T, Schaefer J. Metal and radionuclide bioremediation: issues, considerations and potentials. Curr Opin Microbiol. 2001;4(3):318–323.
- *Mangwani N, Kumari S, Das S. Bacterial biofilms and quorum sensing: fidelity in bioremediation technology. Biotechnol Genet Eng Rev. 2016;32(1–2):43–73.
- Edwards SJ, Kjellerup BV. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl Microbiol Biotechnol. 2013;97(23):9909–9921.
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108.
- Varjani SJ, Gnansounou E, Pandey A. Comprehensive review on toxicity of persistent organic pollutants from petroleum refinery waste and their degradation by microorganisms. Chemosphere. 2017;188:280–291.
- Mosharaf M, Tanvir M, Haque M, et al. Metal-adapted bacteria isolated from wastewaters produce biofilms by expressing proteinaceous Curli Fimbriae and cellulose nanofibers. Front Microbiol. 2018;9:1334.
- Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by pseudomonas aeruginosa. J Bacteriol. 2007;189(6):2531–2539.
- Cao B, Christophersen L, Kolpen M, et al. Diffusion retardation by binding of tobramycin in an alginate biofilm model. PLoS ONE. 2016;11(4):e0153616.
- Mattei MR, Frunzo L, D’Acunto B, et al. Continuum and discrete approach in modeling biofilm development and structure: a review. J Math Biol. 2018;76(4):945–1003.
- Nishitani K, Sutipornpalangkul W, de Mesy Bentley KL, et al. Quantifying the natural history of biofilm formation in vivo during the establishment of chronic implant-associated Staphylococcus aureus osteomyelitis in mice to identify critical pathogen and host factors. J Orthop Res. 2015;33(9):1311–1319.
- Singh R, Paul D, Jain RK. Biofilms: implications in bioremediation. Trends Microbiol. 2006;14(9):389–397.
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633.
- Stalder T, Top E. Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms Microbiomes. 2016;2(1):16022.
- Yin Q, Gu M, Hermanowicz SW, et al. Potential interactions between syntrophic bacteria and methanogens via type IV pili and quorum-sensing systems. Environ Int. 2020;138:105650.
- Wang X, Bi X, Hem LJ, et al. Microbial community composition of a multi-stage moving bed biofilm reactor and its interaction with kinetic model parameters estimation. J Environ Manag. 2018b;218:340–347.
- Yu QS, Huang H, Ren HQ, et al. In situ activity recovery of aging biofilm in biological aerated filter: surfactants treatment and mechanisms study. Bioresour Technol. 2016;219:403–410.
- Lai CY, Dong QY, Chen JX, et al. Role of extracellular polymeric substances in a methane based membrane biofilm reactor reducing vanadate. Environ Sci Technol. 2018;52(18):10680–10688.
- Wang JF, Ren HQ, Li XH, et al. In situ monitoring of wastewater biofilm formation process via ultrasonic time domain reflectometry (UTDR). Chem Eng J. 2018a;334:2134–2141.
- Ren TT, Li XY, Yu HQ. Effect of N-acy-L-homoserine lactones like molecules from aerobic granules on biofilm formation by Escherichia coli K12. Bioresour Technol. 2013;129:655–658.
- Schmid N, Suppiger A, Steiner E, et al. High intracellular c-di-GMP levels antagonize quorum sensing and virulence gene expression in Burkholderia cenocepacia H111. Microbiol-Sam. 2017;163(5):754–764.
- Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A. 2012;109(41):16690–16695.
- Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52(4):917–924.
- Lazazzera BA. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr Opin Microbiol. 2005;8(2):222–227.
- Shukla SK, Rao TS. Effect of calcium on Staphylococcus aureus biofilm architecture: a confocal laser scanning microscopic study. Colloids Surf B. 2013a;103:448–454.
- Otzen D, Nielsen PH. We find them here, we find them there: functional bacterial amyloid. Cell Mol Life Sci. 2008;65(6):910–927.
- Gloag ES, Turnbull L, Huang A, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. PNAS USA. 2013;110(28):11541–11546.
- Zhang W, Sun J, Ding W, et al. Extracellular matrix-associated proteins form an integral and dynamic system during Pseudomonas aeruginosa biofilm development. Front Cell Inf Microbiol. 2015;5:40.
- Mooney JA, Pridgen EM, Manasherob R, et al. Periprosthetic bacterial biofilm and quorum sensing. J Orthop Res. 2018;36(9):2331–2339.
- Passos da Silva D, Schofield MC, Parsek MR, et al. An update on the sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens. 2017;6(4):51.
- Aspiras MB, Ellen RP, Cvitkovitch DG, et al. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol Lett. 2004;238(1):167–174.
- Kumari S, Mangwani N, Das S. Synergistic effect of quorum sensing genes in biofilm development and PAHs degradation by a marine bacterium. Bioengineered. 2016;7(3):205–207.
- Dusane DH, Zinjarde SS, Venugopalan VP, et al. Quorum sensing: implications on rhamnolipid biosurfactant production. Biotechnol Genet Eng Rev. 2010;27(1):159–184.
- Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551(7680):313–320.
- Labbate M, Queck SY, Koh KS, et al. Quorum sensing-controlled biofilm development in serratia liquefaciens MG1. J Bacteriol. 2004;186(3):692–698.
- Maddela NR, Sheng B, Yuan S, et al. Roles of quorum sensing in biological wastewater treatment: a critical review. Chemosphere. 2019;221:616–629.
- Mangwani N, Dash HR, Chauhan A, et al. Bacterial quorum sensing, functional features and potential applications in biotechnology. J Mol Microbiol Biotechnol. 2012;22(4):215–227.
- Churchill ME, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem Rev. 2011;111(1):68–85.
- Xu B, Ng TCA, Huang S, et al. Quorum quenching affects biofilm development in an anaerobic membrane bioreactor (AnMBR): from macro to micro perspective. Bioresour Technol. 2021Oct;25:126183.
- Fetzner S. Quorum quenching enzymes. J Biotechnol. 2015;201:2–14.
- Verbeke F, De Craemer S, Debunne N, et al. Peptides as quorum sensing molecules: measurement techniques and obtained levels in vitro and in vivo. Front Neurosci. 2017;11:183.
- Biswa P, Doble M. Production of acylated homoserine lactone by gram-positive bacteria isolated from marine water. FEMS Microbiol Lett. 2013;343(1):34–41.
- Shukla SK, Mangwani N, Rao TS, et al. Biofilm-mediated bioremediation of polycyclic aromatic hydrocarbons. In: Das S, editors. Microbial biodegradation and bioremediation. 1st ed. Waltham MA: Elsevier; 2014. p. 203–232.
- Gorisa J, Boon N, Lebbe L, et al. Diversity of activated sludge bacteria receiving the 3-chloroaniline-degradative plasmid pC1gfp. FEMS Microbiol Ecol. 2003;46(2):221–230.
- Springael D, Peys K, Ryngaert A, et al. Community shifts in a seeded 3-chlorobenzoate degrading membrane biofilm reactor: indications for involvement of in situ horizontal transfer of the clc-element from inoculum to contaminant bacteria. Environ Microbiol. 2002;4(2):70–80.
- Kalathil S, Khan MM, Lee J, et al. Production of bioelectricity, bio-hydrogen, high value chemicals and bioinspired nanomaterials by electrochemically active biofilms. Biotechnol Adv. 2013;31(6):915–924.
- Mishra B, Varjani S, Agarwal DC, et al. Engineering biocatalytic material for the remediation of pollutants: a comprehensive review. Environ Technol Innovation. 2020;20:101063.
- Mishra B, Varjani S, Iragavarapu GP, et al.Microbial fingerprinting of potential biodegrading organisms. Curr Poll Rep. 2019;1–17. DOI:10.1007/s40726-019-00116-5)
- Lovley DR. Live wires: direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energy Environ Sci. 2011;4(12):4896–4906.
- Saratale GD, Saratale RG, Shahid MK, et al. A comprehensive overview on electro-active biofilms, role of exo-electrogens and their microbial niches in microbial fuel cells (MFCs). Chemosphere. 2017;178:534–547.
- Butti SK, Velvizhi G, Sulonen MLK, et al. Microbial electrochemical technologies with the perspective of harnessing bioenergy: maneuvering towards upscaling. Renewable Sustainable Energy Rev. 2016;53:462–476.
- Rahimnejad M, Najafpour GD, Ghoreyshi AA, et al. Methylene blue as electron promoters in microbial fuel cell. Int J Hydrogen Energy. 2011;36(20):13335–13341.
- Li Y, Li H. Type IV pili of Acidithiobacillus ferrooxidans can transfer electrons from extracellular electron donors. J Bas Microbiol. 2014;54(3):226–231.
- Kumar R, Singh L, Zularisam AW. Exoelectrogens: recent advances in molecular drivers involved in extracellular electron transfer and strategies used to improve it for microbial fuel cell applications. Renewable Sustainable Energy Rev. 2016;56:1322–1336.
- Cusick RD, Kiely PD, Logan BE. A monetary comparison of energy recovered from microbial fuel cells and microbial electrolysis cells fed winery or domestic wastewaters. Int J Hydrogen Energy. 2010;35(17):8855–8861.
- Li Z, Haynes R, Sato E, et al. Microbial community analysis of a single chamber microbial fuel cell using potato wastewater. Water Environ Res. 2014;86(4):324–330.
- Popov A, Michie IS, Kim JR, et al. Enrichment strategy for enhanced bioelectrochemical hydrogen production and the prevention of methanogenesis. Int J Hydrogen Energy. 2016;41(7):4120–4131.
- Wrighton KC, Thrash JC, Melnyk RA, et al. Evidence for direct electron transfer by a gram-positive bacterium isolated from a microbial fuel cell. Appl Environ Microbiol. 2011;77(21):7633–7639.
- Gaur VK, Sharma P, Gupta S, et al. Opportunities and challenges in omics approaches for biosurfactant production and feasibility of site remediation. Strateg Adv Environ Technol Innov. 2022;25:102132.
- Rajmohan KS, Ramya C, Varjani S. Plastic pollutants: waste management for pollution control and abatement. Curr Opin Environ Sci Health. 2019;12:72–84.
- Shrout JD, Nerenberg R. Monitoring bacterial twitter: does quorum sensing determine the behavior of water and wastewater treatment biofilms? Environ Sci Technol. 2012;46(4):1995–2005.
- Mangwani N, Shukla SK, Rao TS, et al. Calcium-mediated modulation of Pseudomonas mendocina NR802 biofilm influences the phenanthrene degradation. Colloids Surf B. 2013;114:301–309.
- Markande AR, Patel D, Varjani S. A review on biosurfactants: properties, applications and current developments. Bioresour Technol. Jun 2021; 330:124963.
- Makkar RS, Rockne KJ. Comparison of synthetic surfactants and biosurfactants in enhancing biodegradation of polycyclic aromatic hydrocarbons. Environ Toxicol Chem. 2003;22(10):2280–2292.
- Varjani S, Upasani VN. Evaluation of rhamnolipid production by a halotolerant novel strain of Pseudomonas aeruginosa. Bioresour Technol. 2019;288:121577.
- Varjani SJ, Upasani VN. Biodegradation of petroleum hydrocarbons by oleophilic strain of Pseudomonas aeruginosa NCIM 5514. Bioresour Technol. 2016;222:195–201.
- Wilson C, Lukowicz R, and Merchant S, et al. Quantitative and qualitative assessment methods for biofilm growth: a mini-review. Res Rev J Eng Technol. 2017;6(4):17.
- Ta-Chen L, Chang JS, Young CC. Exopolysaccharides produced by Gordonia alkanivorans enhance bacterial degradation activity for diesel. Biotechnol Lett. 2008;30(7):1201–1206.
- Siddharth T, Sridhar P, Vinila V, et al. Environmental applications of microbial extracellular polymeric substance (EPS): a review. J Environ Manage. 2021;287:112307.
- Yong YC, Wu XY, Sun JZ, et al. Engineering quorum sensing signaling of Pseudomonas for enhanced wastewater treatment and electricity harvest: a review. Chemosphere. 2015;140:18–25.
- Rathankumar AK, Saikia K, Kumar PS, et al. Surfactant-aided mycoremediation of soil contaminated with polycyclic aromatic hydrocarbon (PAHs): progress, limitation, and countermeasures. J Chem Technol Biot. 2022;97(2):391–408.
- Varjani SJ, Rana DP, Jain AK, et al. Synergistic exsitu biodegradation of crude oil by halotolerant bacterial consortium of indigenous strains isolated from on shore sites of Gujarat, India. Int Biodeterior Biodegrad. 2015;103:116–124.
- Alegbeleye OO, Opeolu BO, Jackson VA. Polycyclic aromatic hydrocarbons: a critical review of environmental occurrence and bioremediation. Environ Manage. 2017;60(4):758–783.
- *Varjani S, Upasani VN, Pandey A. Bioremediation of oily sludge polluted soil employing a novel strain of Pseudomonas aeruginosa and phytotoxicity of petroleum hydrocarbons for seed germination. SciTotal Environ. 2020;737:139766.
- Pacwa-Płociniczak M, Płaza GA, Piotrowska-Seget Z, et al. Environmental applications of biosurfactants: recent advances. Int J Mol Sci. 2011;12(1):633–654.
- Varjani S, Upasani VN. Bioaugmentation of Pseudomonas aeruginosa NCIM 5514 - A novel oily waste degrader for treatment of petroleum hydrocarbons. Bioresour Technol. 2021;319:124240.
- Varjani SJ, Gnansounou E. Microbial dynamics in petroleum oilfields and their relationship with physiological properties of petroleum oil reservoirs. Bioresour Technol. 2017;245:1258–1265.
- Xiao-Hong PEI, Xin-Hua ZHAN, Shi-Mei WANG, et al. Effects of a biosurfactant and a synthetic surfactant on phenanthrene degradation by a Sphingomonas strain. Pedosphere. 2010;20(6):771–779.
- Owsianiak M, Chrzanowski Ł, Szulc A, et al. Biodegradation of diesel/biodiesel blends by a consortium of hydrocarbon degraders: effect of the type of blend and the addition of biosurfactants. Bioresour Technol. 2009;100(3):1497–1500.
- Labrenz M, Druschel GK, Thomsen-Ebert T, et al. Formation of sphalerite (ZnS) deposits in natural biofilms of sulfate-reducing bacteria. Science. 2000;290(5497):1744–1747.
- Kargi F, Eker S. Removal of 2,4-dichlorophenol and toxicity from synthetic wastewater in a rotating perforated tube biofilm reactor. Process Biochem. 2005;40(6):2105–2111.
- Wolfaardt GM, Lawrence JR, Robarts RD. Bioaccumulation of the herbicide diclofop in extracellular polymers and its utilization by a biofilm community during starvation. Appl Environ Microbiol. 1995;61(1):152–158.
- Papa R, Parrilli E, Sannia G. Engineered marine Antarctic bacterium Pseudoalteromonas haloplanktis TAC125: a promising micro-organism for the bioremediation of aromatic compounds. J Appl Microbiol. 2009;106(1):49–56.
- Messina E, Denaro R, Crisafi F, et al. Genome sequence of obligate marine polycyclic aromatic hydrocarbons-degrading bacterium Cycloclasticus sp. 78-ME, isolated from petroleum deposits of the sunken tanker Amoco Milford Haven, Mediterranean Sea. Mar Genomics. 2016;25:11–13.
- Löffler FE, Ritalahti KM, Zinder SH. Dehalococcoides and reductive dechlorination of chlorinated solvents. In: Stroo HF, et al, editors. Bioaugmentation for groundwater remediation. Vol. 5. New York: Springer; 2013. p. 39–88.
- Yoshida S, Ogawa N, Fujii T, et al. Enhanced biofilm formation and 3-chlorobenzoate degrading activity by the bacterial consortium of Burkholderia sp. NK8 and Pseudomonas aeruginosa PAO1. J Appl Microbiol. 2009;106(3):790–800.
- Shimada K, Itoh Y, Washio K, et al. Efficacy of forming biofilms by naphthalene degrading Pseudomonas stutzeri T102 toward bioremediation technology and its molecular mechanisms. Chemosphere. 2012;87(3):226–233.
- Tribedi P, Gupta AD, Sil AK. Adaptation of Pseudomonas sp. AKS2 in biofilm on low-density polyethylene surface: an effective strategy for efficient survival and polymer degradation. Bioresources Bioprocess. 2015;2(14):1–10.
- Hussaini SZ, Shaker M, Iqbal MA. Isolation of bacterial for degradation of selected pesticides. Adv Biores. 2013;4:82–85.
- Albrecht R, Gourry JC, Simonnot MO, et al. Complex conductivity response to microbial growth and biofilm formation on phenanthrene spiked medium. J Appl Geophys. 2011;75(3):558–564.
- Chen Q, Tu H, Luo X, et al. The regulation of para-nitrophenol degradation in Pseudomonas putida DLL-E4. PLoS One. 2016;11(5):e0155485.
- Żur J, Piński A, Marchlewicz A, et al. Organic micropollutants paracetamol and ibuprofen-toxicity, biodegradation, and genetic background of their utilization by bacteria. Environ Sci Pollut Res Int. 2018;25(22):21498–21524.
- Murdoch RW, Hay AG. The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu-1. Biodegradation. 2015;26(2):105–113.
- Marchlewicz A, Guzik U, Hupert-Kocurek K, et al. Toxicity and biodegradation of ibuprofen by Bacillus thuringiensis B1(2015b). Environ Sci Pollut Res. 2017;24(8):7572–7584.
- Krell T, Lacal J, Reyes-Darias JA, et al. Bioavailability of pollutants and chemotaxis. Curr Opin Microbiol. 2013;24:451–456.
- Dussud C, Ghiglione J-F. Bacterial degradation of synthetic plastics. In: Briand F, editor. Marine litter in the mediterranean and black seas. Paris: CIESM Publisher; 2014. p. 180.
- Mahon AM, O’Conell B, Healy MG, et al. Microplastics in sewage sludge: effects of treatment. Environ Sci Technol. 2017;51(2):810–818.
- Bardají DKR, Moretto JAS, Furlan JPR, et al. A mini-review: current advances in polyethylene biodegradation. World J Microbiol Biotechnol. 2020;36(2):32.
- Yang J, Yang Y, Wu WM, et al. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ Sci Technol. 2014;48(23):13776–13784.
- Hossain MR, Jiang M, Wei Q, et al. Microplastic surface properties affect bacterial colonization in freshwater. J Bas Microbiol. 2019;59(1):54–61.
- Yuan J, Ma J, Sun Y, et al. Microbial degradation and other environmental aspects of microplastics/plastics. Sci Total Environ. 2020;715:136968.
- Sekiguchi T, Saika A, Nomura K, et al. Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly(ɛ-caprolactone)-degrading bacteria. Polym Degrad Stab. 2011;96(7):1397–1403.
- Romera-Castillo C, Pinto M, Langer TM, et al. Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat Commun. 2018;9(1):1430.
- Gewert B, Plassmann MM, Macleod M. Pathways for degradation of plastic polymers floating in the marine environment. Environ Sci Process Impacts. 2015;17(9):1513–1521.
- Santo M, Weitsman R, Sivan A. The role of the copper-binding enzyme – laccase – in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int Biodeterior Biodegradation. 2013;84:204–210.
- Dang BT, Bui XT, Tran DPH, et al. Current application of algae derivatives for bioplastic production: a review. Bioresour Technol. 2022;347:126698.
- Valle A, Bailey MJ, Whiteley AS, et al. N-acyl-l-homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ Microbiol. 2004;6(4):424–433.
- Kang YS, Park W. Contribution of quorum-sensing system to hexadecane degradation and biofilm formation in Acinetobacter sp. strain DR1. J Appl Microbiol. 2010;109(5):1650–1659.
- Yong YC, Zhong JJ. Regulation of aromatics biodegradation by rhl quorum sensing system through induction of catechol meta-cleavage pathway. Bioresour Technol. 2013;136:761–765.
- Chong G, Kimyon O, Rice SA, et al. The presence and role of bacterial quorum sensing in activated sludge. Microb Biotechnol. 2012;5(5):621–633.
- Gao J, Duan Y, Liu Y, et al. Long- and short-chain AHLs affect AOA and AOB microbial community composition and ammonia oxidation rate in activated sludge. J Environ Sci. 2018;78:53–62.
- Kim Y, Wang X, Zhang XS, et al. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ Microbiol. 2010;12(5):1105–1121.
- Sun S, Liu X, Ma B, et al. The role of autoinducer-2 in aerobic granulation using alternating feed loadings strategy. Bioresour Technol. 2016;201:58–64.
- Tan CH, Koh KS, Xie C, et al. Community quorum sensing signalling and quenching: microbial granular biofilm assembly. Npj Biofilm Microb. 2015;1(1):15006.
- Tan CH, Koh KS, Xie C, et al. The role of quorum sensing signalling in EPS production and the assembly of a sludge community into aerobic granules. ISME J. 2014;8(6):1186–1197.
- Ma H, Wang X, Zhang Y, et al. The diversity, distribution and function of N-acyl-homoserine lactone (AHL) in industrial anaerobic granular sludge. Bioresour Technol. 2018;247:116–124.
- Cheong WS, Kim SR, Oh HS, et al. Design of quorum quenching microbial vessel to enhance cell viability for biofouling control in membrane bioreactor. J Microbiol Biotechnol. 2014;24(1):97–105.
- Li Y-S, Cao J-S, Li -B-B, et al. Outcompeting presence of acyl-homoserine-lactone (AHL)-quenching bacteria over AHL-producing bacteria in aerobic granules. Environ Sci Technol Lett. 2016;3(1):36–40.
- Kim AL, Park SY, Lee CH, et al. Quorum quenching bacteria isolated from the sludge of a wastewater treatment plant and their application for controlling biofilm formation. J Microbiol Biotechnol. 2014;24(11):1574–1582.
- Hu H, He J, Yu H, et al. A strategy to speed up formation and strengthen activity of biofilms at low temperature. RSC Adv. 2017;7(37):22788–22796.
- Lee K, Lee S, Lee SH, et al. Fungal quorum quenching: a paradigm shift for energy savings in membrane bioreactor (MBR) for wastewater treatment. Environ Sci Technol. 2016;50(20):10914–10922.
- Igiri BE, Okoduwa SIR, Idoko GO, et al. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J Toxicol. 2018;2018:2568038.
- Li C, Li Y, Cheng X, et al. Immobilization of Rhodococcus rhodochrous BX2 (an acetonitrile-degrading bacterium) with biofilm-forming bacteria for wastewater treatment. Bioresour Technol. 2013;131:390–396.
- Kragh KN, Hutchison JB, Melaugh G, et al. Role of multicellular aggregates in biofilm formation. mBio. 2016;7(2):e00237–00216.
- Dey S, Paul AJ. Influence of metal ions on biofilm formation by Arthrobacter sp. SUK 1205 and evaluation of their Cr (VI) removal efficacy. Int Biodeter Biodegrad. 2018;132:122–131.
- Lakshmi S, Suvedha K, Sruthi R, et al. Hexavalent chromium sequestration from electronic waste by biomass of Aspergillus carbonarius. Bioengineered. 2020;11(1):708–717.
- Yong YC, Zhong JJ. N-acylated homoserine lactone production and involvement in the biodegradation of aromatics by an environmental isolate of Pseudomonas aeruginosa. Process Biochem. 2010;45(12):1944–1948.
- Miran W, Jang J, Nawaz M, et al. Mixed sulfate-reducing bacteria-enriched microbial fuel cells for the treatment of wastewater containing copper. Chemosphere. 2017;189:134–142.
- Cydzik-Kwiatkowska A, Zielińska M. Microbial composition of biofilm treating wastewater rich in bisphenol A. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2018;53(4):385–392.
- de Beer DM, Botes M, Cloete TE. The microbial community of a biofilm contact reactor for the treatment of winery wastewater. J Appl Microbiol. 2018;124(2):598–610.
- Nakkeeran E, Varjani S, Goswami S, et al. Chitosan based silver nanocomposite for hexavalent chromium removal from tannery industry effluent using a packed bed reactor. J Environ Eng. 2020;146(6):04020032.
- Nguyen TTD, Bui XT, Nguyen TT, et al. Co-culture of microalgae-activated sludge in sequencing batch photobioreactor systems: effects of natural and artificial lighting on wastewater treatment. Bioresour Technol. 2022;343:126091.
- Wang X, Daigger G, de Vries W, et al. Impact hotspots of reduced nutrient discharge shift across the globe with population and dietary changes. Nat Commun. 2019;10(1):2627.
- Chaali M, Naghdi M, Brar SK, et al. A review on the advances in nitrifying biofilm reactors and their removal rates in wastewater treatment. J Chem Technol Biotechnol. 2018;93(11):3113–3124.
- Leyva-Díaz JC, Monteoliva-García A, Martín-Pascual J, et al. Moving bed biofilm reactor as an alternative wastewater treatment process for nutrient removal and recovery in the circular economy model. Bioresour Technol. 2020;299:122631.
- Tdh V, Bui XT, Dang BT, et al. Influence of organic loading rates on treatment performance of membrane bioreactor treating tannery wastewater. Environ Technol Innovation. 2021;24:101810.
- Di Biase A, Kowalski MS, Devlin TR, et al. Moving bed biofilm reactor technology in municipal wastewater treatment: a review. J Environ Manage. 2019;247:849–866.
- Huang S, Pooi CK, Shi X, et al. Performance and process simulation of membrane bioreactor (MBR) treating petrochemical wastewater. SciTotal Environ. 2020;747:141311.
- Chu L, Wang J, Quan F, et al. Modification of polyurethane foam carriers and application in a moving bed biofilm reactor. Process Biochem. 2014;49(11):1979–1982.
- Guo W, Ngo -H-H, Li J. A mini-review on membrane fouling. Bioresour Technol. 2012;122:27–34.
- *Lade H, Paul D, Kweon JH. Isolation and molecular characterization of biofouling bacteria and profiling of quorum sensing signal molecules from membrane bioreactor activated sludge. Int J Mol Sci. 2014;15(2):2255–2273.
- Shindhal T, Rakholiya P, Varjani S, et al. A critical review on advances in the practices and perspectives for the treatment of dye industry wastewater. Bioengineered. 2021;12(1):70–87.
- Angelidaki I, Karakashev D, Batstone DJ, et al. Biomethanation and its potential. Methods Enzymol. 2011;494:327–351.
- Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci. 2008;1125(1):171–189.
- Leclerc M, Delgènes J-P, Godon -J-J. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ Microbiol. 2004;6(8):809–819.
- Zhang D, Li J, Guo P, et al. Dynamic transition of microbial communities in response to acidification in fixed-bed anaerobic baffled reactors (FABR) of two different flow directions. Bioresour Technol. 2011;102(7):4703–4711.
- Kobayashi T, Yan F, Takahashi S, et al. Effect of starch addition on the biological conversion and microbial community in a methanol-fed UASB reactor during long-term continuous operation. Bioresour Technol. 2011;102(17):7713–7719.
- Zhang G, Zhang F, Ding G, et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 2012;6(7):1336–1344.
- Sasaki K, Haruta S, Ueno Y, et al. Archaeal population on supporting material in methanogenic packed-bed reactor. J Biosci Bioeng. 2006;102(3):244–246.
- Jaiswal S, Shukla P. Alternative strategies for microbial remediation of pollutants via synthetic biology. Front Microbiol. 2020;11:808.
- Mujawar SY, Shamim K, Vaigankar DC, et al. Arsenite biotransformation and bioaccumulation by Klebsiella pneumoniae strain SSSW7 possessing arsenite oxidase (aioA) gene. Biometals. 2019;32(1):65–76.
- Guzik U, Gre´n I, Hupert-Kocurek K, et al. Catechol 1, 2-dioxygenase from the new aromatic compounds–degrading Pseudomonas putida strain N6. Int Biodeterior Biodegrad. 2011;65(3):504–512.
- Gong T, Liu R, Zuo Z, et al. Metabolic engineering of Pseudomonas putida KT2440 for complete mineralization of methyl parathion and γ-Hexachlorocyclohexane. ACS Synth Biol. 2016;5(5):434–442.
- Dvorak P, Bidmanova S, Damborsky J, et al. Immobilized synthetic pathway for biodegradation of toxic recalcitrant pollutant 1, 2, 3- trichloropropane. Environ Sci Technol. 2014;48(12):6859–6866.
- de la Pena Mattozzi M, Tehara SK, Hong T, et al. Mineralization of paraoxon and its use as a sole c and p source by a rationally designed catabolic pathway in Pseudomonas putida. Appl Environ Microbiol. 2006;72(10):6699–6706.
- Zampolli J, Di Canito A, Cappelletti M, et al. Biodegradation of naphthenic acids: identification of Rhodococcus opacus R7 genes as molecular markers for environmental monitoring and their application in slurry microcosms. Appl Microbiol Biotechnol. 2020;104(6):2675–2689.
- Feng S, Hao Ngo H, Guo W, et al. Roles and applications of enzymes for resistant pollutants removal in wastewater treatment. Bioresour Technol. Sep 2021;335:125278.
- Shah SB, Ali F, Huang L, et al. Complete genome sequence of bacillus sp. HBCD-sjtu, an efficient HBCD-degrading bacterium. 3 Biotech. 2018;8(7):291.
- Singh SB. Enzyme catalysis and its role in food processing industries. In: A B, editor. Enzymes in food technology. Law (Singapore: Springer; 2018. p. 143–165.
- Carbajosa G, Cases I. Transcriptional networks that regulate hydrocarbon biodegradation. In: Timmis KN, McGenity T, van der Meer JR, et al, editors. Handbook of hydrocarbon and lipid microbiology. Berlin: Springer; 2010. p. 1399–1410.
- Garg V, Khan S, Dutt K. Systematic analysis of microbial degradation pathway of 1-Naphthyl-N-Methyl carbamate generated by EAWAG biocatalysis/biodegradation database-pathway prediction system. Int J Appl Sci Res Rev. 2014;1:049–055.
- Dangi AK, Sharma B, Hill RT, et al. Bioremediation through microbes: systems biology and metabolic engineering approach. Crit Rev Biotechnol. 2019;39(1):79–98.
- Parellada EA, Igarza M, Isacc P, et al. Squamocin, an annonaceous acetogenin, enhances naphthalene degradation mediated by Bacillus atrophaeus CN4. Rev Argent Microbiol. 2017;49(3):282–288.
- Dellamatrice PM, Silva-Stenico ME, Moraes LABD, et al. Degradation of textile dyes by cyanobacteria. Braz J Microbiol. 2017;48(1):25–31.
- Kang C, Yang JW, Cho W, et al. Oxidative biodegradation of 4-chlorophenol by using recombinant monooxygenase cloned and overexpressed from Arthrobacter chlorophenolicus A6. Bioresour Technol. 2017;240:123–129.
- Yaashika PR, Kumar PS, Varjani S, et al. Rhizoremediation of Cu(II) ions from contaminated soil using plant growth promoting bacteria: an outlook on pyrolysis conditions on plant residues for methylene Orange dye biosorption. Bioengineered. 2020;11(1):175–187.
- *Lade H, Paul D, Kweon JH. Quorum quenching mediated approaches for control of membrane biofouling. Int J Biol Sci. 2014;10(5):547–562.
- *Varjani SJ, Upasani VN. A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int Biodeterior Biodegrad. 2017;120:71–83.